Submitted:
29 January 2025
Posted:
30 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Actinomycete Isolation
2.2. Phenotypic and Physiological Characterization
2.3. MALDI-TOF MS Identification
2.4. DNA Extraction, 16SrRNA Gene Sequencing and Phylogenetic Analysis
2.5. Antimicrobial Testing Activity of Streptomyces sp. ACT158
2.6. Genome sequencing, Assembly, Gene Prediction and Functional Annotation
2.7. Phyogenomics, and Comparative Genome Analysis
3. Results
3.1. Phenotypical Characterization of ACT158
3.2. MALDITOF and 16SrRNA Based Analysis
3.3. Genome Assembly, Annotation and Functional Analysis
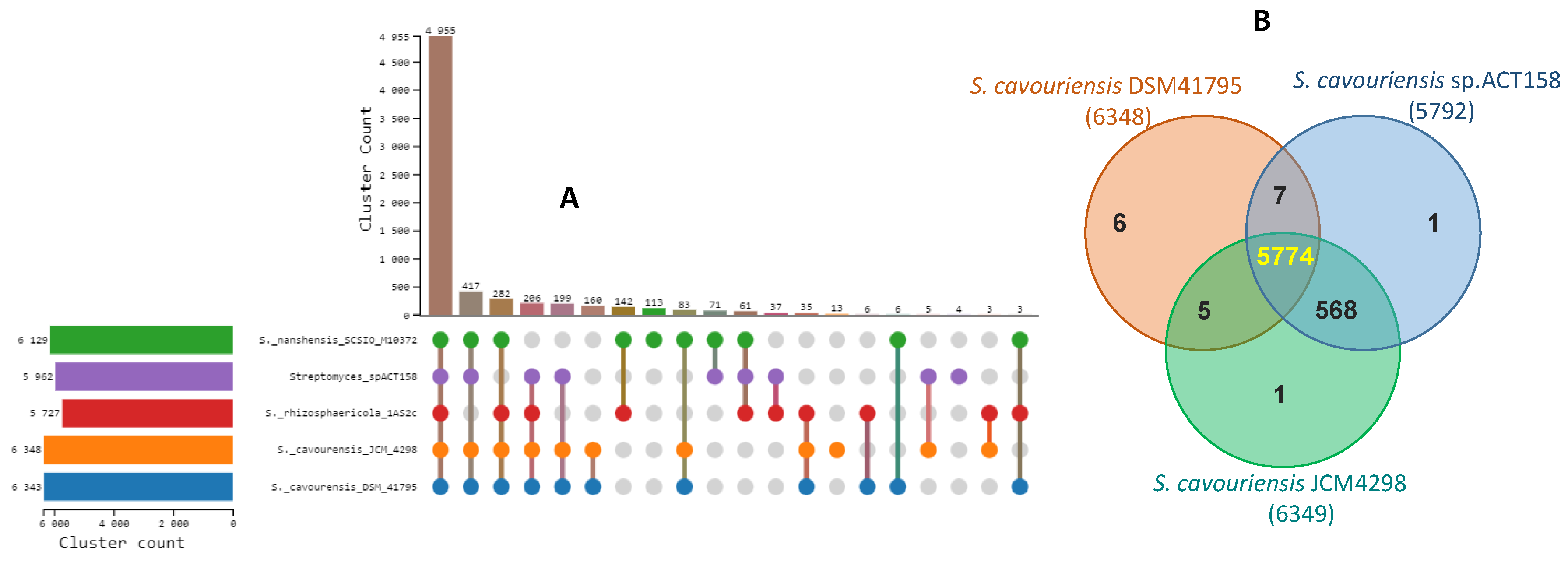
3.5. Comparative Genome Analysis
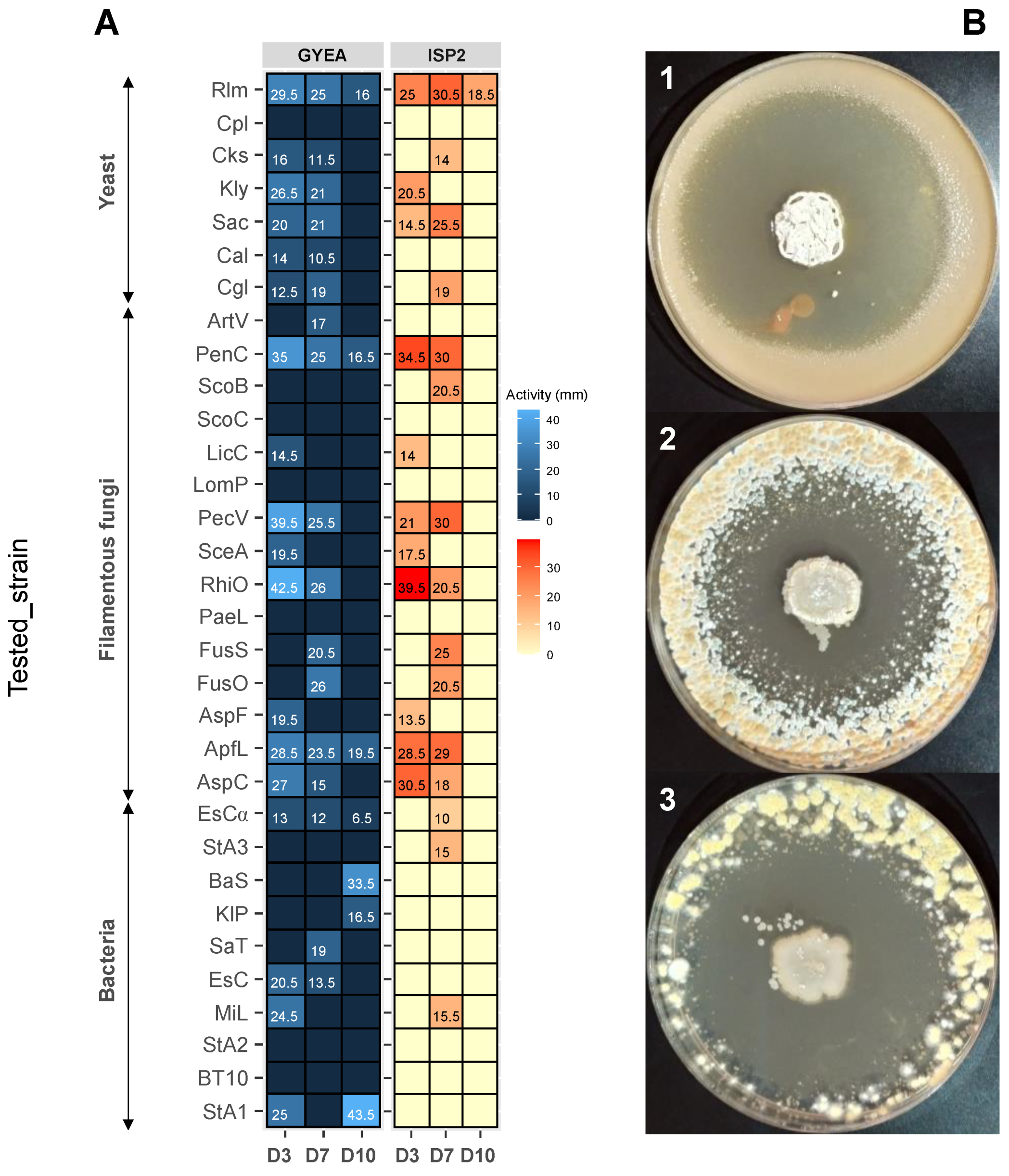
3.6. Antimicrobial Activity of ACT158
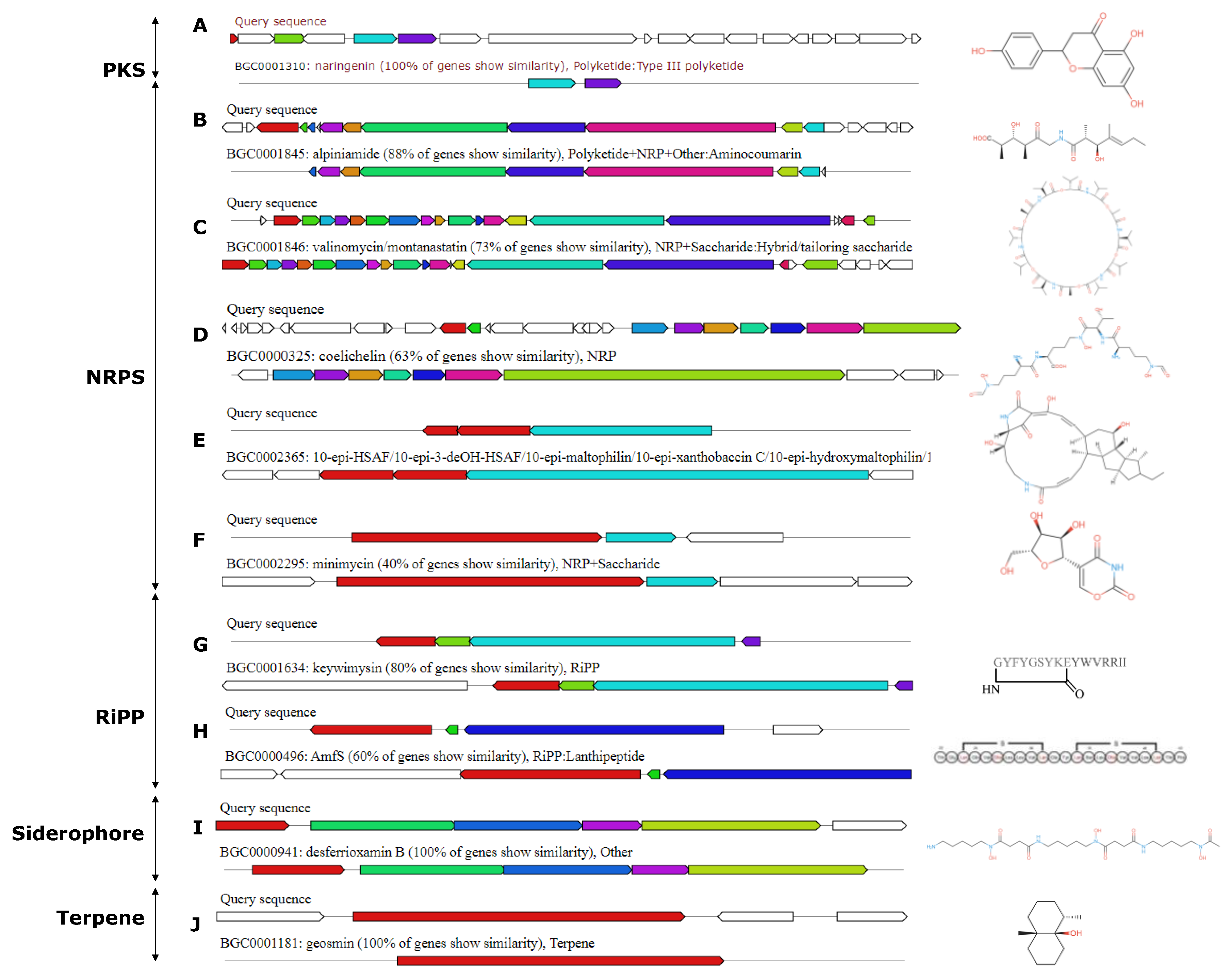
3.7. Analysis of Secondary Metabolic Biosynthetic Gene Clusters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rani, A; Saini, K. C; Bast, F; Mehariya, S; Bhatia, S. K; Lavecchia, R; & Zuorro, A. Microorganisms: a potential source of bioactive molecules for antioxidant applications. Molecules, 2021. 26(4), 1142. [CrossRef]
- Selim, M. S. M; Abdelhamid, S. A; & Mohamed, S. S. Secondary metabolites and biodiversity of actinomycetes. J. Gen. Eng. Biotechnol, 2021. 19(1), 72. [CrossRef]
