Submitted:
23 January 2025
Posted:
24 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
3.1 Antibacterial Activity
3.2 Evaluation of Antibacterial Activity through Efflux Pump Inhibition
3.3 Evaluation of NorA and MepA Efflux Pump Inhibition by Fluorescence Emission
3.2 Molecular Docking Evaluation of Compound IJ28
3. Discussion
4. Materials and Methods
4.1. Material Acquisition
4.2. General Procedure
- 5-(4-Methoxyphenyl)-N-phenyl-6H-1,3,4-thiadiazin-2-amine (7g) IJ26
- 5-(4-Chlorophenyl)-N-phenyl-6H-1,3,4-thiadiazin-2-amine (7c) IJ 27
- 4-(2-(Phenylamino)-6H-1,3,4-thiadiazin-5-yl)phenol (7f) IJ28
- N-phenyl-5-(p-tolyl)-6H-1,3,4-thiadiazin-2-amine (7e) IJ29
4.3. Preparation of Microorganisms and Inoculum for Microdilution
4.4. Preparation of Substances for Microdilution Test
4.5. Determination of Minimum Inhibitory Concentration (MIC)
4.6. Evaluation of Antibacterial Activity through Efflux Pump Inhibition
4.7. Inhibitory Action of Efflux Pumps NorA Assessed by Increased Fluorescence Emission of Ethidium Bromide
4.8. Molecular Docking Evaluation of Compound IJ28
4.9. Statistical Analysis
5 Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alós, J.-I. Antibiotic Resistance: A Global Crisis. Enfermedades Infecciosas y Microbiología Clínica 2015, 33(10), 692–699. [Google Scholar] [CrossRef] [PubMed]
- Cussolim, P. A.; Salvi Junior, A.; Melo, A. L. Mechanisms of Resistance of Staphylococcus aureus to Antibiotics. Revista Faculdade do Saber 2021, 6(12), 831–843. [Google Scholar]
- Reygaert, W. C. An Overview of Bacterial Antimicrobial Resistance Mechanisms. AIMS Microbiology 2018, 4(3), 482–501. [Google Scholar] [CrossRef]
- Martinez, J. L. General Principles of Antibiotic Resistance in Bacteria. Drug Discovery Today Technologies 2014, 11, 33–39. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Strategies for Bacteriostasis. Antibiotics 2022, 11(9), 1215. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L. J. V. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clinical Microbiology Reviews 2006, 19(2), 382–402. [Google Scholar] [CrossRef] [PubMed]
- Neyfakh, A. A.; Borsch, C. M.; Kaatz, G. W. The NorA Fluoroquinolone Resistance Protein of Staphylococcus aureus is a Multidrug Efflux Transporter. Antimicrobial Agents and Chemotherapy 1993, 37(1), 128–129. [Google Scholar] [CrossRef] [PubMed]
- Lekshmi, M.; Ammini, P.; Adjei, J.; Zhang, F.; Cheng, W. Modulation of Antimicrobial Efflux Pumps of the Major Facilitator Superfamily in Staphylococcus aureus. AIMS Microbiology 2018, 4(1), 1–18. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, R. H. S.; Rocha, J. E.; de Freitas, T. S.; Pereira, R. L. S.; Junior, F. N. P.; de Oliveira, M. R. C.; Batista, F. L. A.; Coutinho, H. D. M.; de Menezes, I. R. A. Evaluation of the Antibacterial Activity and Reversal of the NorA and MepA Efflux Pump by Estragole Against Staphylococcus aureus Bacteria. Archives of Microbiology 2021, 203(6), 3551–3555. [Google Scholar] [CrossRef]
- Almeida, R. S.; Freitas, R. P.; Araújo, A. C. J.; Oliveira, I. R.; Santos, E. L.; Oliveira, R. A.; Oliveira, F. F.; Filho, J. R.; Ferreira, V. A.; Silva, A. C. A. GC-MS Profile and Enhancement of Antibiotic Activity by the Essential Oil of Ocotea odorífera and Safrole: Efflux Pump Inhibition in Staphylococcus aureus. Antibiotics 2020, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J. E.; de Freitas, T. S.; da Cunha Xavier, J.; Pereira, R. L. S.; Junior, F. N. P.; Nogueira, C. E. S.; Marinho, M. M.; Bandeira, P. N.; de Oliveira, M. R.; Marinho, E. S.; Teixeira, A. M. R.; Dos Santos, H. S.; Coutinho, H. D. M. Antibacterial and Antibiotic-Modifying Activity, ADMET Study, and Molecular Docking of Synthetic Chalcone (E)-1-(2-Hydroxyphenyl)-3-(2,4-Dimethoxyphenyl)prop-2-en-1-one in Staphylococcus aureus Strains with NorA and MepA Efflux Pumps. Biomedicine & Pharmacotherapy 2021, 140, 111768. [Google Scholar] [CrossRef]
