1. Introduction
Approximately 0.5% of children face chronic medical complexities affecting multiple bodily systems leading to frequent hospitalizations and significant healthcare costs [
1,
2,
3]. These children and their caregivers exhibit remarkable resilience, yet their emotional well-being is often undermined by factors such as stress and disrupted social interactions [
4]. Children with severe motor and communication impairments (SMCI) are particularly affected, as their limited ability to communicate can lead to social isolation and challenges in expressing their needs [
5]. The United Nations Convention on the Rights of the Child emphasizes the importance of respecting every child’s preferences [
6]. In this context, accurately identifying and understanding the emotions of children who are non-communicative is crucial for ensuring their preferences are respected and effectively met especially in healthcare settings.
The current standard for self-reporting emotions is the Self-Assessment Manikin (SAM) method [
7,
8]. This approach uses digitized images depicting various emotional states, asking users to select images across three dimensions from a scale of one to nine: pleasure (happy-1 vs. unhappy-9), arousal (excited-1 vs. calm-9), and dominance (controlled-1 vs. in control-9). While SAM facilitates non-verbal interaction, it still requires participants to have communication skills and a cognitive understanding of emotions to navigate and respond to the images [
7].
Previous researchers only categorize each of pleasure, arousal, and dominance into 2 or 3 categories (High, Low, and Neutral), but to the best of our knowledge no researcher has attempted to categorize into 9 categories (for each of the three PAD dimensions). Nine categories would represent the full spectrum of emotions available when an individual self-identifies using the SAM [
8,
9,
10,
11,
12,
13,
14]. A study by Goshvarpour [
11] used only the pleasure and arousal dimensions while classifying four quadrants of emotion using wavelets for both ECG (electrocardiography) and GSR (galvanic skin response) with 100% and 94% [
11], whereas Zied
et al. [
9] had 95% accuracy by fusing the features from four physiological signals and using a continuous wavelet to identify four emotions [
9].
While PAD scores are recognized within the research environment as emotion identifiers, colloquial terminology that represents emotion is more easily understood by the general population. However, very little research exists that maps PAD scores to emotion. In a previous paper [
15], an Emotion Identification (EI) model was developed that maps the PAD scores to an output of one of six basic emotion clusters from the subjective words representing emotion. By combining the model in the current research with the EI model that links PAD scores to basic emotions, emotions can be determined from the physiological signals.
The objective of the current research was to create a model that identifies emotion using physiological data from participants both Typically Developing (TD) and participants who have SMCI.
