1. Introduction
Urinary tract infections (UTIs) rank among the most prevalent bacterial infections worldwide, affecting millions annually and imposing significant healthcare and economic burdens (Kot et al., 2021; López-Montesinos & Horcajada, 2019)。 Complicated urinary tract infections (cUTIs) are particularly challenging to treat due to their association with risk factors such as anatomical abnormalities, immunosuppression, and the presence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) pathogens (Wagenlehner et al., 2020 & Walkty et al., 2022). MDR is defined as acquired non-susceptibility to ≥ 1 agent in three or more different classes of antimicrobials while XDR is defined as being non-susceptible to ≥ 1 agent in all but remaining susceptible to two or fewer antimicrobial classes (Magiorakos et al., 2012). These infections are predominantly caused by Gram-negative bacilli, including Escherichia coli and Klebsiella pneumoniae, which frequently exhibit varying levels of resistance through mechanisms such as extended-spectrum beta-lactamase (ESBL) production, AmpC beta-lactamase, and carbapenemase activity. (Zilberberg et al., 2020; Ruppé et al., 2015).
The lack of diagnostic precision in identifying the resistance markers often leads to inappropriate antibiotic prescriptions, further fueling resistance. This necessitates routine identification of the resistance mechanisms so that appropriate antimicrobials can be prescribed to ensure optimum patient care while also applying principles of antimicrobial stewardship and utilising reserve antibiotics judiciously. This study evaluated the susceptibility profile of E coli and Klebsiella pneumoniae isolated from cUTI to inform practice, identify cheap, simple and reliable tools to identify ESBL, AmpC and carbapenemase resistant Enterobacterales (CRE) and to understand the circulating MLST and AMR genes in AmpC and carbapenemase producing strains by whole genome sequencing representative isolates.
2. Materials and Methods
2.1. Study Period and Setting
This study was conducted from September 2022 to August 2023 at the Department of Microbiology and Immunology, College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman. It was carried out in collaboration with Clinical Microbiology Laboratories at Sultan Qaboos University Hospital (SQUH) and Suhar Hospital, Oman. Ethical approval was obtained from the Medical Research Ethics Committee (MREC), College of Medicine & Health Sciences, Sultan Qaboos University (REF. NO. SQU-EC/377/2021) and the Health Studies and Research Approval Committee, Ministry of Health (MoH/CSR/21/24496).
2.2. Study Design
The study evaluated the demographic, and antimicrobial susceptibility profiles of E. coli and K. pneumoniae isolates from complicated urinary tract infection (cUTI) cases over a one year period.
2.3. Bacterial Identification and Susceptibility Testing
Bacterial identification was performed using MALDI-TOF MS (Bruker, Germany) and Phoenix™ (BD Diagnostics, USA) automated systems at SQUH, while VITEK 2 (Biomérieux) was employed at Suhar Hospital as per standard guidelines (Patel el al.,2015; Funke et al.,2005; Singhal et al., 2015). Antimicrobial susceptibility testing was performed and interpreted following CLSI guidelines (2022) (CLSI, M100- ed32., 2022). The automated systems identified the isolates as ESBLs. Carbapenem resistant isolates were subjected to GeneXpert ®Carba-R (Cepheid, Sunnyvale, CA) for confirmation of carbapenemase production, while AmpC was estimated on the susceptibility profile.
2.4. Manual Phenotypic Detection of ESBL, AmpC and CRE
ESBL Detection: ESBLs were detected by the double-disk synergy tests. Amoxicillin-clavulanate (AMC, 20/10 μg) (Oxoid, UK) was placed at the center of MHA and the β-lactam antibiotic discs, ceftazidime (CAZ, 30 μg) (Oxoid, UK), cefepime (FEP, 30 μg) (Oxoid, UK), aztreonam (ATM, 30 μg) (Oxoid, UK), cefotaxime (CTX, 30 μg) (Oxoid, UK), cefuroxime (CXM, 30μg) (Oxoid, UK), cefoxitin (FOX, 30 µg) ceftriaxone (CRO, 30 μg) (Oxoid, UK), cefoperazone (CFP, 75 μg) (Liofilchem, Italy) and cefpodoxime (CPD, 30 μg) (Liofilchem, Italy) were placed at a distance of 20mm (edge to edge) from the BL/BLI disk. Plates were incubated at 35 ± 2 °C for 18-24 hours and after incubation, the synergistic activity between BL/BLIs and β-lactam antibiotics were examined. Isolates were phenotypically confirmed as ESBL producers when a clear extension of the edge of the inhibition zone of β-lactam antibiotics was observed towards at least one of the BL/BLI disks or when a zone of inhibition appeared in between β-lactam antibiotic and BL/BLI disk (Waheed et al., 2019; Drieux et al., 2008).
AmpC Detection: Disk approximation test: A ceftazidime disk (CAZ, 30 μg) (Oxoid, UK) was placed in the center of the inoculated MHA and disks of the inducing substrates: imipenem (IPM, 10 µg), amoxicillin-clavulanic acid (AMC 20/10 µg) and piperacillin-tazobactam (TZP 110 µg) were placed at a distance of 20 mm apart (edge to edge) from a ceftazidime disk, while the distance between cefoxitin (FOX, 30 µg) and ceftazidime disk was adjusted to 15mm apart (edge to edge) to obtain better induction effect (Gupta et al., 2014; Al Mamari et al., 2023). The plate was incubated at 35°C ± 2 for 18-24 hours.
Figure 1
AmpC detection set (D69C): This is a commercial AmpC detection set from Mast Group Ltd, UK was used for the detection of AmpC β-lactamase enzyme production in 16 isolates. The set contained three cartridges A, B and C. The disk content of cartridge A is cefpodoxime (10 µg) with AmpC inducer while in cartridge B the disk contains cefpodoxime (10 µg) with AmpC inducer and ESBL inhibitor, and in cartridge D the disk composed of cefpodoxime (10 µg) with AmpC inducer, ESBL inhibitor and AmpC inhibitors. The inducers and inhibitors in each cartridge were unspecified. The procedure was performed and interpreted as per the manufacturer's instructions (Halstead et al., 2012)
Figure 1.
Carbapenemase Detection: Modified carbapenem inactivation method (mCIM), (CLSI, M100- ed33., 2023) and a lateral flow immunochromatographic assay KPC/IMP/NDM/VIM/OXA-48 Combo test kit (Medomics, China) for the five major carbapenemase families (KPC, IMP, NDM, VIM, OXA-48) were used for detection of CRE (
Figure 1).
2.5. Whole Genome Sequencing (WGS)
Representative AmpC and CRE
E. coli and
K. pneumoniae strains were subjected to WGS. Itwas carried out in MicrobesNG by Illumina next-generation sequencing (
https://microbesng.co.uk, Birmingham, United Kingdom). The DNA samples were prepared and then sequenced following the manufacturer’s protocol. After sequencing, the samples were assembled using the De Novo assembly by the SPAdes program (Bankevich et al., 2012). The assembled contigs were then annotated by Prokka (Seemann, 2014). The assembled and annotated sequences were uploaded to the website to carry out further analysis.
Detailed resistance gene profiling, with bioinformatics analysis was conducted using Center for Genomic Epidemiology (CGE) website (
https://www.genomicepidemiology.org/) for identifying multi-locus sequence typing (MLST) using MLST tool (Larsen et al., 2012), plasmids using PlasmidFinder tool (Carattoli et al., 2014) and antibiotic resistance genes using ResFinder tool (Bortolaia et al., 2020).
2.6. Statistical Analysis
Data were analyzed using SPSS (version 23). Continuous data were expressed as mean ± SD, while categorical variables were presented as percentages. Sensitivity and specificity analyses of diagnostic tests were performed, and results were visualized using Microsoft Excel.
