Submitted:
24 January 2025
Posted:
27 January 2025
You are already at the latest version
Abstract
Influenza poses a serious threat to both individual and public health. This study aimed to investigate the virological and epidemiological characteristics of influenza infections and to explore the genetic diversity of the circulating influenza viruses. In total, 1886 nasopharyngeal specimens from patients with acute respiratory illnesses were tested against 13 respiratory viruses using a multiplex real-time PCR. Whole-genome sequencing, phylogenetic, and amino acid analyses of representative influenza strains were performed. At least one respiratory virus was detected in 869 (46.1%) patients, 87 (4.6%) were co-infected with two or three viruses. Influenza A(H1N1)pdm09 was the most prevalent virus (16.1%), followed by rhinoviruses (8.1%) and RSV (6.7%). Hemagglutinin (HA) genes of the 74 influenza A(H1N1)pdm09 viruses were categorized in subclades C.1.8, C.1.9, and C.1 within clade 5a.2a and D1, D.2, and D.3 within clade 5a.2a.1 The A(H3N2) viruses analyzed belonged to clade 2a.3a.1, subclades J.2 and J.1. The sequenced B/Victoria lineage viruses fell into clade V1A.3a.2, subclades C.5.6 and C.5.7. Amino acid substitutions in most viral proteins were identified compared with the vaccine strains, including in the HA antigenic sites. This study demonstrated the dominant distribution of the influenza A(H1N1)pdm09 virus among the respiratory viruses studied and the genetic diversity of the circulating influenza viruses.
Keywords:
1. Introduction
2. Methods
2.1. Patients and Specimen Collection
2.2. Molecular Detection of Respiratory Viruses
- Reverse transcription: 25 °C for 2 min and then 50 °C for 15 min;
- Initial denaturation: 95 °C for 2 min;
- Amplification for 45 cycles: 95 °C for 15 s and then 55 °C for 30 s.
2.3. Sequencing and Phylogenetic Analysis of Influenza Viruses
2.4. Deduced Amino Acid Sequence Analysis and Glycosylation Prediction
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