2. Materials and Methods
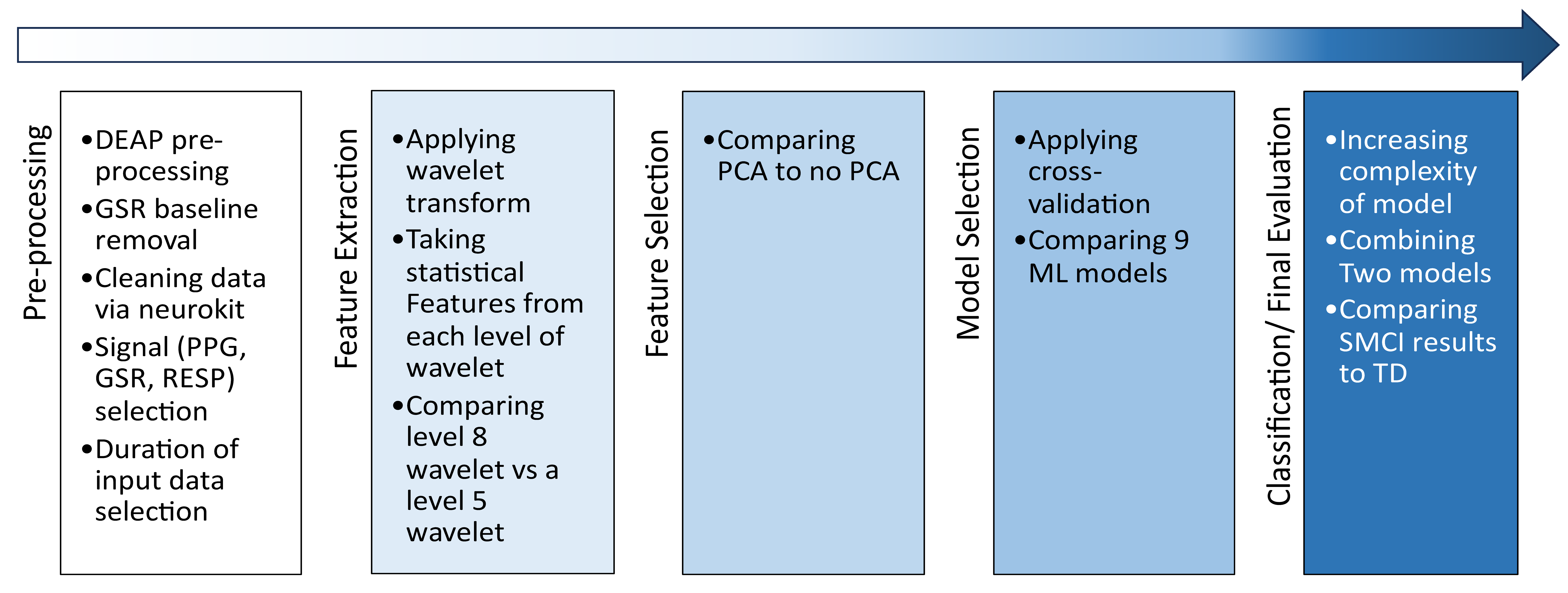
In machine learning, the process involves pre-processing to clean and prepare data, extraction of data characteristics to identify features, selection of features to identify those that are relevant, model selection to determine the best algorithm, and classification to predict categorical labels and evaluate the model’s performance (
Figure 1). The DEAP model was developed using data from the DEAP (Database for Emotion Analysis using Physiological Signals) dataset [
15] of typically developing participants. However, the same protocols could not be used in the collection of data from participants with SMCI. A new protocol was developed for the BDAT dataset (Building and Designing Assistive Technology Lab – author’s lab). The original DEAP machine learning model was adapted to only draw on data that was comparable to the BDAT dataset (SMCI model). The new SMCI model was retrained using the DEAP data and finally tested using the BDAT lab data of persons with SMCI.
2.1. Datasets
Two different datasets were employed in this model: the DEAP dataset and the BDAT dataset.
2.1.1. DEAP Dataset
In the current study, the experimental component of the DEAP dataset was used (
Table 1) [
16]. This dataset was created by having 32 participants view 40 one-minute music video clips online. The participants were asked to record their emotional responses relative to three categories: pleasure, arousal, and dominance. PPG, GSR, and RESP were recorded from the participants while being exposed to the audio stimuli.
2.1.2. BDAT Dataset
There were 10 participants in the BDAT dataset; 9 children with SMCI and 1 matched typically developing (TDM) child. This research was approved by the Queen’s University Health and Research Ethics Board and consent and assent was obtained for all participants. To ensure that the study was accessible to the participant group with SMCI, the study was completed at the participants’ home in most cases.
The IAPS (International Affective Picture System) was used to evoke an emotional response [
17]. The IAPS system was chosen as it is a standardized method that uses the SAM tool to assess emotion. IAPS are a collection of 956 photographs that have been rated by participants for pleasure, arousal, and dominance. The present protocol selected 30 IAPS images that were suitable for children. Participants were shown ten IAPS images for 6 seconds and then self-reported pleasure, arousal and dominance using SAM on a nine-point scale. A grey screen was presented between images, and the timing of this screen depended on when the participant was ready for the next image. The same protocol was followed such that 2 or 3 rounds were conducted, dependent on the willingness of the participant. During the display of images, PPG (photoplethysmography) and GSR (galvanic skin response) data were collected with the Gazepoint GP-3 with included the biometrics system. For children who were unable to self-report their scores on the SAM, information was collected from caregivers of their evaluation of the child’s response.