3. Results
3.1. Demographics and Sample Characteristics
During the study period (September 2022–August 2023), 1,194 non-duplicate urine cultures from patients with suspected complicated urinary tract infections (cUTIs) were analyzed. Of these, 60% were from females (n=712) and 40% from males (n=482). Adults comprised the majority (89%, n=1,066), while 11% (n=128) were pediatric patients. The highest prevalence of cUTIs was observed in patients aged >60 years (47.2%), followed by those aged 30–60 years (32.7%) and ≤13 years (10.7%). Key complicating factors included urinary instrumentation, hypertension, diabetes, chronic kidney disease, dyslipidemia, sickle cell anaemia, malignancy, renal stones, urinary obstruction, benign prostatic hyperplasia and thalassemia.
3.2. Microbial Etiology
A total of 1,233 bacterial isolates were identified amongst which Gram-negative bacilli predominated with E. coli accounting for 406 (33%) and K. pneumoniae 163 (13.5%). In midstream urine (MSU) samples, E. coli was the most common pathogen in both adults (35.5%) and children (49.4%) while in catheter specimen urine (CSU) samples, Candida spp. predominated in adults (41.9%) and E. coli in the pediatric age group (31.1%)
3.3. Antimicrobial Susceptibility Profiles
E. coli exhibited high susceptibility to fosfomycin (100%), nitrofurantoin (96%), piperacillin-tazobactam (95%), gentamicin (87%), amikacin (98%) and meropenem (99%).
K. pneumoniae had lower susceptibility rates than
E. coli, with 38% to nitrofurantoin, fosfomycin 89%, gentamicin (80%), amikacin (84%) and 84% for meropenem susceptibilities, as shown in
Table 1.
Lower susceptibility was observed for fluoroquinolones (ciprofloxacin (44%), levofloxacin (46%) The susceptibility rates to cephalosporins was slightly lower in E. coli (cefazolin (27%), cefuroxime (51%), ceftriaxone (54%), ceftazidime (57%), and cefepime (56%)) than in K. pneumoniae (cefazolin (33%), cefuroxime (55%), ceftriaxone (61%), ceftazidime (60%), and cefepime (63%)). Similar susceptibility to amoxicillin-clavulanate in E. coli and K. pneumoniae was observed: 66% and 65% respectively.
3.4. Prevalence of ESBL, AmpC Beta-Lactamases, and Carbapenemases
ESBL: The overall prevalence of ESBL among
E. coli and
K. pneumoniae (n=569) was 37.2%, with E. coli accounting for 40.4% and
K. pneumoniae 20.2%. (
Table 2)
The ESBL prevalence increased to 43.8% in E. coli and 20.8% when isolates co-harbouring ESBL and AmpC were included.
AmpC: AmpC alone was identified in 16 (2.8%) isolates while AmpC and ESBL co-carriage was found in 15(2.6%) cases.
Carbapenemases: The overall prevalence of carbapenemase producers was 35 (6.2%).
K. pneumoniae predominated as a carbapenemase producer, accounting for 17.2% of the total K. pneumoniae isolated, compared to E. coli 1.7% as seen in
Table 2.
3.5. Detection of Resistance Genes in AmpC Beta Lactamase Producing E. coli and Carbapenemase Producing K. pneumoniae
Diverse antimicrobial resistance genes were detected in the 11 AmpC producing E. coli and 22 carbapenemase producing Klebsiella pneumoniae isolates.
The MLST analysis of the AmpC strains identified the presence of 9 E. coli lineages: ST-10, ST-69, ST-77, ST-131, ST-156, ST-167, ST-361, ST-1125 and ST-2520, with ST-10 and ST-69 being observed in two isolates each.
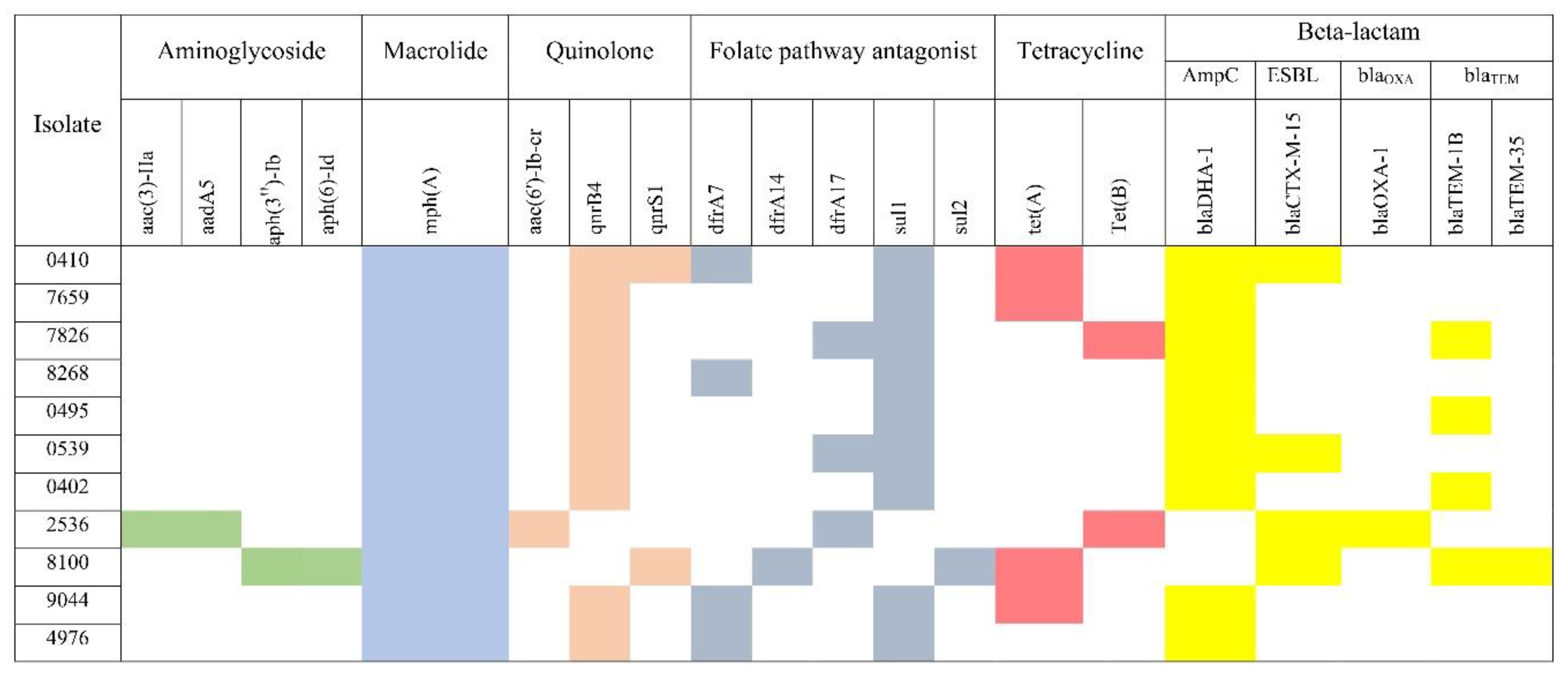
The beta-lactam resistance genes were detected in all isolates as seen in the heat map (
Figure 2). Five different beta-lactam resistance genes were found blaDHA1, blaCTX-M-15, blaOXA-1, blaTEM-1B and blaTEM-35. The AmpC gene blaDHA-1 was detected in 9 isolates. The two isolates, that tested negative for AmpC genes co-harboured blaCTX-M-15 and blaOXA-1, and blaCTX-M-15, blaTEM-1B and blaTEM-35 respectively (
Figure 2). Three were resistant to piperacillin-tazobactam
E. coli 0410, 2536 and 8100 and all co-harboured more than one β-lactamases .However only one carried bla OXA-1. Three different quinolone resistance genes were identified, aac(6')-Ib-cr, qnrB4 and qnrS1. The majority of isolates harboured only one of these quinolone resistance genes and qnrB4 was the predominant gene found in 9 isolates. Regarding ciprofloxacin susceptibility, only five isolates (5/11) remained susceptible and all carried the qnrB4 gene. Among the 11 analysed isolates, only 2 were resistant to cotrimoxazole. The tetracycline resistance genes (tet(A) and tet(B)) were detected in 7 isolates and tet(A) was the most frequent. Only two carried aminoglycoside-resistant genes,
E. coli 2536 (aac(3)-IIa and aadA5) and
E. coli 8100 (aph(3'')-Ib and aph(6)-Id).The marA gene which is responsible for antibiotic efflux as well as reduced cellular permeability to antibiotics was detected in the majority of isolates (10/11). The most frequently detected efflux pump genes belonged to resistance-nodulation-cell division (RND) and major facilitator superfamily (MFS) efflux gene families (
Supplementary Table S1). Overall, H-NS was the predominant efflux gene detected in all isolates which confers resistance to macrolide, fluoroquinolone, cephalosporin, cephamycin, penam, and tetracycline, followed by evgA (10 isolates), TolC (9 isolates) and emrB (9 isolates). The efflux gene qacEdelta1 was only found in one isolate. Four antibiotic target alteration genes were detected (bacA, PmrF, ugd and eptA) of which PmrF was the predominant gene found in 7 isolates.