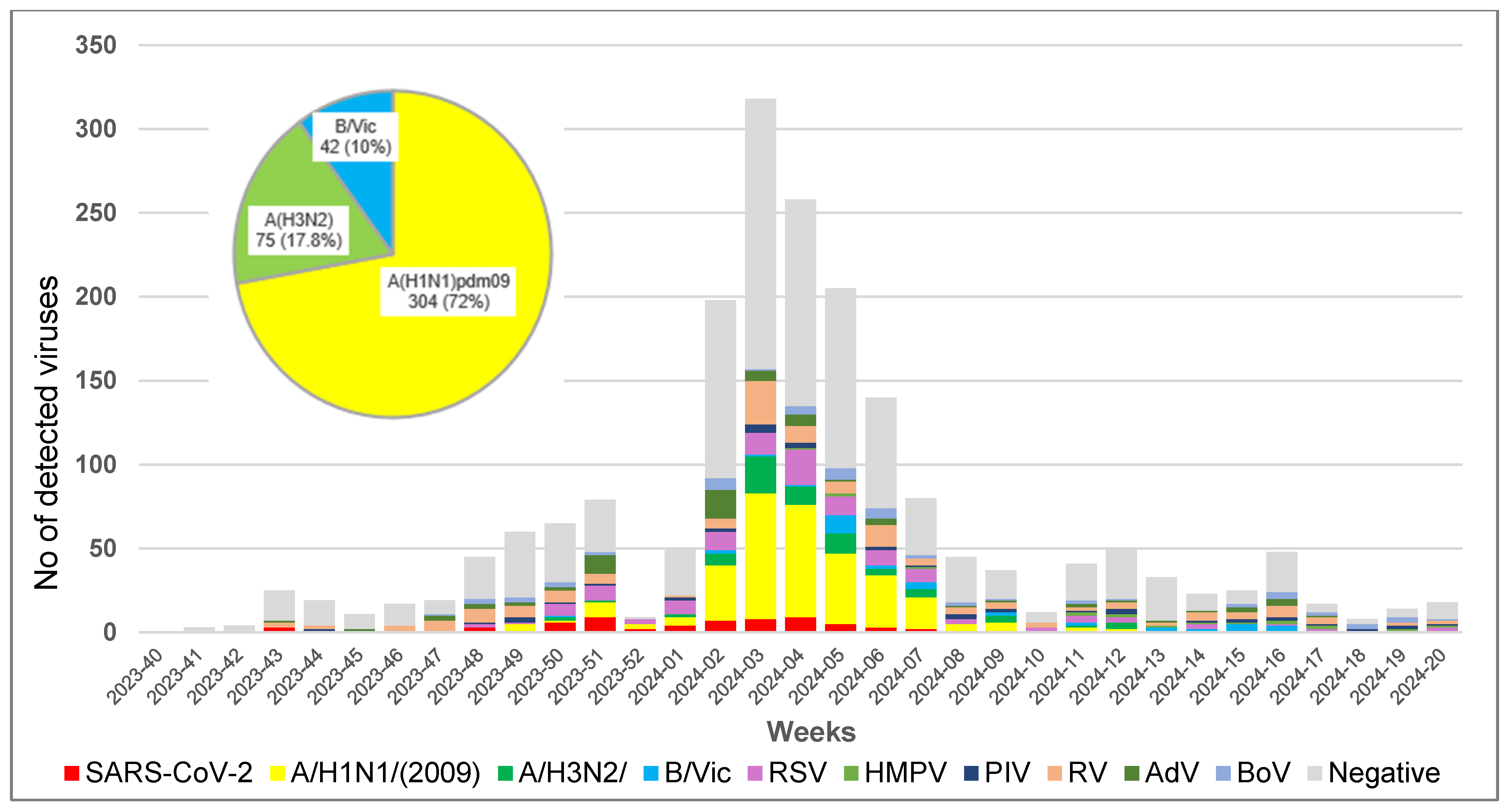
3.2. Virus Detection
3.3. Epidemiological Characteristics
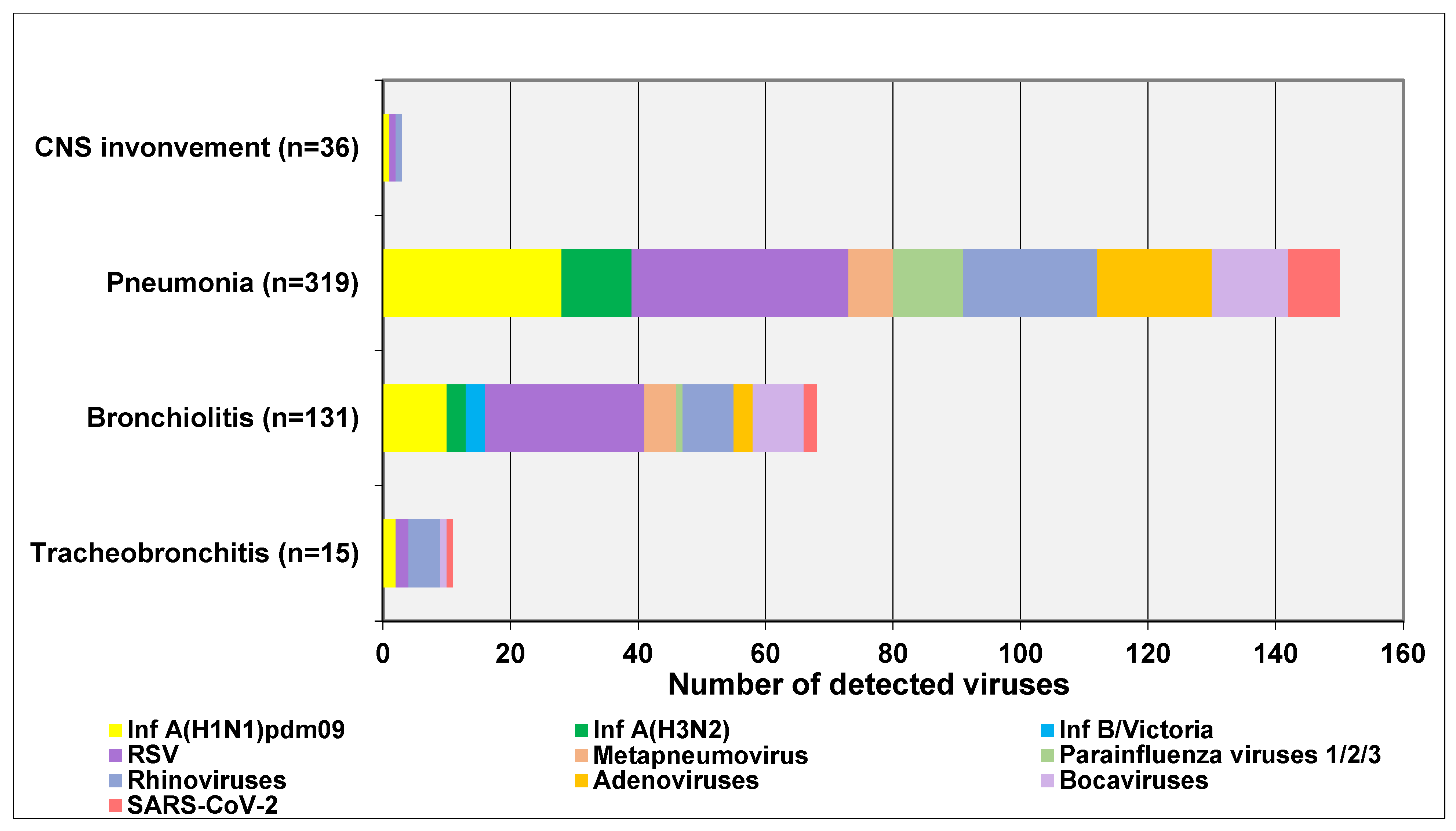
3.4. Clinical Characteristics
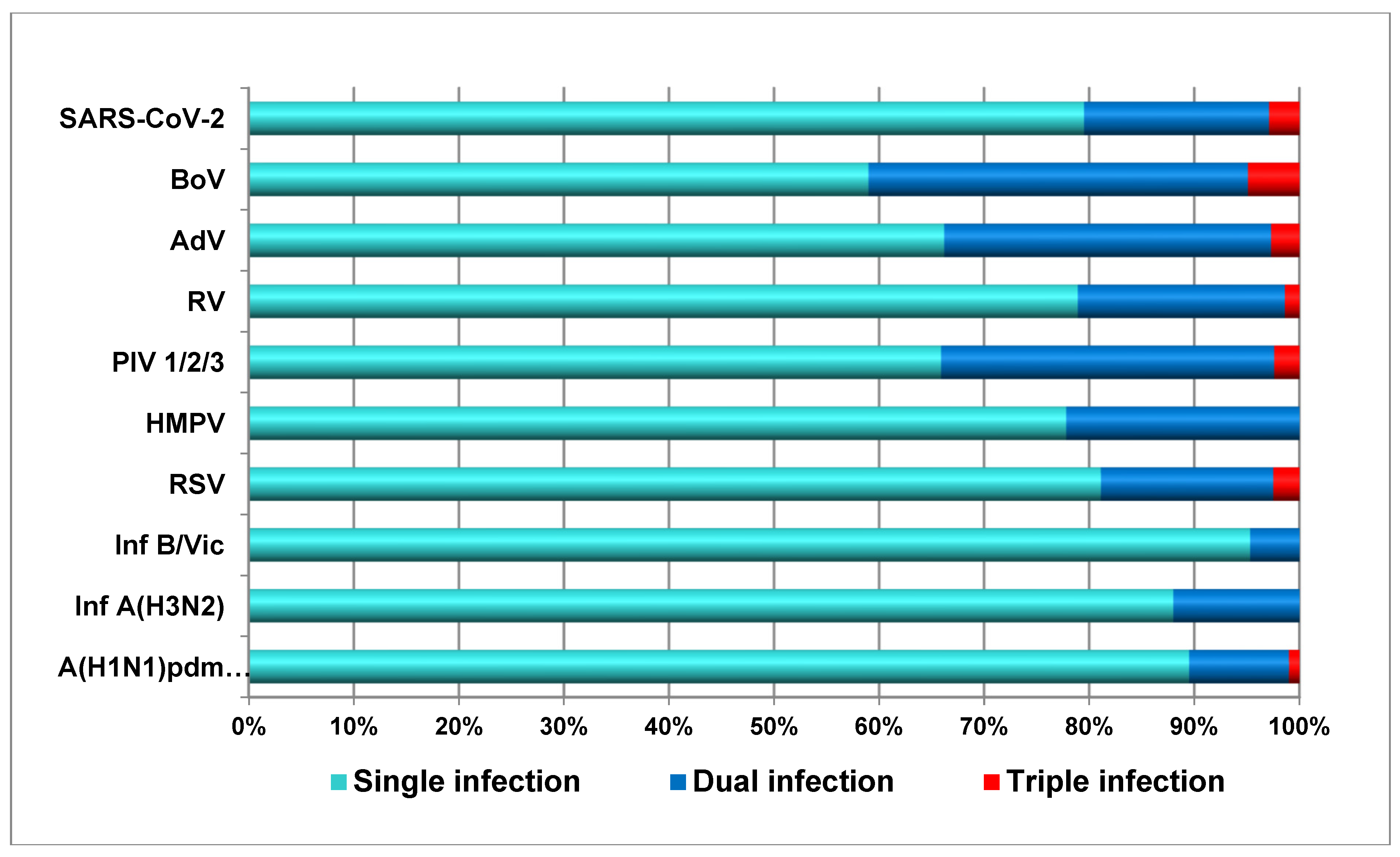
3.5. Incidence of Co-Infections Between Respiratory Viruses
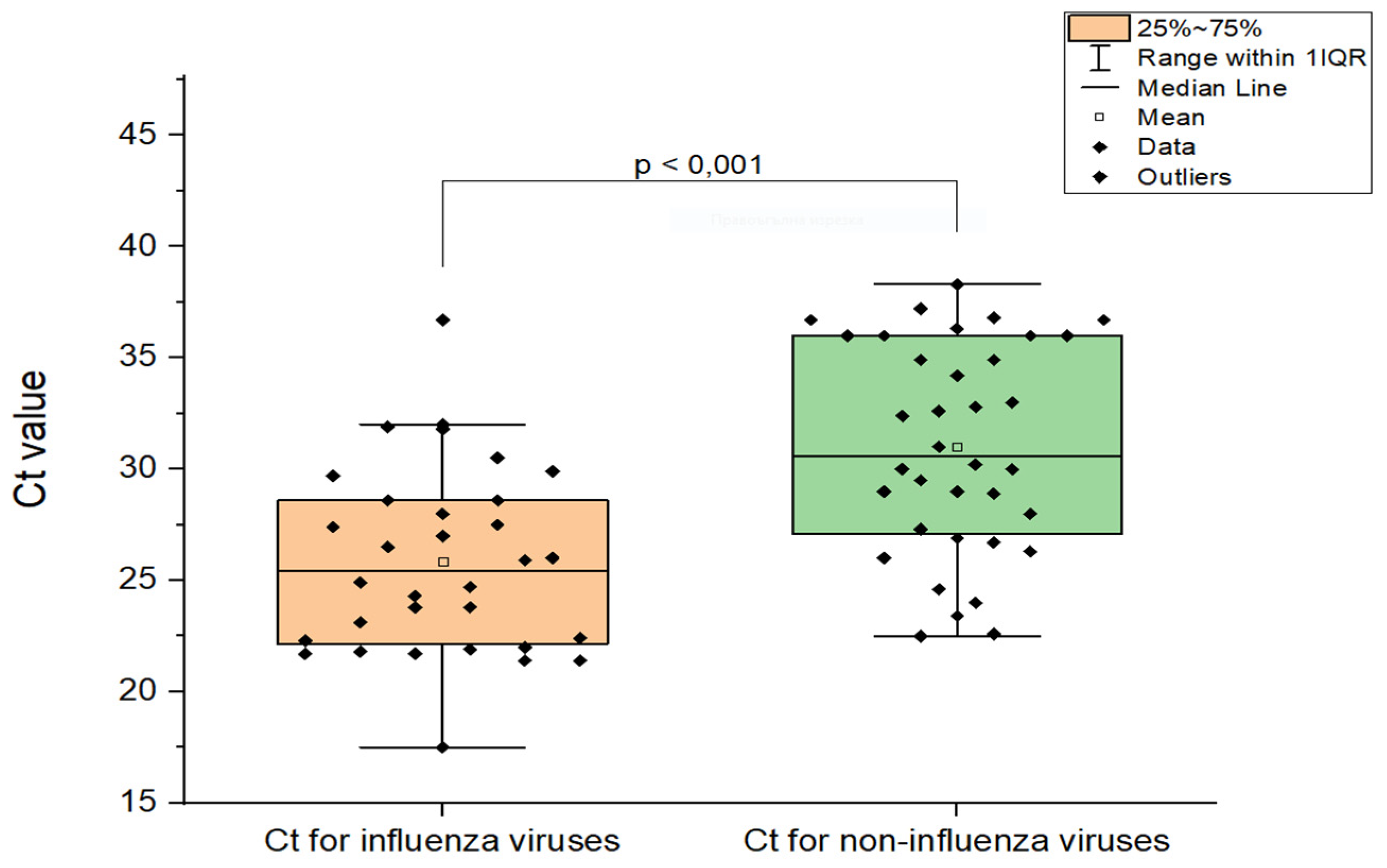
3.6. Comparison of Ct values in Cases of Influenza Virus Coinfections
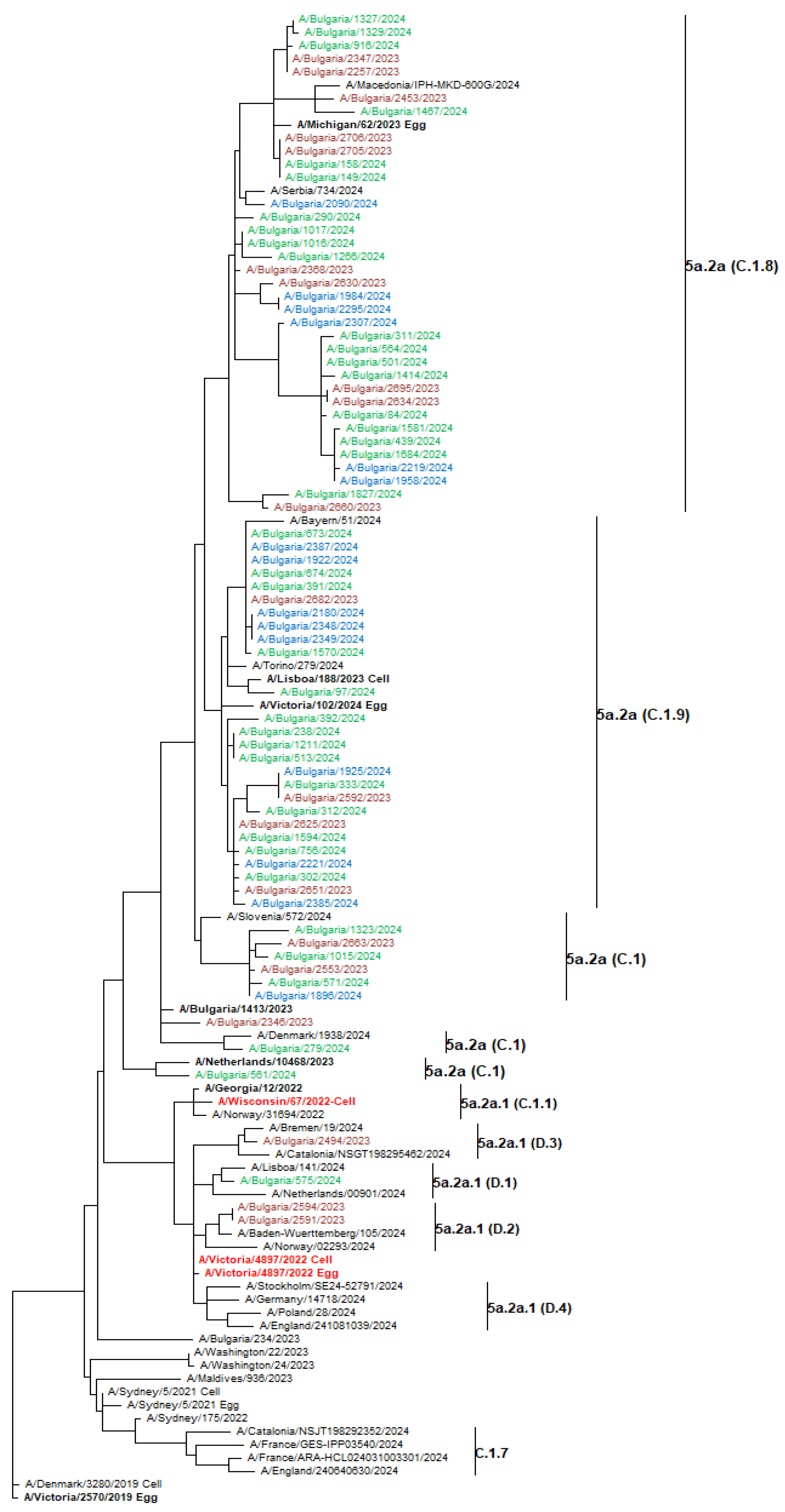
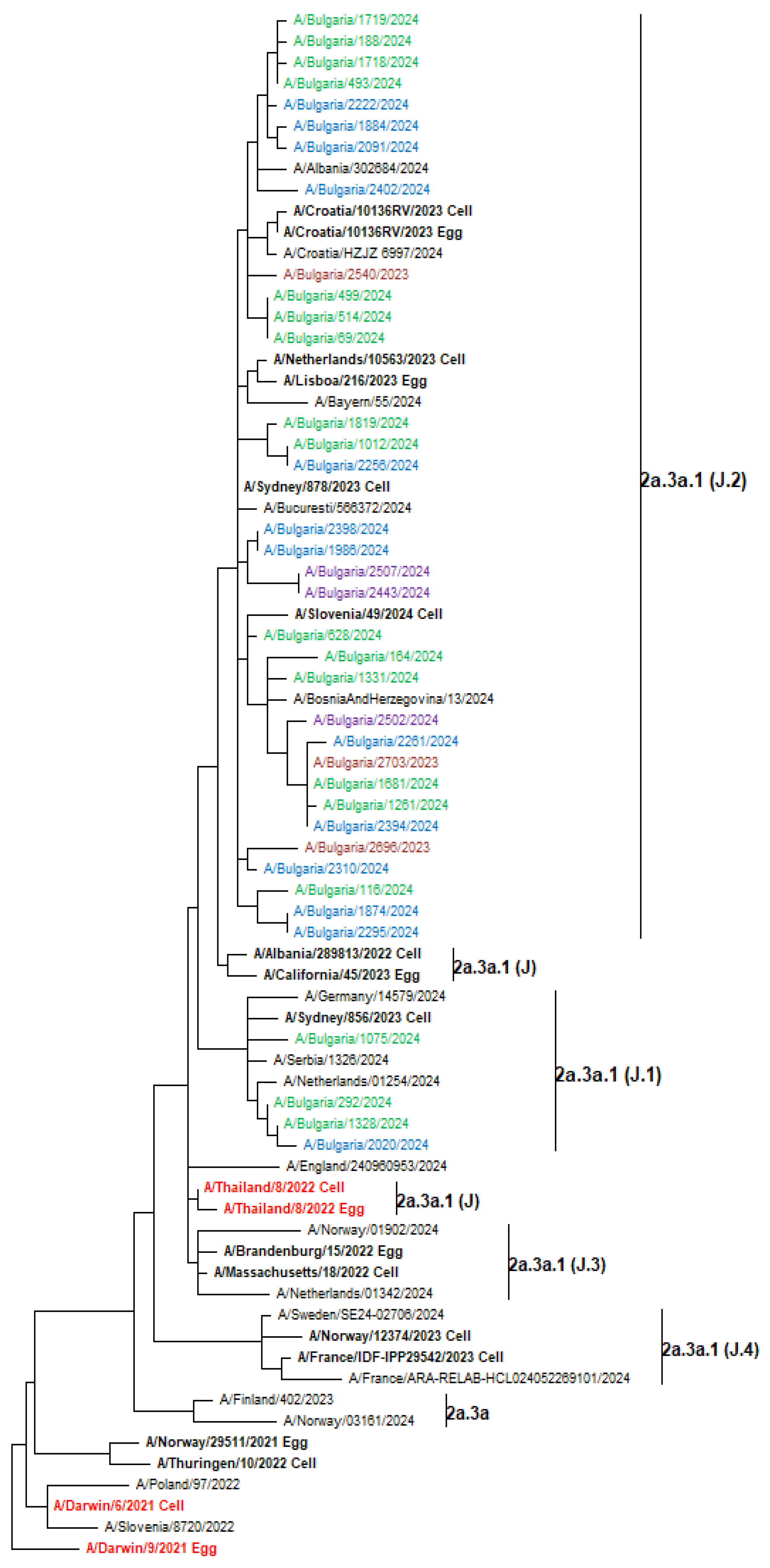
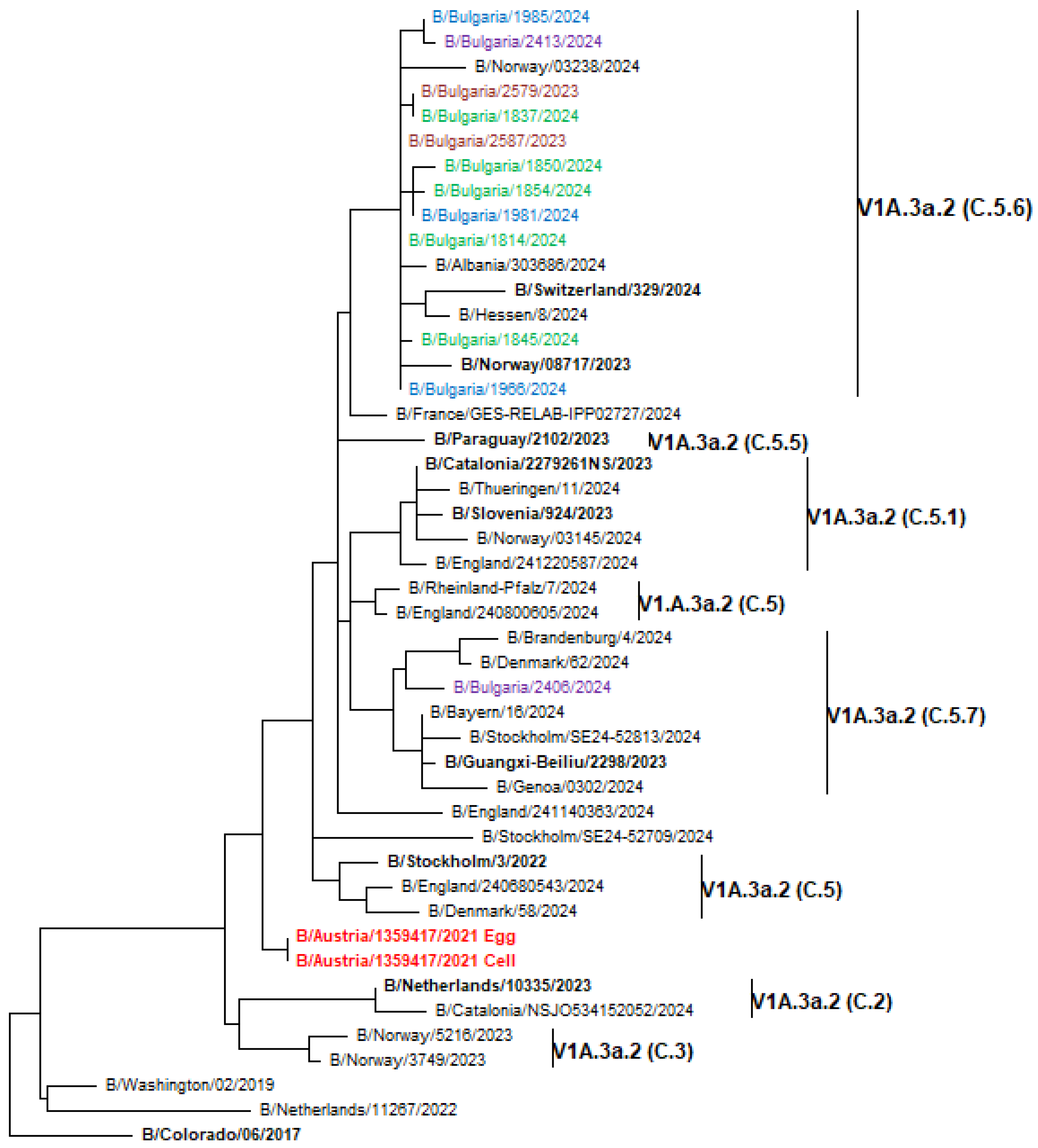
3.7. Phylogenetic Analysis of Influenza Viruses
3.8. Amino Acid Polymorphisms in Viral Proteins
3.8.1. A(H1N1)pdm09
3.8.2. A(H3N2)
3.8.3. B/Victoria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabatabai, J.; Ihling, C.M.; Manuel, B.; Rehbein, R.M.; Schnee, S.V.; Hoos, J.; Pfeil, J.; Grulich-Henn, J.; Schnitzler, P. Viral Etiology and Clinical Characteristics of Acute Respiratory Tract Infections in Hospitalized Children in Southern Germany (2014-2018). Open Forum Infect Dis 2023, 10, ofad110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Xu, H.; Hao, L.; Zhao, B.; Ye, C.; Zhu, W. Prevalence of Respiratory Viruses in Children With Acute Respiratory Infections in Shanghai, China, From 2013 to 2022. Influenza Other Respir Viruses 2024, 18, e13310. [Google Scholar] [CrossRef] [PubMed]
- Babawale, P.I.; Guerrero-Plata, A. Respiratory Viral Coinfections: Insights into Epidemiology, Immune Response, Pathology, and Clinical Outcomes. Pathogens 2024, 13, 316. [Google Scholar] [CrossRef] [PubMed]
- Tokars, J.I.; Olsen, S.J.; Reed, C. Seasonal Incidence of Symptomatic Influenza in the United States. Clin Infect Dis. 2018, 66, 1511–1518. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Hui, D.S.; Zambon, M.; Wentworth, D.E.; Mon, A.St. Influenza. Lancet 2022, 400, 693–706. [Google Scholar] [CrossRef]
- WHO. The burden of influenza. Available online: https://www.who.int/news-room/feature-stories/detail/the-burden-of-influenza (accessed on 16 May 2024).