2.2. Algorithmic Approach (Figure 1)
Using the DEAP dataset, multiple models were iteratively built to improve the accuracy and speed of the model. Since the DEAP dataset used different data collection tools, the DEAP model had to be modified to enable analysis of the BDAT dataset. Unlike the DEAP that was sampled for one minute at 128Hz, the BDAT dataset was sampled at 60Hz for six seconds. Adaptations to the model had to be implemented to account for this difference. Once the adaptions were implemented, the model was retrained with the DEAP dataset before testing it on the SMCI data.
2.2.1. Pre-Processing
All model creation was coded in python using sk.learn [
18]and neurokit libraries [
19]. The data were cleaned using neurokit and the baseline average was subtracted from the GSR signals. The data were prepared to allow testing using an input of all three different signals (PPG, GSR, and RESP) both individually and in combination. The duration of input data was also determined during the pre-processing stage
2.2.2. Feature Extraction
The application of discrete wavelet transforms was used to identify statistical features from each level of the wavelet. A comparison was drawn between the level 8 wavelet method and the level 5 wavelet.
2.2.3. Feature Selection
To decrease the number of features and select the most important ones, principal component analysis (PCA) was performed using all 40 trials from each of the 32 participants of the DEAP dataset. The explained variance ratio for each was used to select using a cutoff of 95% of data. PCA was conducted on each signal independently as well as the combined signals.
2.2.4. Classification
The model was tested using the cross-validation method [
18]. A ten-fold cross-validation was used. The dataset was divided into training and validation data. The training data was used during model development, where the model was trained with nine folds and tested on the tenth set. This process was repeated ten times, with each fold being the test set once. The results were then averaged. The DEAP model, and the new SMCI model with adaptations for the BDAT dataset, were both trained using the cross-validation method with the DEAP dataset for testing.
The features extracted from each independent signal (PPG, GSR, RESP) and the fused features were provided as inputs to nine different machine learning (ML) models. The models encompassed various types of trees (Decision, Random Forest, Gradient Boosting Machines, AdaBoost), support vector machines (RBF, linear, poly, sigmoid), and a naïve Bayes model. Decision trees classify data points using predefined rules. Each node in a Decision Tree represents a prediction based on measurable data features, with branches indicating possible outcomes. A Random Forest model combines multiple decision trees, each trained on random subsets of data and features, and predicts by averaging the outputs of all trees. In a Random Forest, all trees carry equal weight in the final prediction. Gradient Boosting Machines and AdaBoost are akin to Random Forests but differ in how trees contribute to predictions—each tree's impact is weighted, influencing the final prediction differently. Support Vector Machines (SVMs) seek an optimal hyperplane to separate data into classes. By mapping data to a higher-dimensional space, SVM identifies a hyperplane maximizing margin between classes. Different kernel functions (RBF, linear, poly, sigmoid) map data to varied spaces. Naïve Bayes is a probabilistic model using Bayes theorem to predict class probabilities based on conditionally independent features. The ML model with the best performance accuracy was used as a robust DEAP model for future analyses and provided output pleasure, arousal, and dominance scores from the signals.
2.2.5. Evaluation
Affective computing researchers usually decrease the model complexity by grouping the output PAD scores (9 categories in the present study) into two or three categories. The present research tested different levels of complexity. First, all ML models were tested with the cross-validation method described above to find the most accurate in identifying pleasure, arousal, and dominance scores across nine categories (the SAM self-reported integer options). A three-category version was also evaluated by separating the groups into low (1-3), neutral (4-6), and high (7-9) scores, and finally a two-category version of each ML model that split the data into low (1-5) and high (5-9).
In addition to reducing the categories of scores in the arousal and pleasure dimensions, many researchers do not model dominance, the third dimension, further decreasing the complexity of the models and improving the results. However, removal of the dominance axis is inappropriate [
20] so the dominance component was included for all models in this study.
The DEAP dataset consists of two sub-studies: an online dataset in which participants self-reported both PAD scores and an emotional word (from a list of 16), and an experimental study in which participants’ physiological signals were collected and they self-reported only PAD scores.
Once the PAD score is obtained using the ML model, the results can be mapped to words that represent emotion. The current approach maps the complete PAD score and links those PAD scores to six words that represent emotion. These six words were chosen as a result of a mapping evaluation among datasets that include both PAD scores and words that represent emotions [
20].