In the 22 carbapenemase producing
K. pneumoniae, MLST revealed the presence of 5 lineages: ST-2096, ST-231, ST-147, ST-1770 and ST-111 with ST-2096 (12 isolates) and ST-147 (7 isolates) being the most common. The ST-1770 was identified as
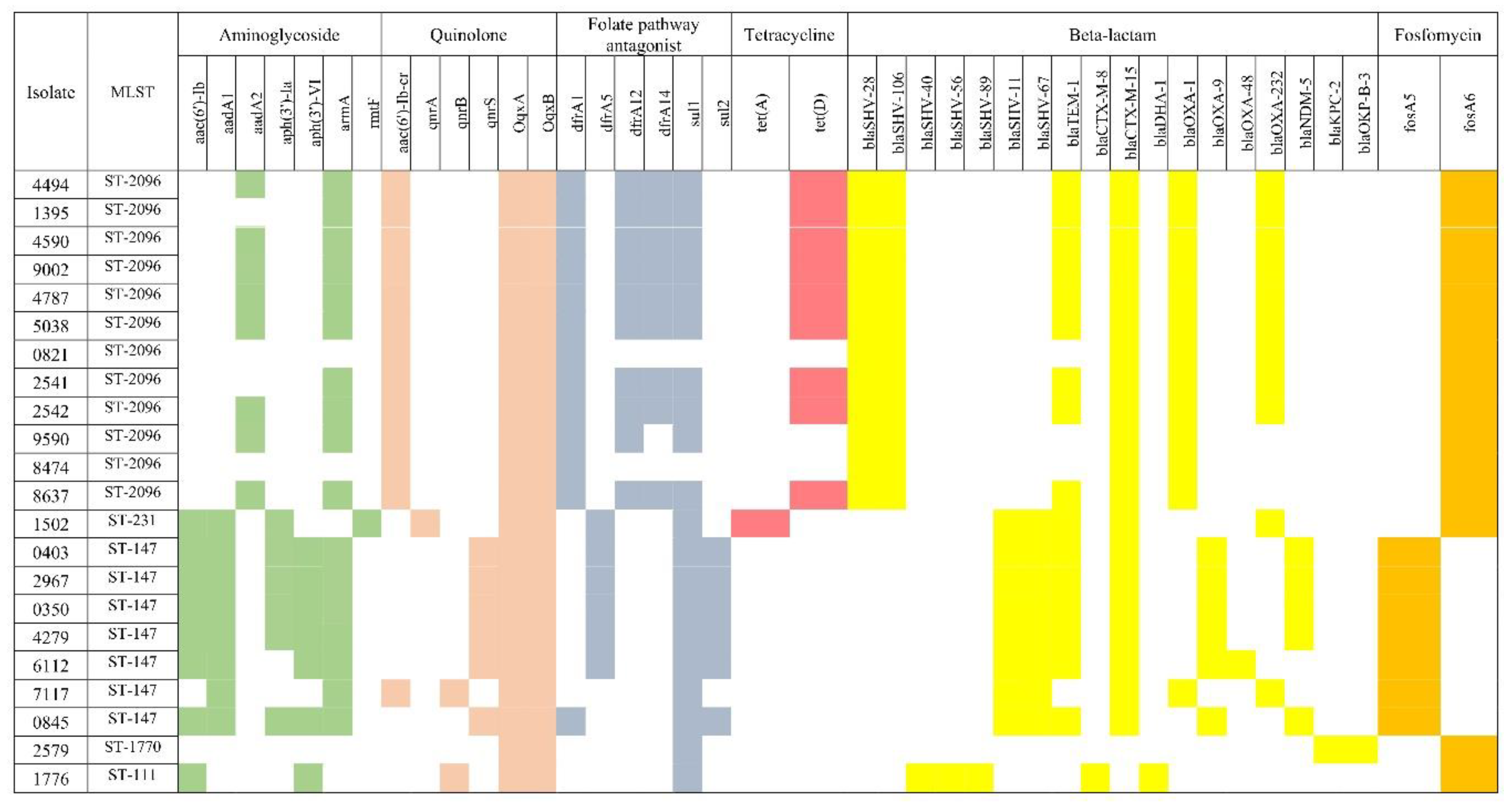
Klebsiella quasipneumoniae.The resistance genes’ heatmap using resfinder is shown in
Figure 3. The β-lactam resistance genes were the most predominant among all identified antibiotic-resistant genes. In total, 18 different β-lactam resistance genes were found (blaSHV-11, blaSHV-28, blaSHV-40, blaSHV-56, blaSHV-67, blaSHV-89, blaSHV-106, blaTEM-1, blaCTX-M-8, blaCTX-M-15, blaDHA-1, blaOXA-1, blaOXA-9, blaOXA-48 ,blaOXA-232, blaNDM-5, blaKPC-2 and blaOKP-B-3). Among SHV genes, blaSHV-28 and blaSHV-106 were the most common as only detected in isolates of ST-2096. All isolates of ST-147 carried the blaSHV-11 and blaSHV-67. The blaTEM-1 detected in 72.7% (16/22) of the isolates. All isolates of ST-2096, ST-147 and ST231 carried the ESBL gene blaCTX-M-15. All isolates of ST-2096 carried the blaOXA-1 while the majority (6/7) of isolates of ST-147 harboured blaOXA-9. The blaDHA-1 and blaCTX-M-8, blaSHV-40, bla-SHV56 and bla-SHV89 detected only once in isolate (
K. pneumoniae 1776) which was negative for carbapenemase. Among the carbapenemase genes, blaOXA-232 was detected in 11 isolates, blaNDM-5 in 5 isolates, blaOXA-48 in 1 isolate and blaKPC-2 in 1 isolate. The one ST-1770 carried KPC-2 carbapenemase. All carbapenemase producing isolates were non-susceptible to ertapenem, imipenem and meropenem except the
K. pneumoniae isolate 7117 which harboured the blaOXA-232. Eight aminoglycoside resistance genes (aac(6')-Ib, aadA1, aadA2, aph(3')-Ia, ph(3')-VI, aph(6)-Id, armA and rmtF) were identified and the majority (17 isolates) carried the armA gene. The rmtF gene was detected only in one isolate. Eighteen isolates resistant to gentamicin, netilmicin and amikacin harboured 16S rRNA methyltransferase (17 isolates carried armA while one carried the rmtF).Six quinolone resistance genes were identified among CRE
K. pneumoniae, aac(6')-Ib-cr , qnrA, qnrB, qnrS, OqxA and OqxB. All isolates harboured the fluoroquinolone resistance genes OqxA and OqxB among which 13 isolates co-harboured aac(6')-Ib-cr. The mdtq gene which is responsible for antibiotic efflux as well as reduced cellular permeability to antibiotics was detected in all isolates except one (
Supplementary Table S2). Four more efflux pumps, lptD,
Klebsiella pneumoniae kpnE,
Klebsiella pneumoniae kpnF and qacEdelta1 were detected by RGI and not detected by ResFinder and
Klebsiella pneumoniae kpnF detected in all isolates while qacEdelta1 in 18 isolates.