- Lee, Y.; Jang, T.S.; Kim, J.K. Effects of Coronavirus Disease 2019 on Prevalence of Acute Respiratory Viruses: Changes during the Pandemic. J Glob Infect Dis 2024, 16, 27–32. [Google Scholar] [CrossRef]
- Wang, X.; Walker, G.; Kim, K.W.; Stelzer-Braid, S.; Scotch, M.; Rawlinson, W.D. The resurgence of influenza A/H3N2 virus in Australia after the relaxation of COVID-19 restrictions during the 2022 season. J Med Virol 2024, 96, e29922. [Google Scholar] [CrossRef]
- CDC. The Pink Book: Course Textbook - 14th Edition (2021). Available online: https://www.cdc.gov/vaccines/pubs/pinkbook/flu.html.
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31, 417–427. [Google Scholar] [CrossRef]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef]
- Krystal, M.; Young, J.F.; Palese, P.; Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc Natl Acad Sci U S A 1983, 80, 4527–4531. [Google Scholar] [CrossRef]
- Skehel, J.J.; Stevens, D.J.; Daniels, R.S.; Douglas, A.R.; Knossow, M.; Wilson, I.A.; Wiley, D.C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A 1984, 81, 1779–1783. [Google Scholar] [CrossRef] [PubMed]
- EU case definitions. Available online: https://ecdc.europa.eu/en/infectious-diseases-public-health/surveillance-and-disease-data/eu-case-definitions.
- Shu, B.; Kirby, M.K.; Davis, W.G.; Warnes, C.; Liddell, J.; Liu, J.; Wu, K.H.; Hassell, N.; Benitez, A.J.; Wilson, M.M.; et al. Multiplex Real-Time Reverse Transcription PCR for Influenza A Virus, Influenza B Virus, and Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis 2021, 27, 1821–1830. [Google Scholar] [CrossRef]
- Shu, B.; Wu, K.H.; Emery, S.; Villanueva, J.; Johnson, R.; Guthrie, E.; Berman, L.; Warnes, C.; Barnes, N.; Klimov, A.; et al. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J Clin Microbiol 2011, 49, 2614–2619. [Google Scholar] [CrossRef] [PubMed]
- Kodani, M.; Yang, G.; Conklin, L.M.; Travis, T.C.; Whitney, C.G.; Anderson, L.J.; Schrag, S.J.; Taylor, T.H.Jr; Beall, B.W.; Breiman, R.F.; et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol 2011, 49, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Worldwide Influenza Centre, WHO-CC for Reference and Research on Influenza. Report prepared for the WHO Consultation on the Composition of Influenza Virus Vaccines for the Southern Hemisphere 2025. Available online: https://www.crick.ac.uk/sites/default/files/2024-10/WIC-VCM-SH2024_v2.pdf.
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2023-2024 northern hemisphere influenza season. 2023:1–11. Available online: https://www.who.int/publications/m/item/recommended-composition-of-influenza-virus-vaccines-for-use-in-the-2023-2024-northern-hemisphere-influenza-season.
- Blackburne, B.P.; Hay, A.J.; Goldstein, R.A. Changing selective pressure during antigenic changes in human influenza H3. PLoS Pathog 2008, 4, e1000058. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.R.; Guo, Z.; Tzeng, W.P.; Garten, R.J.; Xiyan, X.; Blanchard, E.G.; Blanchfield, K.; Stevens, J.; Katz, J.M.; York, I.A. Diverse antigenic site targeting of influenza hemagglutinin in the murine antibody recall response to A(H1N1)pdm09 virus. Virology 2015, 485, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Sriwilaijaroen, N.; Suzuki, Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci 2012, 88, 226–249. [Google Scholar] [CrossRef]
- Govorkova, E.A.; Takashita, E.; Daniels, R.S.; Fujisaki, S.; Presser, L.D.; Patel, M.C.; Huang, W.; Lackenby, A.; Nguyen, H.T.; Pereyaslov, D.; et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2018-2020. Antiviral Res 2022, 200, 105281. [Google Scholar] [CrossRef]
- de Graaf, M.; Fouchier, R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 2014, 33, 823–841. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, F.; Lu, M.; Tian, X.; Ma, J. Crystal structure of unliganded influenza B virus hemagglutinin. J Virol 2008, 82, 3011–3020. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, X.; Chen, X.; Ma, J. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc Natl Acad Sci U S A 2007, 104, 16874–16879. [Google Scholar] [CrossRef]
- Rivas, M.J.; Alegretti, M.; Cóppola, L.; Ramas, V.; Chiparelli, H.; Goñi, N. Epidemiology and Genetic Variability of Circulating Influenza B Viruses in Uruguay, 2012-2019. Microorganisms 2020, 8, 591. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, Y.; Wang, J.; Li, Y.; Wang, Y.; Gao, Y.; Zhao, M.; Zhao, M.; Tan, H.; Tie, Y.; et al. Epidemiology of respiratory pathogens in patients with acute respiratory infections during the COVID-19 pandemic and after easing of COVID-19 restrictions. Microbiol Spectr 2024, 12, e0116124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Tao, R.; Shang, S. Analyzing infections caused by 11 respiratory pathogens in children: Pre- and post-COVID-19 pandemic trends in China. J Med Virol. 2024, 96, e29929. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Gu, Y.; Tang, X.; Jiang, C.; Fang, F.; Chu, W.; Tao, L.; Zhang, X.; Chen, M.; Wu, H.; et al. Whole-genome analysis of circulating influenza A virus (H3N2) strains in Shanghai, China from 2005 to 2023. Emerg Microbes Infect 2024, 13, 2396867. [Google Scholar] [CrossRef]