- Silva, M. O.; Aquino, S. Antimicrobial Resistance: A Review of the Challenges in the Search for New Treatment Alternatives. Revista de Epidemiologia e Controle de Infecção 2018, 8(4), 472–482. [Google Scholar] [CrossRef]
- Souza, M. A.; Comin, T.; Feiden, T.; Fritzen, A. A.; Polina, C. C.; Galvagni, E.; Fisher, B.; Fernandes, I. A.; Souza, R. C.; Backes, G. T.; Cansian, R. L. Alternative Methods for Microbial Control. PERSPECTIVA, Erechim 2019, 43(163), 17–25. Available online: https://www.uricer.edu.br/site/pdfs/perspectiva/163_768.pdf.
- Nascimento, I. J. S. Synthesis, Biological Evaluation, and In Silico Studies of New Thiazolic and Thiadiazine Compounds with Potential Against Trypanosoma cruzi and Leishmania amazonensis. Federal University of Alagoas. Thesis 2022. https://www.repositorio.ufal.br/jspui/handle/123456789/8659.
- Javadpour, M.; Juban, M.; Lo, W.; Bishop, S.; Alberty, J.; Mann, C.; Markhan, J. L. A New Method for Determining the Minimum Inhibitory Concentration of Essential Oils. J. Appl. Microbiol. 1996, 84, 538–544. [Google Scholar]
- Coutinho, H. D.; Costa, J. G.; Lima, E. O.; Falcão-Silva, V. S.; Siqueira-Júnior, J. P. Enhancement of Antibiotic Activity Against Multidrug-Resistant Escherichia coli by Mentha arvensis L. and Chlorpromazine. Chemotherapy 2008, 54, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Santos, R.; Soeiro, P.; Silvestre, S.; Ferreira, S. Resveratrol as an Inhibitor of the NorA Efflux Pump and Resistance Modulator in Staphylococcus aureus. Antibiotics 2023, 12(7), 1168. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M. C.; Schwede, T. Automated Comparative Protein Structure Modeling with SWISS-MODEL and Swiss-PdbViewer: A Historical Perspective. Electrophoresis 2009, 30(S1), S162–S173. [Google Scholar] [CrossRef]
- Araújo-Neto, J. B.; Oliveira-Tintino, C. D. M.; Araújo, G. A.; Alves, D. S.; Ribeiro, F. R.; Brancaglion, G. A.; Carvalho, D. T.; Lima, C. M. G.; Mohammed Ali, H. S. H.; Rather, I. A.; Wani, M. Y.; Emran, T. B.; Coutinho, H. D. M.; Balbino, V. Q.; Tintino, S. R. Substituted Coumarins Inhibit NorA and MepA Efflux Pumps in Staphylococcus aureus. Antibiotics 2023, 12(12), 1739. [Google Scholar] [CrossRef]
- Dos Santos Barbosa, C. R. , et al. Evaluation of the Antibacterial Activity and Inhibition of the MepA Efflux Pump in Staphylococcus aureus by Riparins I, II, III, and IV. Archives of Biochemistry and Biophysics 2023, 748, 109782. [Google Scholar] [CrossRef]
- Morris, G. M.; Huey, R.; Lindstrom, W.; Sanner, M. F.; Belew, R. K.; Goodsell, D. S.; Olson, A. J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. Journal of Computational Chemistry 2009, 30(16), 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Brawley, D. N. , et al. Structural Basis for the Inhibition of the Staphylococcus aureus NorA Efflux Pump. Nature Chemical Biology 2022, 18(7), 706–712. [Google Scholar] [CrossRef] [PubMed]
- Meng, E. C.; Goddard, T. D.; Pettersen, E. F.; Couch, G. S.; Pearson, Z. J.; Morris, J. H.; Ferrin, T. E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Science 2023, 32(11), e4792. [Google Scholar] [CrossRef]
- Pereira, P. S. Anticancer, Antibacterial, and Toxicological Effects of Thiazole and Thiazolidinedione Derivatives. PhD Thesis in Biotechnology - Federal University of Pernambuco, Recife, 2020. https://repositorio.ufpe.br/handle/123456789/37937.