4. Discussion
Patients with cUTIs are at a higher risk of treatment failure, since these infections are frequently caused by MDR Gram-negative bacteria (Li et al., 2017). Mapping susceptibility of uropathogens in cUTI is imperative to institute optimum empiric treatment. E. coli and K. pneumoniae, the most frequently detected pathogens in UTIs are commonly associated with β-lactamases such as ESBLs, AmpC β-lactamases and carbapenemases (Wang & Palasik, 2022). Optimizing their detection is critical for appropriate management of cUTIs and promoting antimicrobial stewardship.
Our study revealed similar as well as key critical differences in their susceptibility rates. E. coli exhibited a higher susceptibility rate to nitrofurantoin (96%) than K. pneumoniae (38%). However both displayed excellent rates to fosfomycin (100% vs 89%). Babiker et al.,(2019) too advocated fosfomycin as a good option for management of UTI. Fosfomycin has a low incidence of collateral damage and a broad antimicrobial spectrum. Both exhibited susceptibility greater than 60% to cotrimoxazole and less than 50% to fluoroquinolones which significantly limits their utility in empiric management. Similar rates were reported by Li et al. (2017). Susceptibility to cephalosporins were slightly lower in E. coli [cefazolin (27%), cefuroxime (51%), ceftriaxone (54%), ceftazidime (57%), and cefepime (56%)] than in K. pneumoniae [cefazolin (33%), cefuroxime (55%), ceftriaxone (61%), ceftazidime (60%), and cefepime (63%)] which indicates higher prevalence of ESBL in E.coli than K. pneumoniae which was corroborated by further testing. Mohamed et al. (2020) reported a similar trend.
Conversely the lower piperacillin-tazobactam, aminoglycoside and carbapenem susceptibility rates in K. pneumoniae [piperacillin-tazobactam 72%, amikacin (84%), gentamicin (80%), ertapenem (81%), imipenem (83%), and meropenem (84%)] compared to E. coli [piperacillin-tazobactam (95%), amikacin (98%) and gentamicin (87%), ertapenem (98%), imipenem (98%), and meropenem (99%)] suggests K. pneumoniae is more likely to carry blaOXA-1, aminoglycoside and carbapenem resistance genes. Fernández-Martínez et al., 2018; Abo-State et al., 2018; Arumugam et al., 2024 corroborated these findings.
An interesting epidemiology emerged. E. coli ESBL prevalence (43.8%) was double that in K. pneumoniae (20.8%). An Egyptian study reported a higher prevalence of ESBL, (62%-69% in E. coli, which too was double that of K. pneumoniae (30%-37%) (Shash et.al., 2019). AmpC prevalence too was higher in E. coli (5.9%) than in K. pneumoniae (4.3%). CRE however predominated among K. pneumoniae (17.2% vs 1.7% in E. coli). AL Mamari et al., 2022 reported similar prevalence in this region. As in other parts of the world, XDR K. pneumoniae is a grave public health concern. (Kaza et al., 2024; Chapelle et al., 2021).
A surprisingly pronounced genetic heterogeneity was observed in the 11 AmpC carrying strains, with 9 MLST types observed (ST-10, ST-69, ST-77, ST-131, ST-156, ST-167, ST-361, ST-1125 and ST-2520), indicating the great flux in the E. coli clades circulating in this region. Surprisingly all AmpC producing E. coli carried blaDHA-1 with only two isolates co-harbouring blaCTX-M-15 and three blaTEM-1B. Only ST-167, ST-361, ST-131, and ST-156 have been reported previously from this region (Al-Farsi et al., 2020). Our study highlights the high prevalence of the plasmid-mediated AmpC gene, blaDHA-1, among FOX-resistant E. coli isolates in cUTI. Similar high prevalence of blaDHA-1 in uropathogenic E. coli have been reported from Egypt, Iran and India although Saudi Arabia reported predominance of CMY-2. Mohamed et al. (2020), (Jomehzadeh et al., 2021), (Mohamudha et al., 2012; Bala et al., 2020) (Abdalhamid et al. 2017).
Surprisingly they did not carry aminoglycoside modifying/resistance genes, supporting the empirical use of gentamicin and amikacin in managing E. coli AmpC mediated cUTI infections. On the other hand quinolones should be prescribed with caution as all bla DHA-1 carried the qnrB4 gene .The predominant gene qnrB4 was detected only in AmpC-producing isolates harbouring the blaDHA-1 gene. Similar findings were reported by Kamruzzaman et al. (2013), who found E. coli isolates carried both blaDHA-1 and qnrB. These findings were in agreement with Mata et al. (2011) who found a close association between blaDHA-1 and qnrB genes. Our results support these findings as all blaDHA-1 isolates carried the qnrB gene.) The aac(6')-Ib-cr is a multifunctional gene that can induce resistance to both ciprofloxacin and aminoglycosides (Eftekhar et al., 2015).
Much lower MLST heterogeneity was observed in the CRE K. pneumoniae, with 5 lineages being identified: ST-2096, ST-231, ST-147, ST-1770 and ST-111 with ST-2096 predominating. These findings are starkly different from a previous study in Oman where the majority of XDR/PDR K. pneumoniae belonged to ST-231(Al-Quraini et al., 2022). Al Fadhli et al., 2023 reported that the ST-14 and ST-231 were the predominant STs in the Arabian Peninsula with ST-231 predominating in Oman and ST-2096 in Saudi Arabia. A study from India found that ST-231 was the most common ST (34.8%) among CRE K. pneumoniae, followed by ST-147 (23.5%) and ST-14 (Nagaraj et al., 2021).Maybe uropathogenic K. pneumoniae are characterised by different MLSTs compared to other invasive K. pneumoniae infections. Further studies are needed in cUTI to delineate any ST predominance.
Among the carbapenemases, blaOXA-232 predominated, followed by blaNDM-5, blaOXA-48 and blaKPC-2 . All isolates of ST-2096 and ST-147 carried the ESBL gene blaCTX-M-15 which reflects the strong association and co-localization of blaOXA232 and blaNDM-5 with blaCTX-M-15 in these clades, an association also observed in other studies. (Shukla et al., 2023; Shankar et al., 2022 ;Sundaresan et al., 2022). ST-231 co-harboured ESBL gene blaCTX-M-15 and bla-OXA232 which was also reported by Al-Quraini et al. (2022) and Mancini et al. (2018). All isolates of ST-2096 carried the blaOXA-1while the majority (6/7) of isolates of ST-147 harboured blaOXA-9. Shankar et al. (2022) found that blaOXA-1 integrated in the chromosome of isolates of ST-2096 and Di Pilato et al. (2022) reported blaOXA-1 and blaOXA-9 in ST-147. K. pneumoniae ST-2096 predominantly carried OXA-232 carbapenemase, a frequent variant of OXA-48-like carbapenemase in Oman while ST-147 predominantly carried NDM-5. Studies from Qatar and India corroborate our findings.(Abid et al., 2021)(Shukla et al., 2023). Surprisingly ST111 co-harboured bla-DHA-1 with blaCTX-M-8 but did not posses carbapenemases. It was interesting to noted that all the other carbapenemase carrying ST types did not carry AmpC beta lactamases. Klebsiella quasipneumoniae ST-1770 carried KPC-2 carbapenemase. A study from China too reported Klebsiella quasipneumoniae ST-1770 from ICU patients (Wang et al., 2023). Klebsiella quasipneumoniae is considered an emerging pathogen in healthcare settings which can acquire carbapenemase plasmids from other CRE isolates (Mathers et al., 2019). Venkitapathi et al., 2022, have reported this pathogen in recurrent UTI in women.
Uniform aminoglycoside resistance was mediated by armA, aac(6')-Ib, aadA1, aadA2, aph(3')-Ia, ph(3')-VI, aph(6)-Id, and rmtF. Alarmingly, majority (17/22 isolates) carried the armA gene. Among 16S rRNA methyltransferase genes, armA and rmtB are the most prevalent in Asia (Shen et al., 2020; Saadatian et al., 2018; Al Sheikh et al., 2014). In contrast, Al-Quraini et al. (2022), detected armA and rmtB genes only in one isolate which co-harboured blaOXA-232 and bla-NDM-5. These findings indicate the increasing prevalence of plasmids carrying the armA gene among the CRE K. pneumoniae in Oman which may be attributed to the excessive antimicrobial use during the COVID-19 pandemic.The findings in the current study indicated the emergence of CRE K. pneumoniae strains harbouring armA and rmtF genes that retain resistance to all currently available aminoglycosides which is a cause of great concern.