- Lv, G.; Shi, L.; Liu, Y.; Sun, X.; Mu, K. Epidemiological characteristics of common respiratory pathogens in children. Sci Rep 2024, 14, 16299. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, H.H.; Fall, A.; Norton, J.M.; Sachithanandham, J.; Yunker, M.; Abdullah, O.; Hanlon, A.; Gluck, L.; Morris, C.P.; Pekosz, A.; et al. Respiratory virus disease and outcomes at a large academic medical center in the United States: a retrospective observational study of the early 2023/2024 respiratory viral season. Microbiol Spectr 2024, 12, e0111624. [Google Scholar] [CrossRef]
- Korsun, N.; Daniels, R.; Angelova, S.; Ermetal, B.; Grigorova, I.; Voleva, S.; Trifonova, I.; Kurchatova, A.; McCauley, J. Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season. J Med Microbiol 2020, 69, 986–998. [Google Scholar] [CrossRef]
- Korsun, N.; Trifonova, I.; Dobrinov, V.; Madzharova, I.; Grigorova, I.; Christova, I. Low prevalence of influenza viruses and predominance of A(H3N2) virus with respect to SARS-CoV-2 during the 2021-2022 season in Bulgaria. J Med Virol 2023, 95, e28489. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Wheatley, A.K.; Laurie, K.; Kent, S.J.; Rockman, S. Influenza lineage extinction during the COVID-19 pandemic? Nat Rev Microbiol 2021, 19, 741–742. [Google Scholar] [CrossRef]
- Tramuto, F.; Maida, C.M.; Randazzo, G.; Previti, A.; Sferlazza, G.; Graziano, G.; Costantino, C.; Mazzucco, W.; Vitale, F. Insights into Genetic and Antigenic Characteristics of Influenza A(H1N1)pdm09 Viruses Circulating in Sicily During the Surveillance Season 2023-2024: The Potential Effect on the Seasonal Vaccine Effectiveness. Viruses 2024, 16, 1644. [Google Scholar] [CrossRef]
- Di Maio, V.C.; Scutari, R.; Forqué, L.; Colagrossi, L.; Coltella, L.; Ranno, S.; Linardos, G.; Gentile, L.; Galeno, E.; Vittucci, A.C.; et al. Presence and Significance of Multiple Respiratory Viral Infections in Children Admitted to a Tertiary Pediatric Hospital in Italy. Viruses 2024, 16, 750. [Google Scholar] [CrossRef] [PubMed]
- Beauté, J.; Zucs, P.; Korsun, N.; Bragstad, K.; Enouf, V.; Kossyvakis, A.; Griškevičius, A.; Olinger, C.M.; Meijer, A.; Guiomar, R.; et al. Age-specific differences in influenza virus type and subtype distribution in the 2012/2013 season in 12 European countries. Epidemiol Infect 2015, 143, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Belazi, S.; Olsen, S.J.; Brown, C.; Green, H.K.; Mook, P.; Nguyen-Van-Tam, J.; Penttinen, P.; Lansbury, L. Spotlight influenza: Laboratory-confirmed seasonal influenza in people with acute respiratory illness: a literature review and meta-analysis, WHO European Region, 2004 to 2017. Euro Surveill 2021, 26, 2000343. [Google Scholar] [CrossRef]
- Siqueira, B.A.; Bredariol, K.O.; Boschiero, M.N.; Marson, F.A.L. Viral co-detection of influenza virus and other respiratory viruses in hospitalized Brazilian patients during the first three years of the coronavirus disease (COVID)-19 pandemic: an epidemiological profile. Front Microbiol 2024, 15, 1462802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Xu, H.; Hao, L.; Zhao, B.; Ye, C.; Zhu, W. Prevalence of Respiratory Viruses in Children With Acute Respiratory Infections in Shanghai, China, From 2013 to 2022. Influenza Other Respir Viruses 2024, 18, e13310. [Google Scholar] [CrossRef] [PubMed]
- Mandelia, Y.; Procop, G.W.; Richter, S.S.; Worley, S.; Liu, W.; Esper, F. Dynamics and predisposition of respiratory viral co-infections in children and adults. Clin Microbiol Infect 2021, 27, 631.e1–631.e6. [Google Scholar] [CrossRef] [PubMed]
- Shirreff, G.; Chaves, S.S.; Coudeville, L.; Mengual-Chuliá, B.; Mira-Iglesias, A.; Puig-Barberà, J.; Orrico-Sanchez, A.; Díez-Domingo, J.; Valencia Hospital Surveillance Network for the Study of Influenza and Other Respiratory Viruses (VAHNSI); Opatowski, L.; Lopez-Labrador, F.X. Seasonality and Co-Detection of Respiratory Viral Infections Among Hospitalised Patients Admitted With Acute Respiratory Illness-Valencia Region, Spain, 2010-2021. Influenza Other Respir Viruses 2024, 18, e70017. [Google Scholar] [CrossRef]
- Mauro, M.V.; Greco, S.; Pellegrini, M.; Campagna, T.; Caprino, F.; Elia, N.; Mastroianni, A.; Greco, F. Epidemiology and Clinical impact of single and multi-viral respiratory infections in post-pandemic era. New Microbiol 2024, 47, 28–32. [Google Scholar]
- Petat, H.; Corbet, S.; Leterrier, B.; Vabret, A.; Ar Gouilh, M. Unravelling the acute respiratory infection landscape: virus type, viral load, health status and coinfection do matter. Front Cell Infect Microbiol 2024, 14, 1380855. [Google Scholar] [CrossRef]