- Alam, K; Mazumder, A; Sikdar, S; Zhao, Y. M; Hao, J; Song, C; ... & Li, A. Streptomyces: The biofactory of secondary metabolites. Front Microbiol, 2022. 13, 968053. [CrossRef]
- Komaki, H. Recent progress of reclassification of the genus Streptomyces. Microorganisms, 2023. 11(4), 831. [CrossRef]
- Shepherdson, E. M; & Elliot, M. A. Cryptic specialized metabolites drive Streptomyces exploration and provide a competitive advantage during growth with other microbes. Proc Nat Acad Sci, 2022. 119(40), e2211052119. [CrossRef]
- Benhadj, M; Gacemi-Kirane, D; Menasria, T; Guebla, K; & Ahmane, Z. Screening of rare actinomycetes isolated from natural wetland ecosystem (Fetzara Lake, northeastern Algeria) for hydrolytic enzymes and antimicrobial activities. J. King Saud Univ. Sci, 2019. 31(4), 706-712. [CrossRef]
- Antonopoulos, V; Hernandez, M; Arias, M; Mavrakos, E; & Ball, A. The use of extracellular enzymes from Streptomyces albus ATCC 3005 for the bleaching of eucalyptus kraft pulp. Appl. Microbiol. Biotechnol, 2001. 57, 92-97 . [CrossRef]
- Benhadj, M; Metrouh, R; Menasria, T; Gacemi-Kirane, D; Slim, F. Z; & Ranque, S. Broad-spectrum antimicrobial activity of wetland-derived Streptomyces sp. ActiF450. EXCLI J, 2020. 19, 360.