Fluoroquinolone resistance was mediated by OqxA and OqxB genes while ST-2096 co-harboured the aac(6')-Ib-cr and ST-147 predominantly co-harboured qnrS. Urooj et al., 2022 reported similar findings. The aac(6′)-Ib-cr gene was found integrated in the chromosome of K. pneumoniae of ST-2096 which was transferred through MGEs (Shankar et al., 2022). A study from India reported qnrB, aac(6′)-Ib-cr, oqxA, oqxB in XDR K. pneumoniae belonging to ST147 (Dey et al., 2020).
The fosA6 was the predominant gene in ST-2096 and ST-231 clones which agrees with previous studies from California and India (Cerón et al.,2023; Boonyasiri et al., 2021). On the other hand, the fosA5 was only found in the ST-147 clone, similar to Al-Quraini et al’s report. (2022). Although all isolates harboured the fosA gene, only four 4 isolates phenotypically expressed resistance to fosfomycin.
Identifying the molecular types of AmpC β-lactamase and CRE at the national level is needed for understanding the burden and epidemiology of resistance and in optimising the antimicrobial therapy as well as in promoting antimicrobial stewardship and greater vigilance in infection control.
5. Conclusions
Fosfomycin emerges as an excellent choice for the management of UTI as it has a broad spectrum of action. Nitrofurantoin remains an excellent oral treatment option for cystitis caused by E. coli but not for K. pneumoniae. Empirical fluoroquinolone therapy is best avoided, only to be prescribed after susceptibility confirmation. ESBL and AmpC prevalence was higher in E. coli while CRE in K. pneumoniae. DHA-1 was the only incriminating AmpC while OXA-232 and NDM-5 were the predominant circulating carbapenemases. In AmpC producing E. coli significant MLST heterogeneity was observed while in CRE Klebsiella pneumoniae ST-2096 and ST-147 predominated. Routine monitoring of the circulating clades is important for tracking the spread of drug-resistant variants in different regions. Concerning emergence of CRE K. pneumoniae strains harbouring armA and rmtF genes that are resistant to all currently available aminoglycosides was observed. Tracking the extent of dissemination of these stains at the national level is essential along with the implementation of effective infection prevention control measures.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, M.R. and N.A.S; data curation, N.A.A. and Z.A.J.; formal analysis, M.R. and N.A.S; investigation, N.A.S. and Z.A.J.; methodology, M.R.; writing—original draft preparation, N.A.S.; writing—review and editing, T.A.L, Z.A.I, F.K, S.R, Z.A.M, H.S and M.R.; supervision, M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Sultan Qaboos University internal grant (IG/MED/MICR/24/01)
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman (SQU-EC/377/2021).
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Kot, B., Grużewska, A., Szweda, P., Wicha, J., & Parulska, U. (2021). Antibiotic resistance of uropathogens isolated from patients hospitalized in district hospital in central Poland in 2020. Antibiotics, 10(4). [CrossRef]
- López-Montesinos, I., & Horcajada, J. P. (2019). Oral and intravenous fosfomycin in complicated urinary tract infections. Revista espanola de quimioterapia: publicacion oficial de la Sociedad Espanola de Quimioterapia, 32(1), 37–44. https://api.semanticscholar.org/CorpusID:167210853.
- Wagenlehner, F. M. E., Bjerklund Johansen, T. E., Cai, T., Koves, B., Kranz, J., Pilatz, A., & Tandogdu, Z. (2020). Epidemiology, definition and treatment of complicated urinary tract infections. In Nature Reviews Urology, 17 (10), 586–600. [CrossRef]
- Walkty, A., Karlowsky, J. A., Lagace-Wiens, P., Baxter, M. R., Adam, H. J., & Zhanel, G. G. (2022). Antimicrobial resistance patterns of bacterial pathogens recovered from the urine of patients at Canadian hospitals from 2009 to 2020. JAC-Antimicrobial Resistance, 4(6), dlac122. [CrossRef]
- Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., & Monnet, D. L. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281.
- Zilberberg, M. D., Nathanso, B. H., Sulham, K., & Shor, A. F. (2020). Antimicrobial susceptibility and cross-resistance patterns among common complicated urinary tract infections in U.S. hospitals, 2013 to 2018. Antimicrobial Agents and Chemotherapy, 64(8). [CrossRef]
- Ruppé, É., Woerther, P. L., & Barbier, F. (2015). Mechanisms of antimicrobial resistance in Gram-negative bacilli. In Annals of Intensive Care, 5 (1), 61. [CrossRef]
- Patel, R. (2013). MALDI-TOF MS for the diagnosis of infectious diseases. Clinical Chemistry, 59(2), 340-342.
- Funke, G., & Funke-Kissling, P. (2005). Evaluation of the new VITEK 2 card for identification of medically relevant gram-positive cocci. Journal of Clinical Microbiology, 43(1), 84-88.
- Singhal, N., Kumar, M., Kanaujia, P. K., & Virdi, J. S. (2015). MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Frontiers in Microbiology, 6, 791.
- Waheed, A., Saleem, S., Shahzad, N., Akhtar, J., Saeed, M., Jameel, I., Rasheed., F, Jahan., S. (2019). Prevalence of Extended Spectrum [beta]-lactamase SHV and OXA Producing Gram Negative Bacteria at Tertiary Care Hospital of Lahore, Pakistan. Pakistan Journal of Zoology. 51(6):2345. http://dx.doi.org/10.17582/journal.pjz/2019.51.6.2345.2351.
- Drieux, L., Brossier, F., Sougakoff, W., & Jarlier, V. (2008). Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 14 Suppl 1, 90–103.