- Freitas, P. R.; Araújo, A. C. J.; Araújo, I. M.; Siqueira, G. M.; Alves, D. S.; Borges, J. A. O.; Miranda, G. M.; Nascimento, I. J. S.; Araújo-Júnior, J. X.; Silva-Júnior, E. F.; de Aquino, T. M.; Junior, F. J. B. M.; Marinho, E. S.; Santos, H. S. D.; Tintin, S. R.; Lima, C. M. G.; Emran, T. B.; Ahmad, S. F.; Attia, S. M.; Coutinho, H. D. M. ADMET Analysis, Antibacterial Activities, and Antibiotic Modifications of Thiadiazine Derivatives. J. Biol. Regul. Homeost. Agents 2024, 38(1), 123–135. [Google Scholar] [CrossRef]
- De Araújo, A. C. J.; Freitas, P. R.; Araújo, I. M.; et al. Enhancement of Antibiotic Activity and Analysis of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Properties of Synthetic Thiadiazines Against Multidrug-Resistant (MDR) Strains. Fundamental & Clinical Pharmacology 2023, 1–15. [Google Scholar] [CrossRef]
- Jain, V. S.; Vora, D. K.; Ramma, C. S. Thiazolidine-2,4-dione: Progress in Multifaceted Applications. Bioorg. Med. Chem. 2013, 21, 1599–1620. [Google Scholar] [CrossRef]
- Aufort, M.; Herscovici, J.; Bouhours, P.; Moreau, N.; Girard, C. Synthesis and Antibiotic Activity of a Library of Small Molecules Derived from 1,2,3-Triazole. Bioorganic & Medicinal Chemistry Letters 2008, 18(3), 1195–1198. [Google Scholar] [CrossRef]
- Martins, M.; Couto, I.; Viveiros, M.; Amaral, L. Identification of Multidrug Resistance Mediated by Efflux in Clinical Bacterial Isolates by Two Simple Methods. Methods Mol. Biol. 2010, 642, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Santana, L. M. D. The Potential of Antimicrobial Photodynamic Therapy in the Development of Resistance in Candida albicans. Master's Thesis in Oral Rehabilitation – São Paulo State University, School of Dentistry, 2020. 137 pages. http://hdl.handle.net/11449/192633.