- LPSN (List of Prokaryotic names with Standing in Nomenclature) https://lpsn.dsmz.de/genus/streptomyces (accessed on 20 December 2024).
- Zaatout, N; Al-Mustapha, A. I; Bouaziz, A; Ouchene, R; & Heikinheimo, A. Prevalence of AmpC, ESBL, and colistin resistance genes in Enterobacterales isolated from ready-to-eat food in Algeria. Brazilian J. Microbiol, 2023. 54(3), 2205-2218. [CrossRef]
- Boukoucha, M; Menasria, T; & Bouguerra, N. Phenotypic characterization and genotypic subtyping of Salmonella enterica Serovars enteritidis and Gallinarum isolated from human and poultry-related samples. Food. Biotechnol, 2018. 32(3), 206-221. [CrossRef]
- Menasria, T; Monteoliva-Sánchez, M; Benhadj, M; Benammar, L; Boukoucha, M; & Aguilera, M. Unraveling the enzymatic and antibacterial potential of rare halophilic actinomycetes from Algerian hypersaline wetland ecosystems. J. Basic Microbiol, 2022. 62(10), 1202-1215. [CrossRef]
- Soldatou, S; Eldjárn, G. H; Ramsay, A; Van Der Hooft, J. J; Hughes, A. H; Rogers, S; & Duncan, K. R. Comparative metabologenomics analysis of polar actinomycetes. Marine drugs, 2021. 19(2), 103 . [CrossRef]
- Kim, J. H; Lee, N; Hwang, S; Kim, W; Lee, Y; Cho. Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture. J. Indust. Microbiol. Biotechnol, 2021. 48(3-4), kuaa001. [CrossRef]
- Lee, N; Hwang, S; Kim, J; Cho, S; Palsson, B; & Cho, B. K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Structural Biotechnol. J, 18, 2020. 23 1548-1556. [CrossRef]
- Menasria, T; Monteoliva-Sánchez, M; Benammar, L; Benhadj, M; Ayachi, A; Hacène, H; ... & Aguilera, M. Culturable halophilic bacteria inhabiting Algerian saline ecosystems: a source of promising features and potentialities. World J. Microbiol Biotechnol, 2019. 35, 1-16. [CrossRef]
- Menasria, T; Aguilera, M; Hocine, H; Benammar, L; Ayachi, A; Bachir, A. S; ... & Monteoliva-Sánchez, M. Diversity and bioprospecting of extremely halophilic archaea isolated from Algerian arid and semi-arid wetland ecosystems for halophilic-active hydrolytic enzymes. Microbiol. Res, 2018. 207, 289-298. [CrossRef]
- Benhadj, M; Gacemi-Kirane, D; Toussaint, M; Hotel, L; Bontemps, C; Duval, R. E; ... & Leblond, P. Diversity and antimicrobial activities of Streptomyces isolates from Fetzara Lake, north eastern Algeria. Annal Biol Clinique 2018. 76, No. 1). [CrossRef]
- Shirling, E. T; & Gottlieb, D. Methods for characterization of Streptomyces species. Inter. J. Syst. Evol. Microbiol, 1966. 16(3), 313- 340. [CrossRef]
- Gordon, R. E; Barnett, D. A; Handerhan, J. E; & Pang, C. H. N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. International Journal of Systematic and Evolutionary Microbiology, 1974. 24(1), 54-63.
- Cassagne C., Normand A.C., Bonzon L., L'Ollivier C., Gautier M., Jeddi F., Ranque S., & Piarroux R. Routine identification and mixed species detection in 6,192 clinical yeast isolates. Med Mycol, 2016. 54(3):256-65. [CrossRef]
- Kieser, T. B. M. J; Bibb, M. J; Buttner, M. J; Chater, K. F; & Hopwood, D. A. Practical Streptomyces. Genetics, 2000. 59.
- Weisburg, W. G; Barns, S. M; Pelletier, D. A; & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol, 1991. 173(2), 697-703. [CrossRef]
- Yoon, S. H; Ha, S. M; Lim, J; Kwon, S; & Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek, 2017. 110, 1281-1286. [CrossRef]
- Larkin, M. A; Blackshields, G; Brown, N. P; Chenna, R; McGettigan, P. A; McWilliam, H et al. Clustal W and Clustal X version 2.0. Bioinformatics, 2007. 23(21), 2947-2948. [CrossRef]
- Andrews, S. FastQC: a quality control tool for high throughput sequence data. 2010.