- Gupta, G., Tak, V., & Mathur, P. (2014). Detection of AmpC β Lactamases in Gram-negative Bacteria. Journal of laboratory physicians, 6(1), 1–6. [CrossRef]
- Al Mamari, A. M. K., Al Jabri, Z., Sami, H., Rizvi, S. G. A., Chan, M. F., Al Siyabi, T., Al Muharrmi, Z., & Rizvi, M. (2023). Evaluation of six commercial and in-house phenotypic tests for detection of AmpC β-lactamases: is routine detection possible?. JAC-antimicrobial resistance, 5(5), dlad101. [CrossRef]
- Halstead, F. D., Vanstone, G. L., & Balakrishnan, I. (2012). An evaluation of the Mast D69C AmpC Detection Disc Set for the detection of inducible and derepressed AmpC β-lactamases. The Journal of antimicrobial chemotherapy, 67(9), 2303–2304. [CrossRef]
- Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A., & Pevzner, P. A. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology : a journal of computational molecular cell biology, 19(5), 455–477. [CrossRef]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England), 30(14), 2068–2069. [CrossRef]
- Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., Jelsbak, L., Sicheritz-Pontén, T., Ussery, D. W., Aarestrup, F. M., & Lund, O. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. Journal of clinical microbiology, 50(4), 1355–1361. [CrossRef]
- Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., Møller Aarestrup, F., & Hasman, H. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial agents and chemotherapy, 58(7), 3895–3903. [CrossRef]
- Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., Philippon, A., Allesoe, R. L., Rebelo, A. R., Florensa, A. F., Fagelhauer, L., Chakraborty, T., Neumann, B., Werner, G., Bender, J. K., Stingl, K., Nguyen, M., Coppens, J., Xavier, B. B., Malhotra-Kumar, S., Aarestrup, F. M. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. The Journal of antimicrobial chemotherapy, 75(12), 3491–3500. [CrossRef]
- Li, X., Chen, Y., Gao, W., Ye, H., Shen, Z., Wen, Z., & Wei, J. (2017). A 6-year study of complicated urinary tract infections in southern China: prevalence, antibiotic resistance, clinical and economic outcomes. Therapeutics and clinical risk management, 13, 1479–1487. [CrossRef]
- Wang, H., & Palasik, B. N. (2022). Combating antimicrobial resistance with cefiderocol for complicated infections involving the urinary tract. In Therapeutic Advances in Urology, 14, 17562872211065570. [CrossRef]
- Babiker, A., Clarke, L., Doi, Y., & Shields, R. K. (2019). Fosfomycin for treatment of multidrug-resistant pathogens causing urinary tract infection: A real-world perspective and review of the literature. Diagnostic microbiology and infectious disease, 95(3), 114856. [CrossRef]
- Mohamed, A., Mohamud, M., & Mohamud, H. (2020). Epidemiology and Antimicrobial Susceptibility Pattern of Uropathogens in Patients with the Community- and Hospital-Acquired Urinary Tract Infections at a Tertiary Hospital in Somalia. Jundishapur Journal of Microbiology, 13(9), e107453. [CrossRef]
- Al Mamari, Y., Sami, H., Siddiqui, K., Tahir, H. B., Al Jabri, Z., Al Muharrmi, Z., Rizvi, S. G. A., & Rizvi, M. (2022). Trends of antimicrobial resistance in patients with complicated urinary tract infection: Suggested empirical therapy and lessons learned from a retrospective observational study in Oman. Urology annals, 14(4), 345–352. [CrossRef]
- Shash, R. Y., Elshimy, A. A., Soliman, M. Y., & Mosharafa, A. A. (2019). Molecular Characterization of Extended-Spectrum β-Lactamase Enterobacteriaceae Isolated from Egyptian Patients with Community- and Hospital-Acquired Urinary Tract Infection. The American journal of tropical medicine and hygiene, 100(3), 522–528. [CrossRef]
- Fernández-Martínez, M., Ruiz Del Castillo, B., Lecea-Cuello, M. J., Rodríguez-Baño, J., Pascual, Á., Martínez-Martínez, L., & Spanish Network for the Research in Infectious Diseases (REIPI) and the Spanish Group for Nosocomial Infections (GEIH) (2018). Prevalence of Aminoglycoside-Modifying Enzymes in Escherichia coli and Klebsiella pneumoniae Producing Extended Spectrum β-Lactamases Collected in Two Multicenter Studies in Spain. Microbial drug resistance (Larchmont, N.Y.), 24(4), 367–376. [CrossRef]
- Abo-State, M.,Saleh, Y., Ghareeb,H. (2018). Prevalence and sequence of aminoglycosides modifying enzymes genes among E.coli and Klebsiella species isolated from Egyptian hospitals. Journal of Radiation Research and Applied Sciences, 11(4), 408-415. [CrossRef]
- Arumugam, K., Karande, G. S., & Patil, S. R. (2024). Prevalence of Carbapenemase Production Among Klebsiella and Escherichia coli Isolated From Urinary Tract Infections. Cureus, 16(10), e70918. [CrossRef]
- Kaza, P., Xavier, B. B., Mahindroo, J., Singh, N., Baker, S., Nguyen, T. N. T., Mavuduru, R. S., Mohan, B., & Taneja, N. (2024). Extensively Drug-Resistant Klebsiella pneumoniae Associated with Complicated Urinary Tract Infection in Northern India. Japanese journal of infectious diseases, 77(1), 7–15. [CrossRef]
- Chapelle, C., Gaborit, B., Dumont, R., Dinh, A., & Vallée, M. (2021). Treatment of UTIs Due to Klebsiella pneumoniae Carbapenemase-Producers: How to Use New Antibiotic Drugs? A Narrative Review. Antibiotics, 10(11), 1332. [CrossRef]
- Al Farsi, H. M., Camporeale, A., Ininbergs, K., Al-Azri, S., Al-Muharrmi, Z., Al-Jardani, A., & Giske, C. G. (2020). Clinical and molecular characteristics of carbapenem non-susceptible Escherichia coli: A nationwide survey from Oman. PloS one, 15(10), e0239924. [CrossRef]
- Mohamed, E. S., Khairy, R. M. M., & Abdelrahim, S. S. (2020). Prevalence and molecular characteristics of ESBL and AmpC β -lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrobial resistance and infection control, 9(1), 198. [CrossRef]
- Jomehzadeh, N., Ahmadi, K., & Rahmani, Z. (2021). Prevalence of plasmid-mediated AmpC β-lactamases among uropathogenic Escherichia coli isolates in southwestern Iran. Osong public health and research perspectives, 12(6), 390–395. [CrossRef]
- Mohamudha, P. R., Harish, B. N., & Parija, S. C. (2012). Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. The Indian journal of medical research, 135(1), 114–119. [CrossRef]
- Bala, R., Singh, V. A., Gupta, N., & Rakshit, P. (2020). Prevalence, multidrug-resistance and risk factors for AmpC β-lactamases producing Escherichia coli from hospitalized patients. Journal of infection in developing countries, 14(12), 1466–1469. [CrossRef]
- Abdalhamid, B., Albunayan, S., Shaikh, A., Elhadi, N., & Aljindan, R. (2017). Prevalence study of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible ampC from Saudi hospitals. Journal of medical microbiology, 66(9), 1286–1290. [CrossRef]
- Kamruzzaman, M., Shoma, S., Bari, S. M., Ginn, A. N., Wiklendt, A. M., Partridge, S. R., Faruque, S. M., & Iredell, J. R. (2013). Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagnostic microbiology and infectious disease, 76(2), 222–226. [CrossRef]
- Mata, C., Miró, E., Toleman, M., Rivera, M. A., Walsh, T. R., & Navarro, F. (2011). Association of bla(DHA-1) and qnrB genes carried by broad-host-range plasmids among isolates of Enterobacteriaceae at a Spanish hospital. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 17(10), 1514–1517.
- Eftekhar, F., PhD, & Seyedpour, S. M., MSc (2015). Prevalence of qnr and aac(6')-Ib-cr Genes in Clinical Isolates of Klebsiella Pneumoniae from Imam Hussein Hospital in Tehran. Iranian journal of medical sciences, 40(6), 515–521. https://api.semanticscholar.org/CorpusID:16193718.