- Carmello, J. C.; Alves, F. G.; Basso, F.; de Souza Costa, C. A.; Bagnato, V. S.; Mima, E. G. de O.; Pavarina, A. C. Treatment of Oral Candidiasis Using Photodynamic Therapy Mediated by Photodithazine® In Vivo. PLOS ONE 2016, 11(6), e0156947. [Google Scholar] [CrossRef] [PubMed]
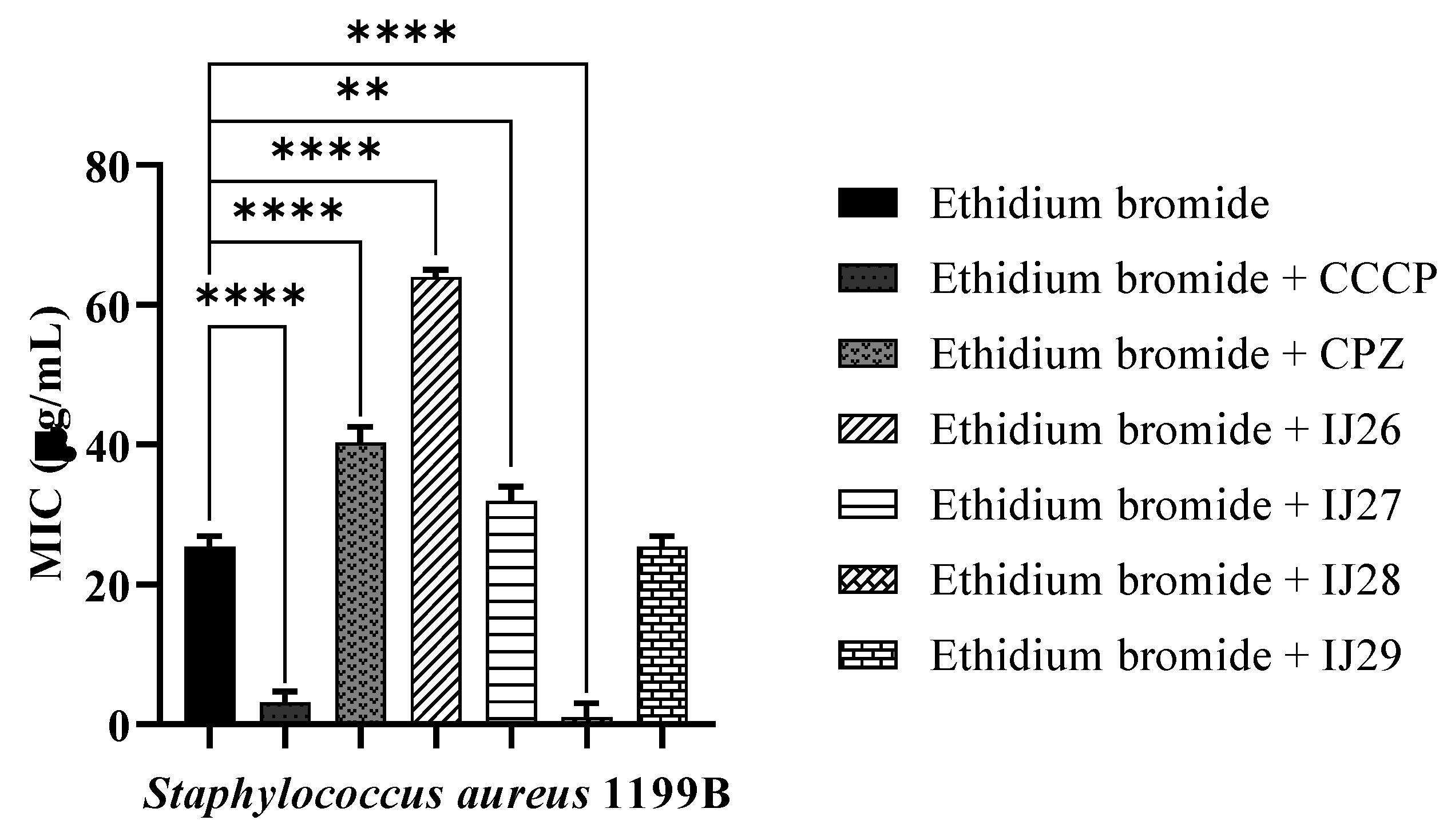
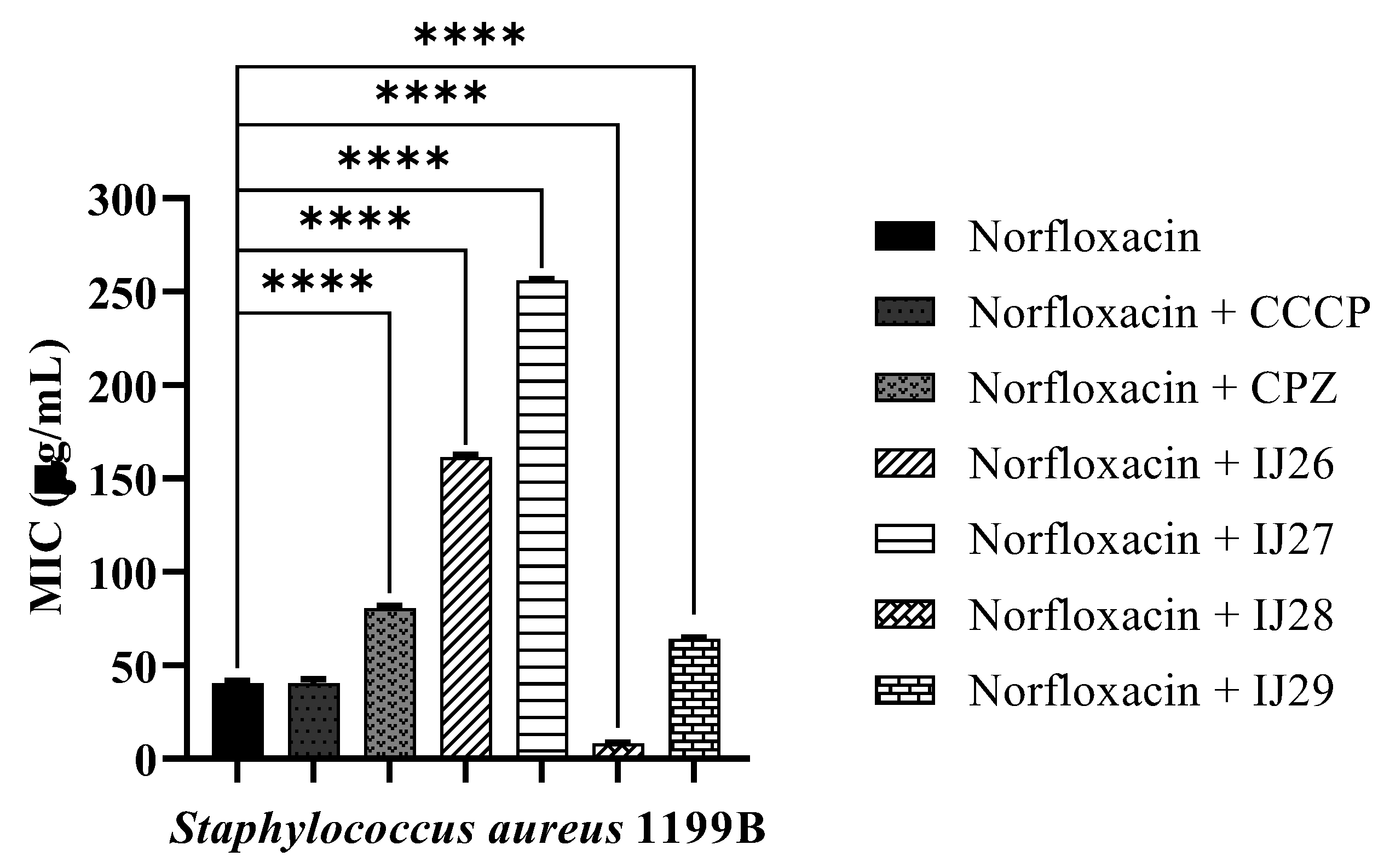
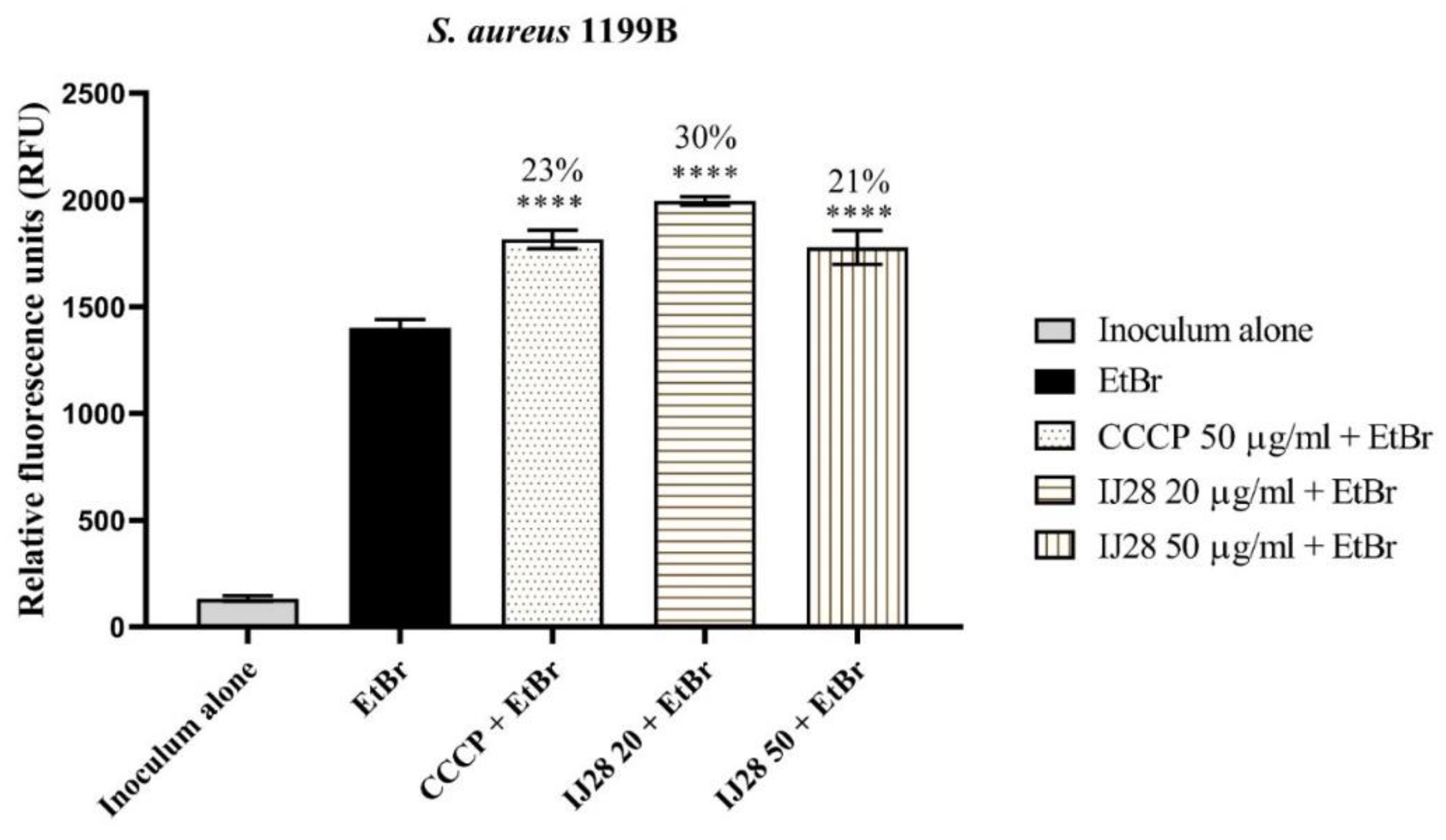
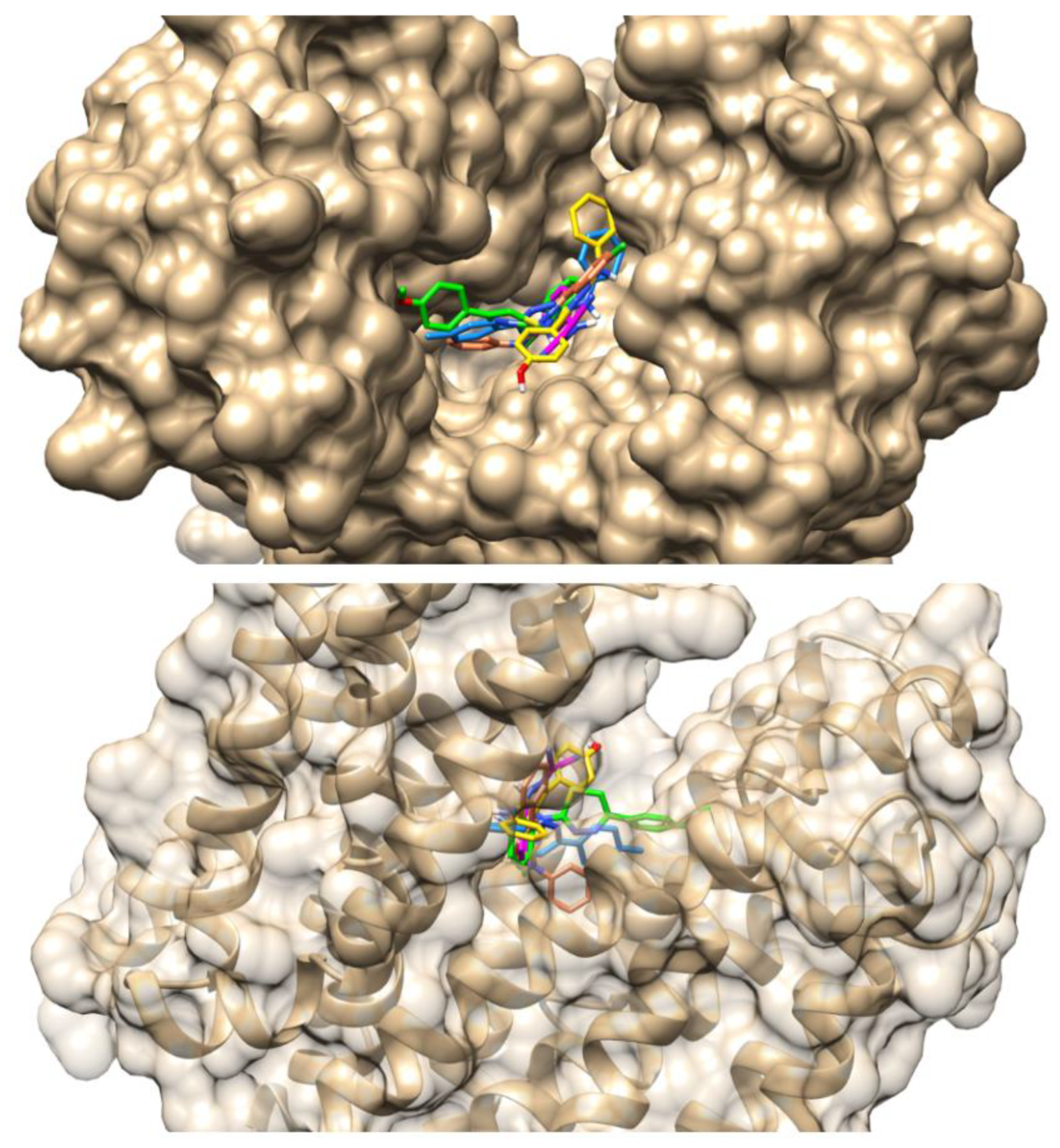