- Goka, E.A.; Vallely, P.J.; Mutton, K.J.; Klapper, P.E. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect 2015, 143, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.L.; Drysdale, S.B.; Snape, M.D.; O’Connor, D.; Brown, A.; MacIntyre-Cockett, G.; Mellado-Gomez, E.; de Cesare, M.; Ansari, M.A.; Bonsall, D.; et al. Targeted metagenomics reveals association between severity and pathogen co-detection in infants with respiratory syncytial virus. Nat Commun 2024, 15, 2379. [Google Scholar] [CrossRef] [PubMed]
- Malveste Ito, C.R.; Moreira, A.L.E.; Silva, P.A.N.D.; Santos, M.O.; Santos, A.P.D.; Rézio, G.S.; Brito, P.N.; Rezende, A.P.C.; Fonseca, J.G.; Peixoto, F.A.O.; et al. Viral Coinfection of Children Hospitalized with Severe Acute Respiratory Infections during COVID-19 Pandemic. Biomedicines 2023, 11, 1402. [Google Scholar] [CrossRef]
- Scotta, M.C.; Chakr, V.C.; de Moura, A.; Becker, R.G.; de Souza, A.P.; Jones, M.H.; Pinto, L.A.; Sarria, E.E.; Pitrez, P.M.; Stein, R.T.; et al. Respiratory viral coinfection and disease severity in children: A systematic review and meta-analysis. J Clin Virol 2016, 80, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Asner, S.A.; Science, M.E.; Tran, D.; Smieja, M.; Merglen, A.; Mertz, D. Clinical disease severity of respiratory viral co-infection versus single viral infection: a systematic review and meta-analysis. PLoS One 2014, 9, e99392. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Viral Interference between Respiratory Viruses. Emerg Infect Dis 2022, 28, 273–281. [Google Scholar] [CrossRef]
- Teluguakula, N.; Chow, V.T.K.; Pandareesh, M.D.; Dasegowda, V.; Kurrapotula, V.; Gopegowda, S.M.; Radic, M. SARS-CoV-2 and Influenza Co-Infection: Fair Competition or Sinister Combination? Viruses 2024, 16, 793. [Google Scholar] [CrossRef]
- Yunker, M.; Villafuerte, D.A.; Fall, A.; Norton, J.M.; Abdullah, O.; Rothman, R.E.; Fenstermacher, K.Z.J.; Morris, C.P.; Pekosz, A.; Klein, E.; et al. Genomic evolution of influenza during the 2023-2024 season, the Johns Hopkins health system. J Clin Virol 2024, 174, 105718. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Lemus, N.; Lai, T.Y.; Mishra, M.; González-Domínguez, I.; Puente-Massaguer, E.; Loganathan, M.; Francis, B.; Samanovic, M.I.; Krammer, F.; et al. The immunodominance of antigenic site Sb on the H1 influenza virus hemagglutinin increases with high immunoglobulin titers of the cohorts and with young age, but not sex. Vaccine 2024, 42, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Koel, B.F.; Mögling, R.; Chutinimitkul, S.; Fraaij, P.L.; Burke, D.F.; van der Vliet, S.; de Wit, E.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; et al. Identification of amino acid substitutions supporting antigenic change of influenza A(H1N1)pdm09 viruses. J Virol 2015, 89, 3763–3775. [Google Scholar] [CrossRef] [PubMed]
- Broberg, E.K.; Vukovikj, M.; Svartström, O.; Hasibra, I.; Riess, M.; Melidou, A.; Members of the ERLI-Net network. Antigenic changes in influenza A(H3N2) driven by genetic evolution: Insights from virological surveillance, EU/EEA, week 40/2023 to week 9/2024. Euro Surveill 2024, 29, 2400395. [Google Scholar] [CrossRef]
- Jin, H.; Zhou, H.; Liu, H.; Chan, W.; Adhikary, L.; Mahmood, K.; Lee, M.S.; Kemble, G. Two residues in the hemagglutinin of A/Fujian/411/02-like influenza viruses are responsible for antigenic drift from A/Panama/2007/99. Virology 2005, 336, 113–119. [Google Scholar] [CrossRef]
- Eshaghi, A.; Duvvuri, V.R.; Li, A.; Patel, S.N.; Bastien, N.; Li, Y.; Low, D.E.; Gubbay, J.B. Genetic characterization of seasonal influenza A (H3N2) viruses in Ontario during 2010-2011 influenza season: high prevalence of mutations at antigenic sites. Influenza Other Respir Viruses 2014, 8, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Chambers, B.S.; Parkhouse, K.; Ross, T.M.; Alby, K.; Hensley, S.E. Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014-2015 Influenza Season. Cell Rep 2015, 12, 1–6. [Google Scholar] [CrossRef]
- Popova, L.; Smith, K.; West, A.H.; Wilson, P.C.; James, J.A.; Thompson, L.F.; Air, G.M. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 2012, 7, e41895. [Google Scholar] [CrossRef] [PubMed]
- Koel, B.F.; Burke, D.F.; Bestebroer, T.M.; van der Vliet, S.; Zondag, G.C.; Vervaet, G.; Skepner, E.; Lewis, N.S.; Spronken, M.I.; Russell, C.A.; et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013, 342, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Bedford, T.; Suchard, M.A.; Lemey, P.; Dudas, G.; Gregory, V.; Hay, A.J.; McCauley, J.W.; Russell, C.A.; Smith, D.J.; Rambaut, A. Integrating influenza antigenic dynamics with molecular evolution. Elife 2014, 3, e01914. [Google Scholar] [CrossRef] [PubMed]
- Perofsky, A.C.; Huddleston, J.; Hansen, C.L.; Barnes, J.R.; Rowe, T.; Xu, X.; Kondor, R.; Wentworth, D.E.; Lewis, N.; Whittaker, L.; et al. Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States. Elife 2024, 13, RP91849. [Google Scholar] [CrossRef] [PubMed]