- Antipov, D; Korobeynikov, A; McLean, J. S; & Pevzner, P. A. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics, 2016. 32(7), 1009- 1015. [CrossRef]
- Tatusova, T; DiCuccio, M; Badretdin, A; Chetvernin, V; Nawrocki, E. P; Zaslavsky, L; ... & Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res, 2016. 44(14), 6614-6624. [CrossRef]
- Aziz, R. K; Bartels, D; Best, A. A; DeJongh, M; Disz, T; Edwards, R. A; ... & Zagnitko, O. The RAST Server: rapid annotations using subsystems technology. BMC genomics, 2008. 9, 1-15. [CrossRef]
- Overbeek, R; Olson, R; Pusch, G. D; Olsen, G. J; Davis, J. J; Disz, T; ... & Stevens, R. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res, 2014. 42(D1), D206-D214. [CrossRef]
- Kanehisa, M; Sato, Y; & Morishima, K. BlastKOALA and GhostKOALA. KEGG tools for functional characterization of genome and metagenome sequences. J. Mol Biol, 2016. 428(4), 726-731. [CrossRef]
- Lombard, V; Golaconda Ramulu, H; Drula, E; Coutinho, P. M; & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res, 2014. 42(D1), D490-D495. [CrossRef]
- Blin, K; Shaw, S; Augustijn, H. E; Reitz, Z. L; Biermann, F; Alanjary, M; ... & Weber, T. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res, 2023. 51(W1), W46-W50. [CrossRef]
- Grant, J. R; Enns, E; Marinier, E; Mandal, A; Herman, E. K; Chen, C. Y; ... & Stothard, P. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res, 2023. 51(W1), W484-W492 . [CrossRef]
- Meier-Kolthoff, J. P; & Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature communications, 2019. 10(1), 2182. [CrossRef]
- Meier-Kolthoff, J. P; Auch, A. F; Klenk, H. P; & Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC bioinformatics, 2013.14, 1-14. [CrossRef]
- Yoon, S. H; Ha, S. M; Lim, J; Kwon, S; & Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek, 2017. 110, 1281-1286. [CrossRef]
- Sun, J; Lu, F; Luo, Y; Bie, L; Xu, L; & Wang, Y. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res, 2023. 51(W1), W397-W403. [CrossRef]
- Del Carratore, F; Hanko, E. K; Breitling, R; & Takano, E. Biotechnological application of Streptomyces for the production of clinical drugs and other bioactive molecules. Curr Opin Biotechnol, 2022. 77, 102762. [CrossRef]
- Ouchene, R; Zaatout, N; & Suzuki, M. T. An Overview on Nocardiopsis Species Originating From North African Biotopes as a Promising Source of Bioactive Compounds and In Silico Genome Mining Analysis of Three Sequenced Genomes. J Basic Microbiol, 2024. 64(9), e2400046. [CrossRef]
- Benammar, L; Menasria, T; & Dibi, A. R. Deciphering the geochemical influences on bacterial diversity and communities among two Algerian hot springs. Environ Sci Poll Res, 2024. 31(32), 44848-44862. [CrossRef]
- Sarmiento-Tovar, A. A; Silva, L; Sanchez-Suarez, J; & Diaz, L. Streptomyces-derived bioactive pigments: ecofriendly source of bioactive compounds. Coatings, 2022. 12(12), 1858. [CrossRef]
- Bentley, S. D; Chater, K. F; Cerdeño-Tárraga, A. M; Challis, G. L; Thomson, N. R; James, K. D; ... & Hopwood, D. A. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3. Nature, 2002. 417(6885), 141-147. [CrossRef]
- Ouchene, R; Intertaglia, L; Zaatout, N; Kecha, M; & Suzuki, M. T. Selective isolation, antimicrobial screening and phylogenetic diversity of marine actinomycetes derived from the Coast of Bejaia City (Algeria), a polluted and microbiologically unexplored environment. J. Appl. Microbiol, 2022 132(4), 2870-2882. [CrossRef]
- Dekak, A; Menasria, T; Benhizia, Y; & Chenchouni, H. Endophytic passenger bacteria associated with Genista cinerea nodules growing in North African drylands. Rhizosphere, 2020. 14, 100205. [CrossRef]
- Abdelgalil, S. A; Soliman, N. A; Abo-Zaid, G. A; & Abdel-Fattah, Y. R. Bioprocessing strategies for cost-effective large-scale production of bacterial laccase from Lysinibacillus macroides LSO using bio-waste. Inter. J. Environ. Sci. Technol, 2020. 1-20. [CrossRef]
- Singhania, R. R; Ruiz, H. A; Awasthi, M. K; Dong, C. D; Chen, C. W; & Patel, A. K. Challenges in cellulase bioprocess for biofuel applications. Renewable Sustain Ener Rev, 2021. 151, 111622 . [CrossRef]
- Farooq, M. A; Ali, S; Hassan, A; Tahir, H. M; Mumtaz, S; & Mumtaz, S. Biosynthesis and industrial applications of α-amylase: A review. Arch Microbiol, 2021. 203, 1281-1292. [CrossRef]
- Naveed, M; Nadeem, F; Mehmood, T; Bilal, M; Anwar, Z; & Amjad, F. Protease—a versatile and ecofriendly biocatalyst with multi-industrial applications: an updated review. Catalysis Lett, 2021. 151, 307-323. [CrossRef]
- Vu, H. N. T; Nguyen, D. T; Nguyen, H. Q; Chu, H. H; Chu, S. K; Chau, M. V; & Phi, Q. T. Antimicrobial and cytotoxic properties of bioactive metabolites produced by Streptomyces cavourensis YBQ59 isolated from Cinnamomum cassia Prels in Yen Bai Province of Vietnam. Curr Microbiol, 2018. 75, 1247-1255. [CrossRef]
- Wibowo, J. T; Kellermann, M. Y; Köck, M; Putra, M. Y; Murniasih, T; Mohr, K. I; .. & Schupp, P. J. Anti-infective and antiviral activity of valinomycin and its analogues from a sea cucumber-associated bacterium, Streptomyces sp. SV 21. Marine drugs, 2021. 19(2), 81.
- Riesco, R; & Trujillo, M. E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Inter J Syst Evol Microbiol, 2024. 74(3), 006300 . [CrossRef]
- Mispelaere, M; De Rop, A. S; Hermans, C; De Maeseneire, S. L; Soetaert, W. K; De Mol, M. L; & Hulpiau, P. Whole genome–based comparative analysis of the genus Streptomyces reveals many misclassifications. Appl Microbiol Biotechnol, 2024. 108(1), 453.