- Al Quraini, M., Rizvi, M., Al-Jabri, Z., Sami, H., Al-Muzahmi, M., Al-Muharrmi, Z., Taneja, N., Al-Busaidi, I., & Soman, R. (2022). Assessment of In-Vitro Synergy of Fosfomycin with Meropenem, Amikacin and Tigecycline in Whole Genome Sequenced Extended and Pan Drug Resistant Klebsiella Pneumoniae: Exploring A Colistin Sparing Protocol. Antibiotics (Basel, Switzerland), 11(2), 153. [CrossRef]
- Al Fadhli, A. H., Mouftah, S. F., Jamal, W. Y., Rotimi, V. O., & Ghazawi, A. (2023). Cracking the Code: Unveiling the Diversity of Carbapenem-Resistant Klebsiella pneumoniae Clones in the Arabian Peninsula through Genomic Surveillance. Antibiotics (Basel, Switzerland), 12(7), 1081. [CrossRef]
- Nagaraj, G., Shamanna, V., Govindan, V., Rose, S., Sravani, D., Akshata, K. P., Shincy, M. R., Venkatesha, V. T., Abrudan, M., Argimón, S., Kekre, M., Underwood, A., Aanensen, D. M., Ravikumar, K. L., & NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. (2021). High-Resolution Genomic Profiling of Carbapenem-Resistant Klebsiella pneumoniae Isolates: A Multicentric Retrospective Indian Study. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, 73(4), 300–307. [CrossRef]
- Shukla, S., Desai, S., Bagchi, A., Singh, P., Joshi, M., Joshi, C., Patankar, J., Maheshwari, G., Rajni, E., Shah, M., & Gajjar, D. (2023). Diversity and Distribution of β-Lactamase Genes Circulating in Indian Isolates of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics (Basel, Switzerland), 12(3), 449. [CrossRef]
- Shankar, C., Vasudevan, K., Jacob, J. J., Baker, S., Isaac, B. J., Neeravi, A. R., Sethuvel, D. P. M., George, B., & Veeraraghavan, B. (2022). Hybrid Plasmids Encoding Antimicrobial Resistance and Virulence Traits Among Hypervirulent Klebsiella pneumoniae ST2096 in India. Frontiers in cellular and infection microbiology, 12, 875116. [CrossRef]
- Sundaresan, A. K., Vincent, K., Mohan, G. B. M., & Ramakrishnan, J. (2022). Association of sequence types, antimicrobial resistance and virulence genes in Indian isolates of Klebsiella pneumoniae: A comparative genomics study. Journal of global antimicrobial resistance, 30, 431–441. [CrossRef]
- Mancini, S., Poirel, L., Tritten, M. L., Lienhard, R., Bassi, C., & Nordmann, P. (2018). Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. The Journal of antimicrobial chemotherapy, 73(3), 821–823. [CrossRef]
- Di Pilato, V., Henrici De Angelis, L., Aiezza, N., Baccani, I., Niccolai, C., Parisio, E. M., Giordano, C., Camarlinghi, G., Barnini, S., Forni, S., Righi, L., Mechi, M. T., Giani, T., Antonelli, A., Rossolini, G. M., & Tuscan Clinical Microbiology Laboratory Network (2022). Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. The Lancet. Microbe, 3(3), e224–e234. [CrossRef]
- Abid, F. B., Tsui, C. K. M., Doi, Y., Deshmukh, A., McElheny, C. L., Bachman, W. C., Fowler, E. L., Albishawi, A., Mushtaq, K., Ibrahim, E. B., Doiphode, S. H., Hamed, M. M., Almaslmani, M. A., Alkhal, A., Butt, A. A., & Omrani, A. S. (2021). Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology, 40(8), 1779–1785. [CrossRef]
- Wang, S., Wang, L., Jin, J., Li, G., Shao, H., Song, Y., Sun, Y., Zhang, Y., Cheng, J., & Li, L. (2023). Genomic Epidemiology and Characterization of Carbapenem-Resistant Klebsiella pneumoniae in ICU Inpatients in Henan Province, China: a Multicenter Cross-Sectional Study. Microbiology spectrum, 11(3), e0419722. [CrossRef]
- Venkitapathi, S., Wijesundara, Y. H., Cornelius, S. A., Herbert, F. C., Gassensmith, J. J., Zimmern, P. E., & De Nisco, N. J. (2022). Conserved FimK Truncation Coincides with Increased Expression of Type 3 Fimbriae and Cultured Bladder Epithelial Cell Association in Klebsiella quasipneumoniae. Journal of bacteriology, 204(9), e0017222. [CrossRef]
- Mathers, A. J., Crook, D., Vaughan, A., Barry, K. E., Vegesana, K., Stoesser, N., Parikh, H. I., Sebra, R., Kotay, S., Walker, A. S., & Sheppard, A. E. (2019). Klebsiella quasipneumoniae Provides a Window into Carbapenemase Gene Transfer, Plasmid Rearrangements, and Patient Interactions with the Hospital Environment. Antimicrobial agents and chemotherapy, 63(6), e02513-18. [CrossRef]
- Shen, X., Liu, L., Yu, J., Ai, W., Cao, X., Zhan, Q., Guo, Y., Wang, L., & Yu, F. (2020). High Prevalence of 16S rRNA Methyltransferase Genes in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates Associated with Bloodstream Infections in 11 Chinese Teaching Hospitals. Infection and drug resistance, 13, 2189–2197. [CrossRef]
- Saadatian Farivar, A., Nowroozi, J., Eslami, G., & Sabokbar, A. (2018). RAPD PCR Profile, Antibiotic Resistance, Prevalence of armA Gene, and Detection of KPC Enzyme in Klebsiella pneumoniae Isolates. The Canadian journal of infectious diseases & medical microbiology, 2018, 6183162. [CrossRef]
- Al Sheikh, Y. A., Marie, M. A., John, J., Krishnappa, L. G., & Dabwab, K. H. (2014). Prevalence of 16S rRNA methylase genes among β-lactamase-producing Enterobacteriaceae clinical isolates in Saudi Arabia. The Libyan journal of medicine, 9(1), 24432. [CrossRef]
- Dey, S., Gaur, M., Sahoo, R. K., Das, A., Jain, B., Pati, S., & Subudhi, E. (2020). Genomic characterization of XDR Klebsiella pneumoniae ST147 co-resistant to carbapenem and colistin - The first report in India. Journal of global antimicrobial resistance, 22, 54–56. [CrossRef]
- Cerón, S., Salem-Bango, Z., Contreras, D. A., Ranson, E. L., & Yang, S. (2023). Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016-2022. Microorganisms, 11(7), 1717. [CrossRef]
- Boonyasiri, A., Jauneikaite, E., Brinkac, L. M., Greco, C., Lerdlamyong, K., Tangkoskul, T., Nguyen, K., Thamlikitkul, V., & Fouts, D. E. (2021). Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC infectious diseases, 21(1), 142. [CrossRef]
Figure 1.
Disk Approximation test, D69C AmpC detection set for AmpC detection (A), Modified Carbapenem Inactivation Method (mCIM) and Combo test kit for carbapenemase detection (B). Disk Approximation test (A, i), Strong and weak induction (Red arrows) towards the ceftazidime (CAZ) as substrate and imipenem (IPM), cefoxitin (FOX), amoxicillin/clavulanic acid (AMC) and piperacillin/tazobactam (TZP) as inducers. The potentiation between drugs is indicated by (Yellow arrows), (A, ii) Isolate exhibits resistance to ceftazidime rendering AmpC production undetectable by the disk approximation method. Synergistic activity between the substrate (ceftazidime) and amoxicillin-clavulanate is indicated by (Yellow arrows). (A, iii) D69C AmpC detection set. (A) AmpC positive. (B and C) AmpC negative (exhibiting other type(s) of resistance). Disk A (cefpodoxime 10 μg + AmpC inducer), disk B (cefpodoxime 10μg + AmpC inducer + ESBL inhibitor), disk C (cefpodoxime 10 μg + AmpC inducer + ESBL inhibitor + AmpC inhibitors). Modified Carbapenem Inactivation Method (mCIM) (B, i), Identification of carbapenemase-producing strains via mCIM test. The control strain E. coli ATCC 25922 was carbapenemase negative, having a zone size of > 19 mm. Two target isolates carried carbapenemase, which broke down the 10 μg meropenem after incubation for 4 h ± 15 min at 35 °C. Combo test kit for carbapenemase detection (B, ii), From left to right – carbapenemase not detected (C= control), OXA 48 detected (O= OAX-48), NDM detected (N= NDM), KPC detected (K=KPC), (V=VIM), (I=IMP).
Figure 1.
Disk Approximation test, D69C AmpC detection set for AmpC detection (A), Modified Carbapenem Inactivation Method (mCIM) and Combo test kit for carbapenemase detection (B). Disk Approximation test (A, i), Strong and weak induction (Red arrows) towards the ceftazidime (CAZ) as substrate and imipenem (IPM), cefoxitin (FOX), amoxicillin/clavulanic acid (AMC) and piperacillin/tazobactam (TZP) as inducers. The potentiation between drugs is indicated by (Yellow arrows), (A, ii) Isolate exhibits resistance to ceftazidime rendering AmpC production undetectable by the disk approximation method. Synergistic activity between the substrate (ceftazidime) and amoxicillin-clavulanate is indicated by (Yellow arrows). (A, iii) D69C AmpC detection set. (A) AmpC positive. (B and C) AmpC negative (exhibiting other type(s) of resistance). Disk A (cefpodoxime 10 μg + AmpC inducer), disk B (cefpodoxime 10μg + AmpC inducer + ESBL inhibitor), disk C (cefpodoxime 10 μg + AmpC inducer + ESBL inhibitor + AmpC inhibitors). Modified Carbapenem Inactivation Method (mCIM) (B, i), Identification of carbapenemase-producing strains via mCIM test. The control strain E. coli ATCC 25922 was carbapenemase negative, having a zone size of > 19 mm. Two target isolates carried carbapenemase, which broke down the 10 μg meropenem after incubation for 4 h ± 15 min at 35 °C. Combo test kit for carbapenemase detection (B, ii), From left to right – carbapenemase not detected (C= control), OXA 48 detected (O= OAX-48), NDM detected (N= NDM), KPC detected (K=KPC), (V=VIM), (I=IMP).