- Lugovtsev, V.Y.; Vodeiko, G.M.; Strupczewski, C.M.; Ye, Z.; Levandowski, R.A. Generation of the influenza B viruses with improved growth phenotype by substitution of specific amino acids of hemagglutinin. Virology 2007, 365, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Holmes, E.C. The evolutionary dynamics of human influenza B virus. J Mol Evol 2008, 66, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Holmes, E.C.; Joseph, U.; Fourment, M.; Su, Y.C.; Halpin, R.; Lee, R.T.; Deng, Y.M.; Gunalan, V.; Lin, X.; et al. The contrasting phylodynamics of human influenza B viruses. Elife 2015, 4, e05055. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Wang, Y.; Mai, H.; Chen, Y.; Zhang, Y.; Ji, Y.; Cong, X.; Gao, Y. Clinical characteristics of outpatients with influenza-B-associated pneumonia and molecular evolution of influenza B virus in Beijing, China, during the 2021-2022 influenza season. Arch Virol 2024, 169, 30. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Jang, Y.H.; Kwon, S.B.; Lee, C.M.; Han, G.; Seong, B.L. Glycosylation of Hemagglutinin and Neuraminidase of Influenza A Virus as Signature for Ecological Spillover and Adaptation among Influenza Reservoirs. Viruses 2018, 10, 183. [Google Scholar] [CrossRef] [PubMed]
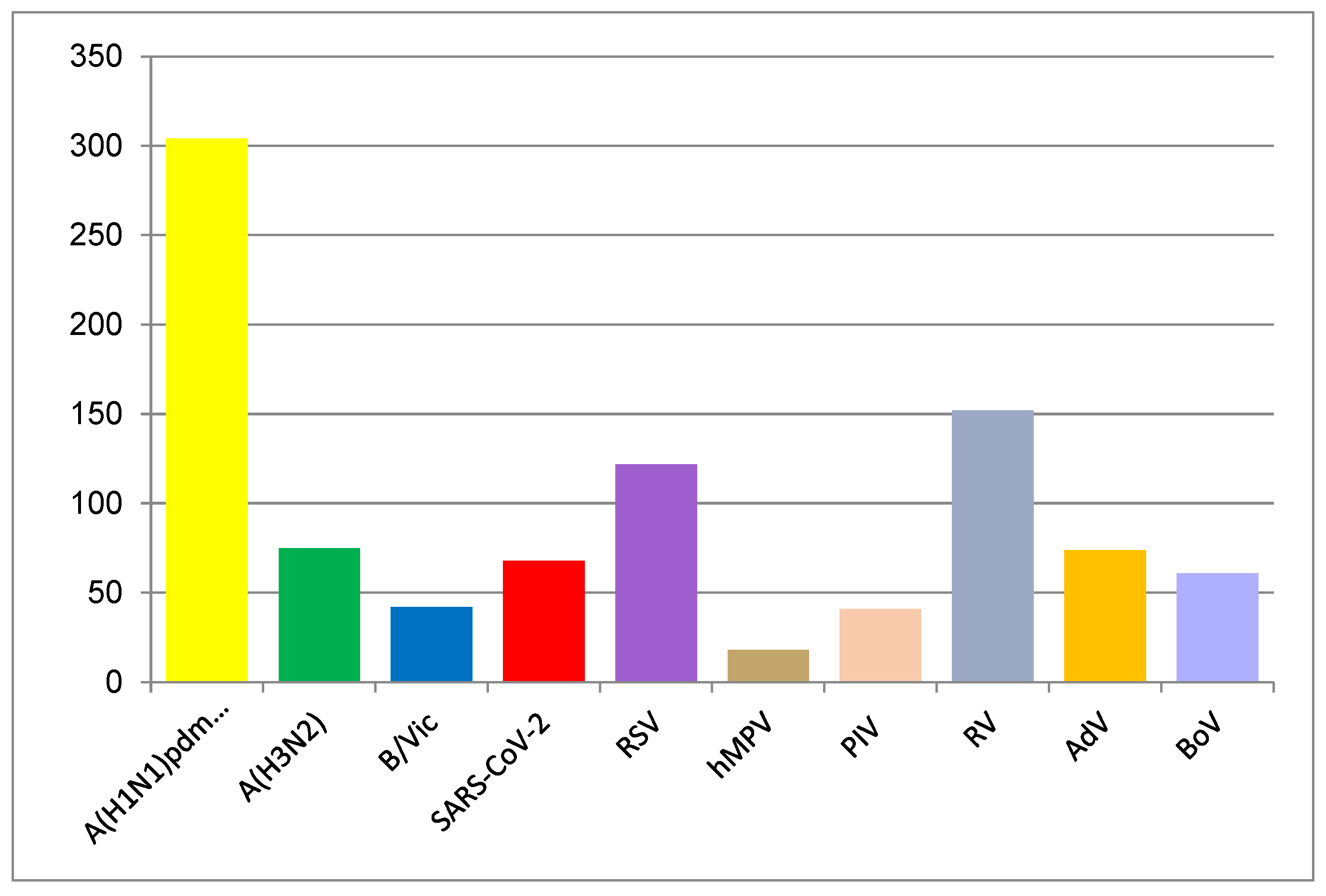
| Age group (years) | Total positive (%) | Mono-infections | Dual infections | Triple infections |
| 0-4 y (n = 816) | 456 (55.9) | 394 | 59 | 3 |
| 5-14 y (n = 799) | 312 (39) | 293 | 17 | 2 |
| 15-29 y (n = 131) | 47 (35.9) | 46 | 1 | - |
| 30-64 y (n = 94) | 38 (40.4) | 34 | 4 | - |
| ≥ 65 y (n = 36) | 11 (30.6) | 11 | - | - |
| without data (10) | 4 | 3 | 1 | - |
| Total (n = 1886) | 869 (46.1) | 781 (41.4%) | 82 (4.3%) | 5 (0.3%) |
| Viruses | Outpatients | Inpatients | Viruses | Outpatients | Inpatients | ||||
| No positive | No (%) of co-infections | No positive | No (%) of co-infections | No positive | No (%) of co-infections | No positive | No (%) of co-infections | ||
| A(H1N1) pdm09 |
123 | 7 (5.7%) | 181 | 24 (13.3%) p < 0,05 |
Parainfluenza type 1 | 0 | 0 (0%) | 1 | 0 (0%) |
| A(H3N2) | 33 | 3 (9.1%) | 42 | 6 (14.3%) | Parainfluenza type 2 | 9 | 3 (33.3%) | 4 | 1 (25%) |
| B/Vic | 14 | 1 (7.1%) | 29 | 1 (3.4%) | Parainfluenza type 3 | 7 | 0 (0%) | 20 | 10 (50%) p < 0,05 |
| SARS-CoV-2 | 27 | 5 (18.5%) | 41 | 9 (22%) | Rhinoviruses | 78 | 7 (9%) | 74 | 25 (33.8%) p < 0,05 |
| RSV | 59 | 3 (5.1%) | 63 | 20 (31.7%) p < 0,05 |
Adenoviruses | 30 | 6 (20%) | 44 | 19 (43.2%) p < 0,05 |
| Metapneumo virus |
10 | 1 (10%) | 8 | 3 (37.5%) | Bocaviruses | 23 | 4 (17.4%) | 38 | 21 (55.3%) p < 0,05 |
| Viruses/genetic groups | AA substitutions | Antigenic sites | Number of strains (%) |
| A(H1N1)pdm09 | |||
| All strains | R223Q | 74 (100) | |
| C.1.9 | N38D | 10 (13.5) | |
| D.1 | R45K | 1 (1.4) | |
| C.1.8 | V47I | 35 (47.3) | |
| C.1.9 | S83F | 11 (14.9) | |
| C.1.8 | I96T | 32 (43.2) | |
| D.2 | R113K | 2 (2.7) | |
| C.1.8, C.1.9, and D.3 | T120A | 62 (83.8) | |
| C.1.8 and C.1.9 | S137P | Ca2 | 61 (82.4) |
| C.8, C.9, and C.1 | R142K | Ca2 | 70 (94.6) |
| C.1.8 | K154R | Sa | 13 (17.6) |
| C.1.9 | K169Q | Ca1 | 26 (35.1) |
| C.1.8 | K208R | 13 (17.6) | |
| C.8, C.9, and C.1 | A216T | 71 (95.9) | |
| C.8, C.9, and C.1 | E260D | 71 (95.9) | |
| C.8, C.9, and C.1 | A277T | 71 (95.9) | |
| C.8, C.9, and C.1 | D356E | 71 (95.9) | |
| D.3 | I372V | 1 (1.4) | |
| C.8, C.9, and C.1 | H451N | 71 (95.9) | |
| D.2 | V427I | 2 (2.7) | |
| C.1.9 | K480R | 10 (13.5) | |
| C.1.8 | V527I | 6 (8.1) | |
| A(H3N2) | |||
| All strains | E50K | C | 37 (100) |
| D53N | C | 37 (100) | |
| N96S | +CHO | 37 (100) | |
| I140K | 37 (100) | ||
| N186D | B | 37 (100) | |
| I192F | B 190-helix | 37 (100) | |
| I223V | 37 (100) | ||
| G225D | 37 (100) | ||
| J.2 | T10M | 5 (13.5) | |
| J.2 | P21S | 6 (16.2) | |
| J.1 | I25V | 4 (10.8) | |
| J.2 | F79L | E | 8 (21.6) |
| J.2 | R92K | 3 (8.1) | |
| J.2 | N122D | A -CHO | 33 (89.2) |
| J.2 | V166L | 5 (13.5) | |
| J.2 | P239S | 8 (21.6) | |
| J.2 | K276E | 33 (89.2) | |
| J.1 | V347M | 3 (8.1) | |
| J.1 and J.2 | N378S | 21 (56.8) | |
| J.2 | L409I | 8 (21.6) | |
| J.1 | I418V | 4 (10.8) | |
| B/Victoria lineage | |||
| All strains | D197E | 190-helix | 12 (100) |
| C.5.6 | D129N | 120-loop | 1 (8.3) |
| C.5.7 | E128G | 120-loop | 12 (100) |
| Influenza viruses |
Vaccine strains |
Antigenicsites | N-glycosylation motifs | ||||||
| HA | NA | ||||||||
| A(H1N1)pdm09 | A/Victoria/4897/2022 | Sa | Sb | Ca1 | Ca2 | Cb | 190-helix | 8 | 8 |
| 1 | - | 1 | 2 | - | - | ||||
| A(H3N2) | A/Darwin/9/2021 | A | B | C | D | E | 190-helix | 12 | 9 |
| 1 | 2 | 2 | - | 1 | 1 | ||||
| B/Victoria lineage | B/Austria/1358417/2021 | 120-loop | 150-loop | 160-loop | 190-helix | 11 | 4 | ||
| 2 | - | - | 1 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