- Mahato, N. K; Gupta, V; Singh, P; Kumari, R; Verma, H; Tripathi, C; ... & Lal, R. Microbial taxonomy in the era of OMICS: application of DNA sequences, computational tools and techniques. Antonie Van Leeuwenhoek, 2017. 110, 1357-1371. [CrossRef]
- Otani, H; Udwary, D. W; & Mouncey, N. J. Comparative and pangenomic analysis of the genus Streptomyces. Scientific rep, 2022. 12(1), 18909 . [CrossRef]
- Skinnider, M. A; Johnston, C. W; Gunabalasingam, M; Merwin, N. J; Kieliszek, A. M; MacLellan, R. J; ... & Magarvey, N. A. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nature communications, 2020. 11(1), 6058 . [CrossRef]
- Ejigu, G. F; & Jung, J. Review on the computational genome annotation of sequences obtained by next-generation sequencing. Biology, 2020. 9(9), 295. [CrossRef]
- Guan, N; Li, J; Shin, H. D; Du, G; Chen, J; & Liu, L. Microbial response to environmental stresses: from fundamental mechanisms to practical applications. Appl Microbiol Biotechnol 2017. 101, 3991-4008. [CrossRef]
- Chandra, P; Enespa, Singh, R; & Arora, P. K. Microbial lipases and their industrial applications: a comprehensive review. Microbial cell factories, 2020. 19, 1-42. [CrossRef]
- Barber, E. A; Liu, Z; & Smith, S. R. Organic contaminant biodegradation by oxidoreductase enzymes in wastewater treatment. Microorganisms, 2020. 8(1), 122 . [CrossRef]
- Huang, X; Pinto, D; Fritz, G; & Mascher, T. Environmental sensing in Actinobacteria: a comprehensive survey on the signaling capacity of this phylum. J Bacteriol, 2015. 197(15), 2517-2535. [CrossRef]
- Qin, S; Feng, W. W; Wang, T. T; Ding, P; Xing, K; & Jiang, J. H. Plant growth promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant and Soil, 2017. 416, 117-132.
- Undabarrena, A; Ugalde, J. A; Seeger, M; & Cámara, B. Genomic data mining of the marine actinobacteria Streptomyces sp. H-KF8 unveils insights into multi-stress related genes and metabolic pathways involved in antimicrobial synthesis. PeerJ, 2017. 5, e2912 . [CrossRef]
- Li, S; Li, Z; Pang, S; Xiang, W; & Wang, W. Coordinating precursor supply for pharmaceutical polyketide production in Streptomyces. Curr Opin Biotechnol, 2021. 69, 26-34. [CrossRef]
- Cheng, M; Chen, D; Parales, R. E; & Jiang, J. Oxygenases as powerful weapons in the microbial degradation of pesticides. Ann Rev Microbiol, 2022. 76(1), 325-348. [CrossRef]
- Palazzotto, E; & Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr Opin Microbiol, 2018. 109-116. [CrossRef]
- van Bergeijk, D. A; Terlouw, B. R; Medema, M. H; & van Wezel, G. P. Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nature Rev Microbiol, 2020. 18(10), 546-558.
- Vicente, C. M; Thibessard, A; Lorenzi, J. N; Benhadj, M; Hôtel, L; Gacemi-Kirane, D; ... & Aigle, B. Comparative genomics among closely related Streptomyces strains revealed specialized metabolite biosynthetic gene cluster diversity. Antibiotics, 2018. 7(4), 86. [CrossRef]
- Vijayakumar, R; Panneerselvam, K; Muthukumar, C; Thajuddin, N; Panneerselvam, A; & Saravanamuthu, R. Optimization of antimicrobial production by a marine actinomycete Streptomyces afghaniensis VPTS3-1 isolated from Palk Strait, East Coast of India. Indian J Microbiol, 2012. 52, 230-239. [CrossRef]
- Sánchez, S; Chávez, A; Forero, A; García-Huante, Y; Romero, A; Sánchez, M; ... & Ruiz, B. Carbon source regulation of antibiotic production. J Antibiotics, 2010. 63(8), 442-459. [CrossRef]
- Skarbek, J. D; & Brady, L. R. Streptomyces cavourensis sp. nov.(nom. rev.) and Streptomyces cavourensis subsp. washingtonensis subsp. nov; a chromomycin-producing subspecies. Inter J Syst Evol Microbiol, 1978. 28, 45-53.
- Xu, W; Zhang, D; Si, C; & Tao, L. Antifungal macrolides from Streptomyces cavourensis YY01-17. Chemistry of Natural Compounds, 2013. 49, 988-989 . [CrossRef]
- Pan, H. Q; Yu, S. Y; Song, C. F; Wang, N; Hua, H. M; Hu, J. C; & Wang, S. J. Identification and characterization of the antifungal substances of a novel Streptomyces cavourensis NA4. J Microbiol Biotechnol, 2015. 25(3) . [CrossRef]
- Robertsen, H. L; & Musiol-Kroll, E. M. Actinomycete-derived polyketides as a source of antibiotics and lead structures for the development of new antimicrobial drugs. Antibiotics, 2019. 8(4), 157 . [CrossRef]
- Álvarez-Álvarez, R; Botas, A; Albillos, S. M; Rumbero, A; Martín, J. F; & Liras, P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microbial Cell factories, 2015. 14, 1-12. [CrossRef]
- Martín, J. F; & Liras, P. Comparative molecular mechanisms of biosynthesis of naringenin and related chalcones in actinobacteria and plants: Relevance for the obtention of potent bioactive metabolites. Antibiotics, 2022. 11(1), 82. [CrossRef]
- Salehi, B; Fokou, P. V. T; Sharifi-Rad, M; Zucca, P; Pezzani, R; Martins, N; & Sharifi- Rad, J. The therapeutic potential of naringenin: a review of clinical trials. Pharmaceuticals, 2019. 12(1), 11. [CrossRef]
- Risdian, C; Mozef, T; & Wink, J. (2019). Biosynthesis of polyketides in Streptomyces. Microorganisms, 7(5), 124. [CrossRef]
- Rajesh, T; Tiwari, M. K; Thiagarajan, S; Nair, P. S; & Jeya, M. Type III polyketide synthases: current state and perspectives. Microbial Technology for the Welfare of Society, 2019.183-200.