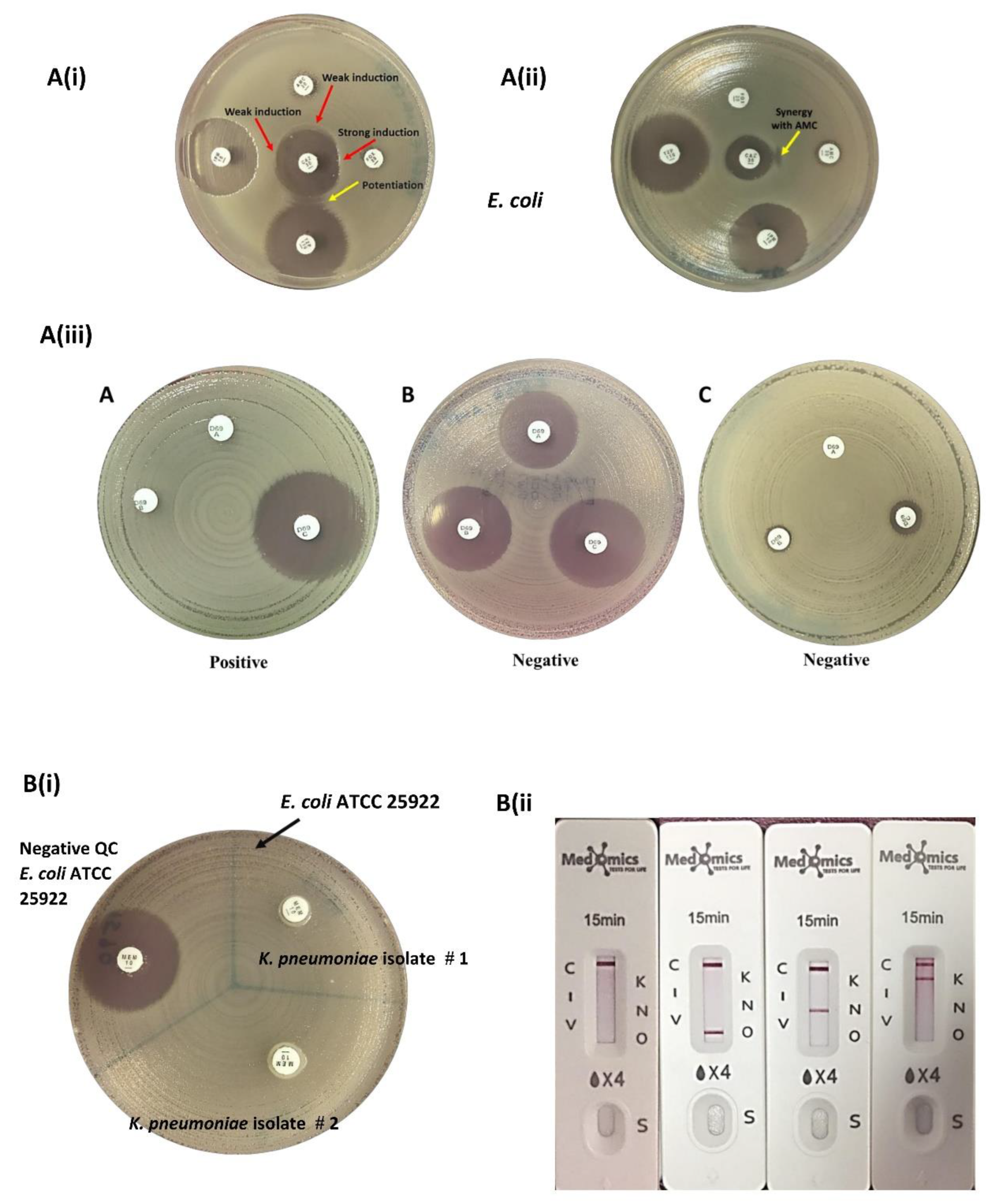
Figure 2.
Heat map of antibiotic resistance genes detected within the FOX-resistant E. coli using ResFinder. The figure shows the distribution of antibiotic resistance genes to each antibiotic class. The coloured boxes to the right of each strain demonstrate the distribution of antimicrobial resistance genes as follows: aminoglycoside (green), macrolide (light blue), quinolone (peach), folate pathway antagonist (dark gray), tetracycline (dark pink), beta-lactam (yellow) and fosfomycin (gold). White boxes indicate absence of these genes
Figure 2.
Heat map of antibiotic resistance genes detected within the FOX-resistant E. coli using ResFinder. The figure shows the distribution of antibiotic resistance genes to each antibiotic class. The coloured boxes to the right of each strain demonstrate the distribution of antimicrobial resistance genes as follows: aminoglycoside (green), macrolide (light blue), quinolone (peach), folate pathway antagonist (dark gray), tetracycline (dark pink), beta-lactam (yellow) and fosfomycin (gold). White boxes indicate absence of these genes
Figure 3.
Heat map of antibiotic resistance genes detected within the CRE K. pneumoniae using ResFinder. The figure shows the distribution of antibiotic resistance genes to each antibiotic class. The coloured boxes to the right of each strain demonstrate the distribution of antimicrobial resistance gene as follows: aminoglycoside (green), quinolone (peach), folate pathway antagonist (dark gray), tetracycline (dark pink), beta-lactam (yellow) and Fosfomycin (gold). White boxes indicate the absence of the genes.
Figure 3.
Heat map of antibiotic resistance genes detected within the CRE K. pneumoniae using ResFinder. The figure shows the distribution of antibiotic resistance genes to each antibiotic class. The coloured boxes to the right of each strain demonstrate the distribution of antimicrobial resistance gene as follows: aminoglycoside (green), quinolone (peach), folate pathway antagonist (dark gray), tetracycline (dark pink), beta-lactam (yellow) and Fosfomycin (gold). White boxes indicate the absence of the genes.
Table 1.
Susceptibility profile of E. coli and Klebsiella pneumoniae isolated from patients with cUTI.
Table 1.
Susceptibility profile of E. coli and Klebsiella pneumoniae isolated from patients with cUTI.
| |
Nitrofurantoin |
Cotrimoxazole |
Fosfomycin |
Ampicillin |
Cefazolin |
Cefuroxime |
Ceftriaxone |
Ceftazidime |
Cefepime |
Amoxicillin-clavulanate |
Ampicillin-sulbactam |
Piperacillin-tazobactam |
Gentamicin |
Amikacin |
Ciprofloxacin |
Levofloxacin |
Ertapenem |
Imipenem |
Meropenem |
E. coli
n=406
|
96% |
63% |
100% |
29% |
27% |
51% |
54% |
57% |
56% |
66% |
65% |
95% |
87% |
98% |
44% |
46% |
98% |
98% |
99% |
K. pneumoniae
n=163
|
38%
(157)
|
65%
(162) |
89%
(119) |
|
33%
(92) |
55%
(161)
|
61%
(161) |
60%
(162) |
63%
(163) |
65%
(77) |
52%
(103) |
72%
(163) |
80%
(163) |
84%
(163) |
47%
(133) |
48%
(136) |
81%
(146) |
83%
(163) |
84%
(163) |
Table 2.
Prevalence of ESBL, AmpC and CRE among E. coli and K. pneumoniae in patients with cUTI.
Table 2.
Prevalence of ESBL, AmpC and CRE among E. coli and K. pneumoniae in patients with cUTI.
| Phenotype |
E. coli,
n (%) |
K. pneumoniae,
n (%) |
Total prevalence among E. coli (n=406) and
K. pneumoniae (n=163) |
| ESBL |
164 (40.4%) |
33 (20.2%) |
197 (34.6%) |
| AmpC |
10 (2.5%) |
6 (3.7%) |
16 (2.8%) |
| ESBL+AmpC |
14 (3.4%) |
1 (0.6%) |
15 (2.6%) |
| CRE |
7 (1.7%) |
28 (17.2%) |
35 (6.2%) |
| Total |
195 |
68 |
263 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).