| Compounds | Binding Energy (Kcal/mol) | Inhibition Constant (µM) | Ligand Efficiency |
| IJ26 | -5.30 | 129.52 | -0.25 |
| IJ27 | -5.46 | 99.54 | -0.27 |
| IJ28 | -5.55 | 84.96 | -0.28 |
| IJ29 | -5.43 | 104.31 | -0.27 |
| CCCP | -4.63 | 403.17 | -0.33 |
| Compounds | vdW | Hydrogen Bond | Hidrophobic | Electrostatic |
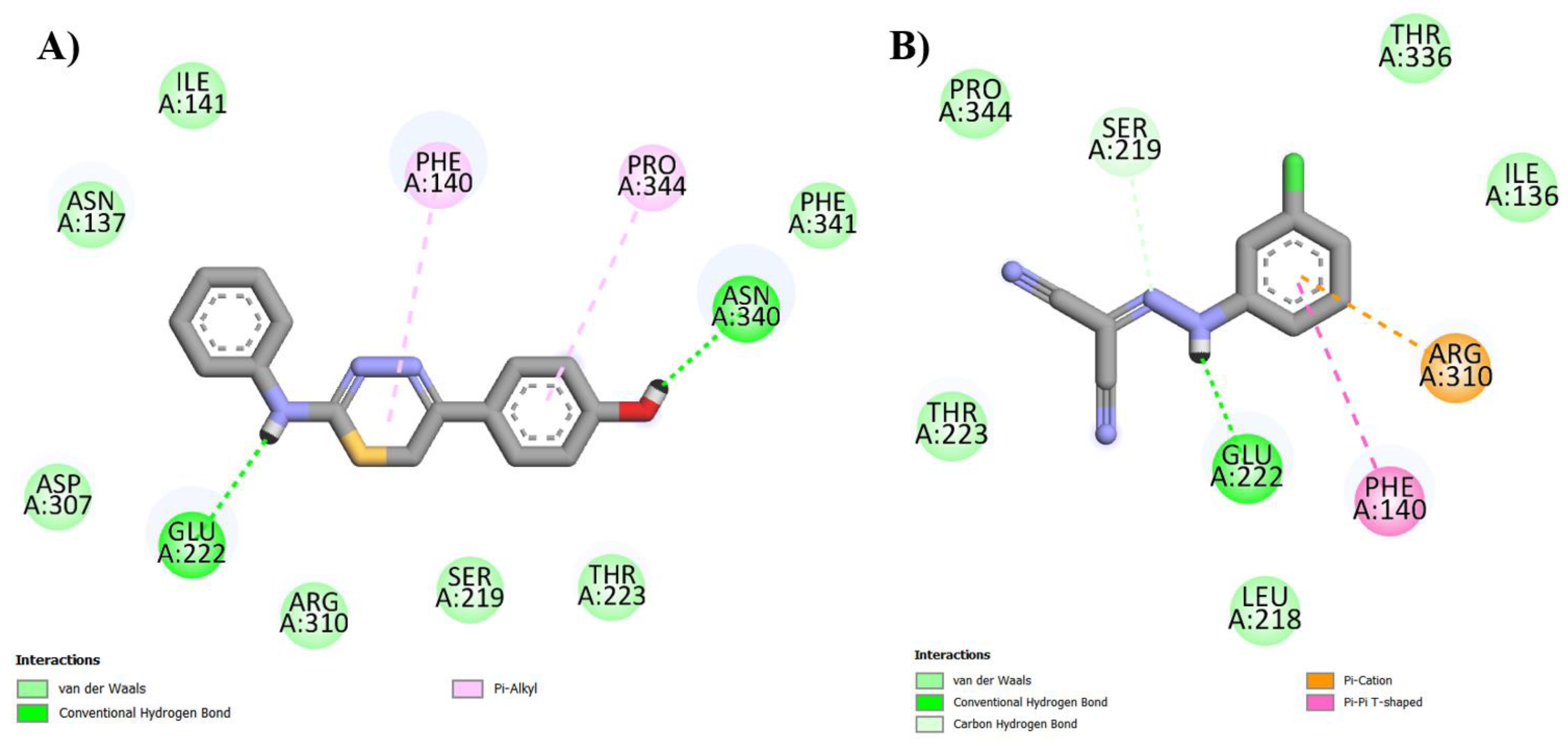
| IJ28 | Arg310, Asn137, Asp307, Ile141, Phe341, Ser219, Thr223 |
Asn340 (1.77 Å) Glu222 (2.23 Å) | Phe140 (5.28 Å) Pro344 (5.29 Å) |
- |
| CCCP | Ile136, Leu218, Pro344, Thr223, Thr336 |
Glu222 (1.74 Å) Ser219 (3.55 Å) | Phe140 (4.88 Å) | Arg310 (3.96 Å) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