- Santamaría, R. I; Martínez-Carrasco, A; Sánchez de la Nieta, R; Torres-Vila, L. M; Bonal, R; Martín, J; ... & Díaz, M. (2020). Characterization of actinomycetes strains isolated from the intestinal tract and feces of the larvae of the longhorn beetle Cerambyx welensii. Microorganisms, 2013. 8(12),. [CrossRef]
- Krause, J. Applications and restrictions of integrated genomic and metabolomic screening: an accelerator for drug discovery from actinomycetes ?. Molecules, 2021. 26(18), 5450. [CrossRef]
- Al-Quwaie, D. A. The role of Streptomyces species in controlling plant diseases: a comprehensive review. Australasian Plant Pathology, 2024. 53(1), 1-14. [CrossRef]
- Hudson, G. A; & Mitchell, D. A. RiPP antibiotics: biosynthesis and engineering potential. Curr Opin Microbiol, 2018. 45, 61-69 . [CrossRef]
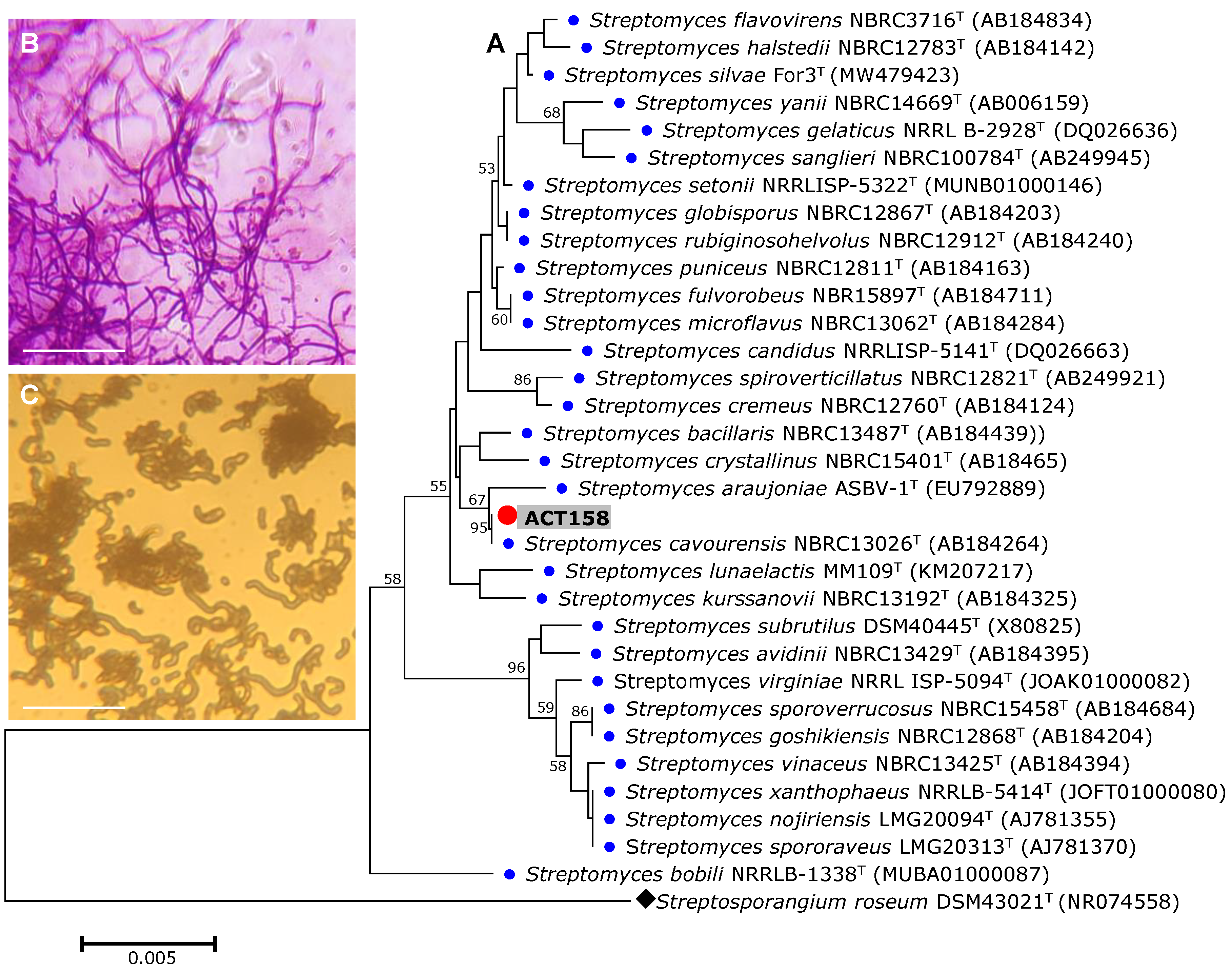
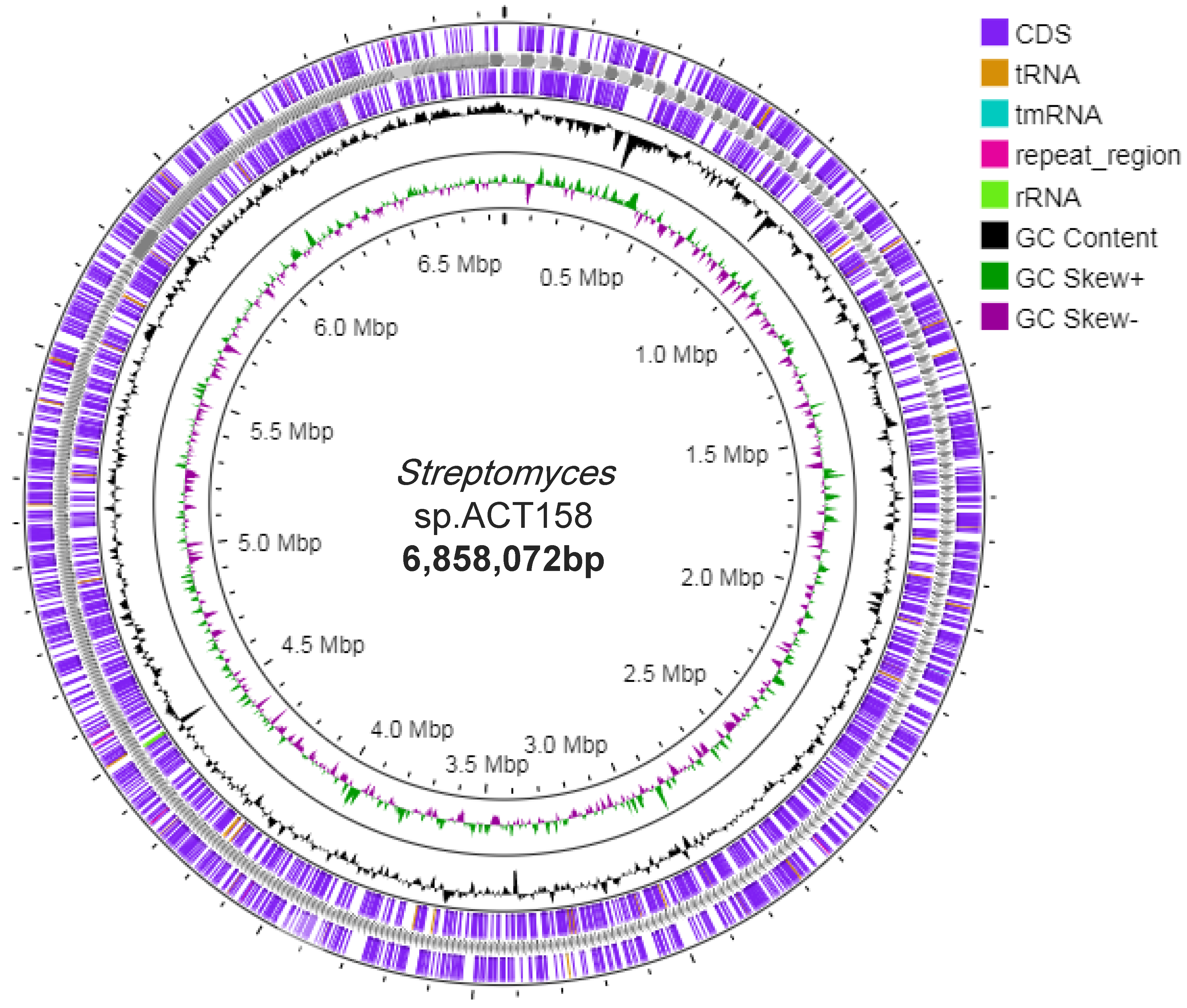
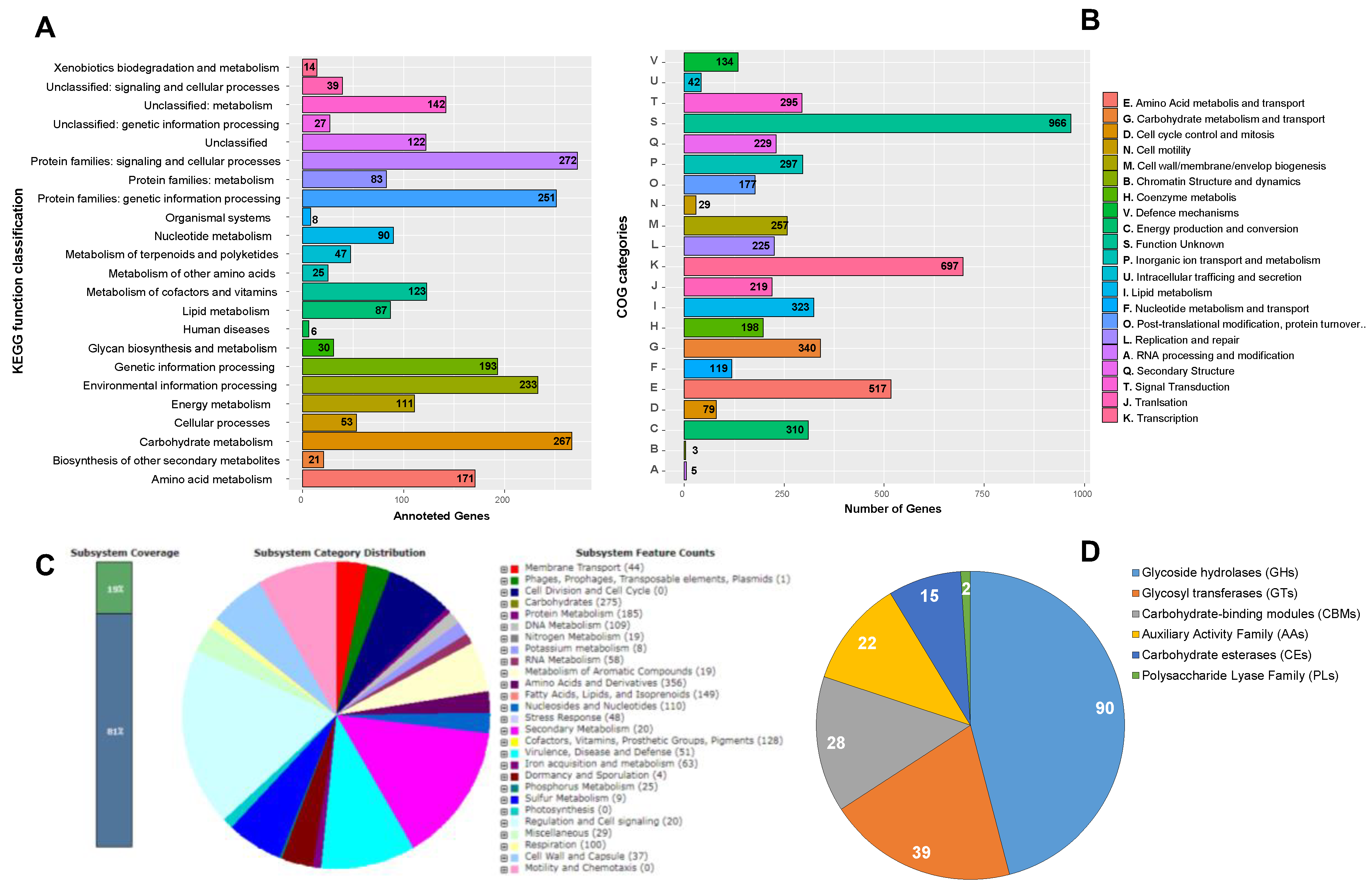
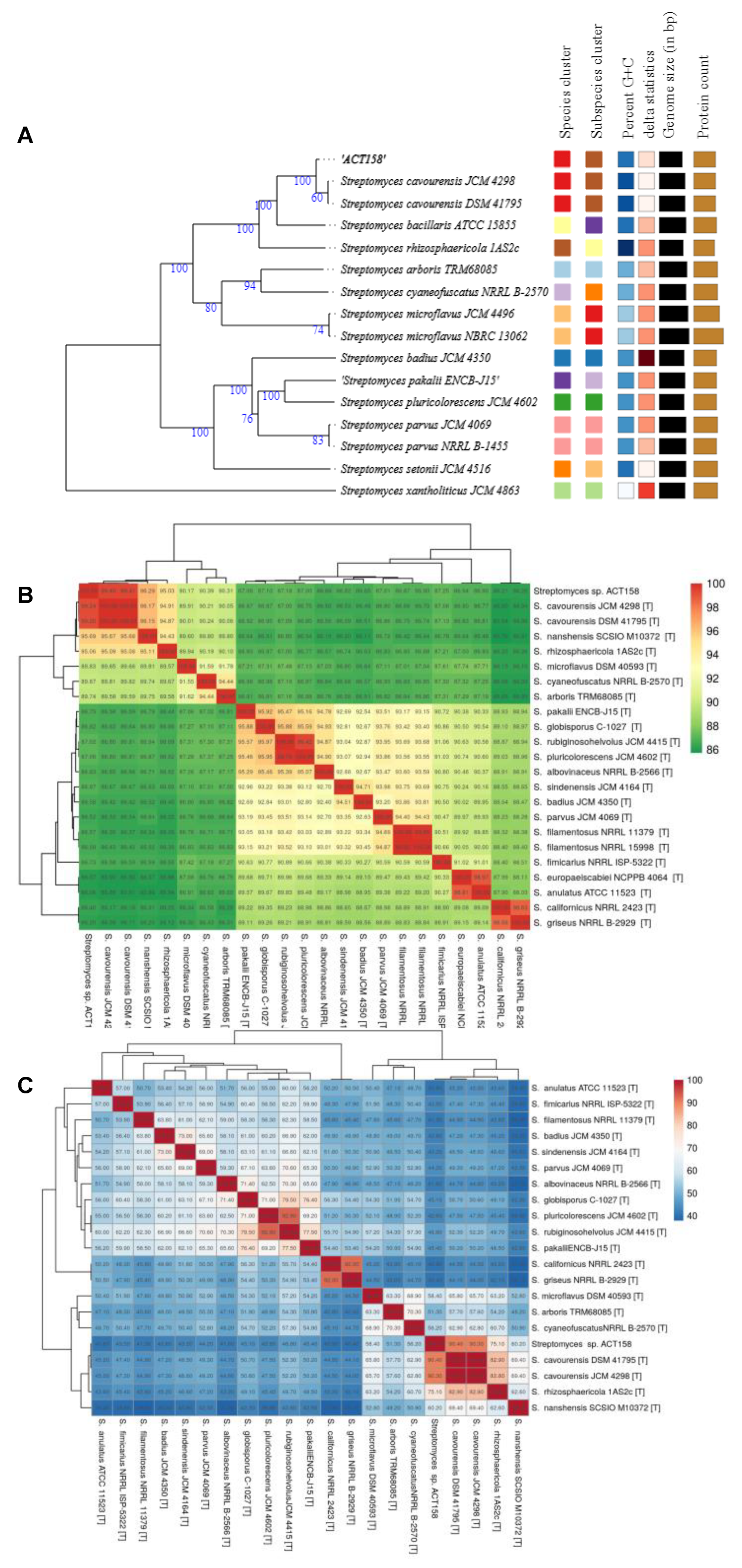
| Characteristics | Values |
| Total Length (bp) | 6,858,072 |
| Number of coding sequences | 6831 |
| GC Content (%) | 71.8% |
| N50 | 8,070 |
| Gap Ratio (%) | 0.0% |
| No. of CDSs | 5,122 |
| No. of rRNA | 3 |
| No. of tRNA | 54 |
| No. of CRISPRS | 8 |
| Coding Ratio (%) | 70.1% |
| Type | MIBiG accession |
Known cluster blast (Biosynthetic gene) |
Similarity | Potential activity |
| NRPS-T1PKS | BGC0001310 | naringenin | 100% | Antifungal, antiviral |
| NRPS-T1PKS | BGC0002591 | aurachin C | 20% | Antibacterial |
| NRPS-T1PKS | BGC0000236 | kinamycin | 13% | Antitumor |
| NRPS-T1PKS | BGC0000273 | steffimycin D | 16% | Antitumor |
| NRPS-T1PKS | BGC0000028 | bafilomycin B1 | 27% | Antibacterial, antitumor |
| NRPS-T1PKS | BGC0001477 | lydicamycin | 32% | Antibacterial |
| NRPS-T1PKS | BGC0001348 | JBIR-100 | 38% | Antimicrobial |
| NRPS-T1PKS | BGC0001700 | niphimycins C-E | 29% | Antimicrobial |
| NRPS-T2PKS | BGC0000234 | jadomycin | 42% | Antimicrobial |
| NRPS-T3PKS | BGC0000282 | alkylresorcinol | 100% | Antibacterial |
| NRPS | BGC0002001 | crochelin | 16% | Antimicrobial, antitumor |
| NRPS | BGC0001845 | alpiniamide | 88% | Antimicrobial |
| NRPS | BGC0000453 | valinomycin/montanastatin | 73% | Antimicrobial |
| NRPS | BGC0000325 | coelichelin | 63% | / |
| NRPS | BGC0001368 | JBIR-126 | 7% | Antimicrobial |
| NRPS | BGC0000368 | griseobactin | 30% | Antibacterial |
| NRPS | BGC0002441 | o-dialkylbenzene 1 | 12% | / |
| NRPS | BGC0002295 | minimycin | 40% | Antimicrobial |
| NRPS | BGC0002365 | heat-stable antifungal factor | 50% | Antifungal |
| NRPS | BGC0000375 | indigoidine | 40% | Antibacterial, antioxydant |
| RiPP | BGC0000583 | linaridin | 20% | Antibacterial |
| RiPP | BGC0000496 | AmfS | 60% | Antimicrobial |
| RiPP | BGC0000606 | lactazole | 22% | Antimicrobial, antitumor |
| RiPP | BGC0001634 | keywimysin | 80% | Antibacterial |
| Siderophore | BGC0000941 | desferrioxamin B | 100% | Antimicrobial,Iron Chelation |
| butyrolactone | BGC0001778 | showdomycin | 29% | Antimicrobial |
| butyrolactone | BGC0000038 | coelimycin | 8% | Antibacterial |
| Ectoine | BGC0000858 | ectoine | 75% | Osmolyte |
| Terpene | BGC0001181 | geosmine | 100% | / |
| Terpene | BGC0000664 | isorenieratene | 42% | / |
| Terpene | BGC0000663 | hopene | 46% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