Using the physiological signals collected from typically developing individuals (the DEAP dataset with PAD scores from auditory stimuli), and from persons with SMCI (the BDAT dataset with IAPS PAD scores), the accuracy of identifying emotions was evaluated.
3. Results
Each model, DEAP model and SMCI adapted model, was evaluated using the five steps listed in
Figure 1. Comparisons of accuracy using various machine learning models that mapped physiological signals to pleasure, arousal, and dominance scores were evaluated.
3.1. DEAP Model
3.1.1. Preprocessing
The DEAP dataset included already preprocessed and reordered data with down sampling to 128Hz. The first three seconds at the beginning of the trial were removed.
3.1.2. Feature Extraction
Discrete wavelet transforms were used to extract different frequency sub-bands from the DEAP database’s PPG, GSR, and RESP signals. For PPG and GSR signals, a Daubechies 4 level 8 wavelet was used. For the RESP signal a symlet 2 level 6 wavelet was evaluated. From each sub-band, 12 features were extracted: entropy, percentiles (5th, 25th, 50th, 75th, 95th), mean, standard deviation, variance, root mean square value, zero crossing, and mean crossing rate. After wavelet transform, 108, 108, and 84 features were extracted from PPG, GSR, and RESP signals respectively, to create the DEAP ML models.
3.1.3. Feature Selection
Principal Component Analysis was performed on all input features from each signal independently as well as a combined model. Using PCA decreased the time to fit the model. For example, the time to process using the gradient boosting model decreased by 26 seconds, the largest processing time decrease of the models evaluated.
When applying PCA to each of the signals independently, 21, 24, and 14 features were selected for PPG, GSR, and RESP, respectively. Applying PCA independently to each of the signals and then summing the total resulted in 59 selected features. However, when PCA was applied to all three signals combined, 54 features were identified (
Table 2). This could indicate that the combined PCA removes some features that are already identified within another principal component.
3.1.4. Classification
Although nine different ML models were compared, only the results from the models with the highest accuracy (and lowest fitting time) for each signal or grouping of signals is shown in
Table 3. These models were trained using 60 seconds of DEAP data and used PCA in feature identification. There was a slight improvement when using multiple signals as compared to independent signals of PPG, GSR, and RESP (23% accuracy as compared to 21%). Overall, SVM-rbf had the best accuracy along with the lowest fitting times, while Gradient Boosting and SVM poly models had the worst fitting times and inferior accuracies.
3.1.5. Evaluation- DEAP Model
The output accuracy of scores for pleasure, arousal, and dominance, for each of the ML models and each level of complexity were calculated (
Table 4 reports the accuracy from the best ML model for pleasure, arousal, and dominance scores relative to the level of complexity). For levels of complexity of nine, three (high, neutral, low), and two categories (high, low) of PAD scores, to ensure the model is performing above chance, an accuracy higher than 11%, 33%, and 50% respectively is required. All models out-performed chance and the most accurate ML model was an SVM-rbf in all cases except two-category arousal. However, the accuracy was not sufficient for identifying the complexities of emotions especially across nine categories. Using nine categories, accuracies were 20%, 21%, and 24% for pleasure, arousal, and dominance, respectively.
To apply this research to emotion recognition, the mapping of PAD scores to emotion is an important output. The EI model described earlier [
15] was used to map the participant self-reported PAD scores to terminology that represents emotion. The physiological signals were used to identify the PAD scores which were then mapped to emotion. These results were compared to the EI model. If the output emotion from the physiological signal matched the output emotion from the EI model, it was considered accurate. Using the DEAP dataset with TD participants the model correctly predicted the emotion with an accuracy level of 24%.
3.2. SMCI Model Adaptations
Once a reliable model was developed using data from persons of the typically developing population (DEAP dataset), it could also be applied to data collected from the population of persons with SMCI. Four of the participants with SMCI were able to use an alternative communication device to provide a self-reported emotion word to express how they were feeling after each image was shown. This provided ground truth data for the model.
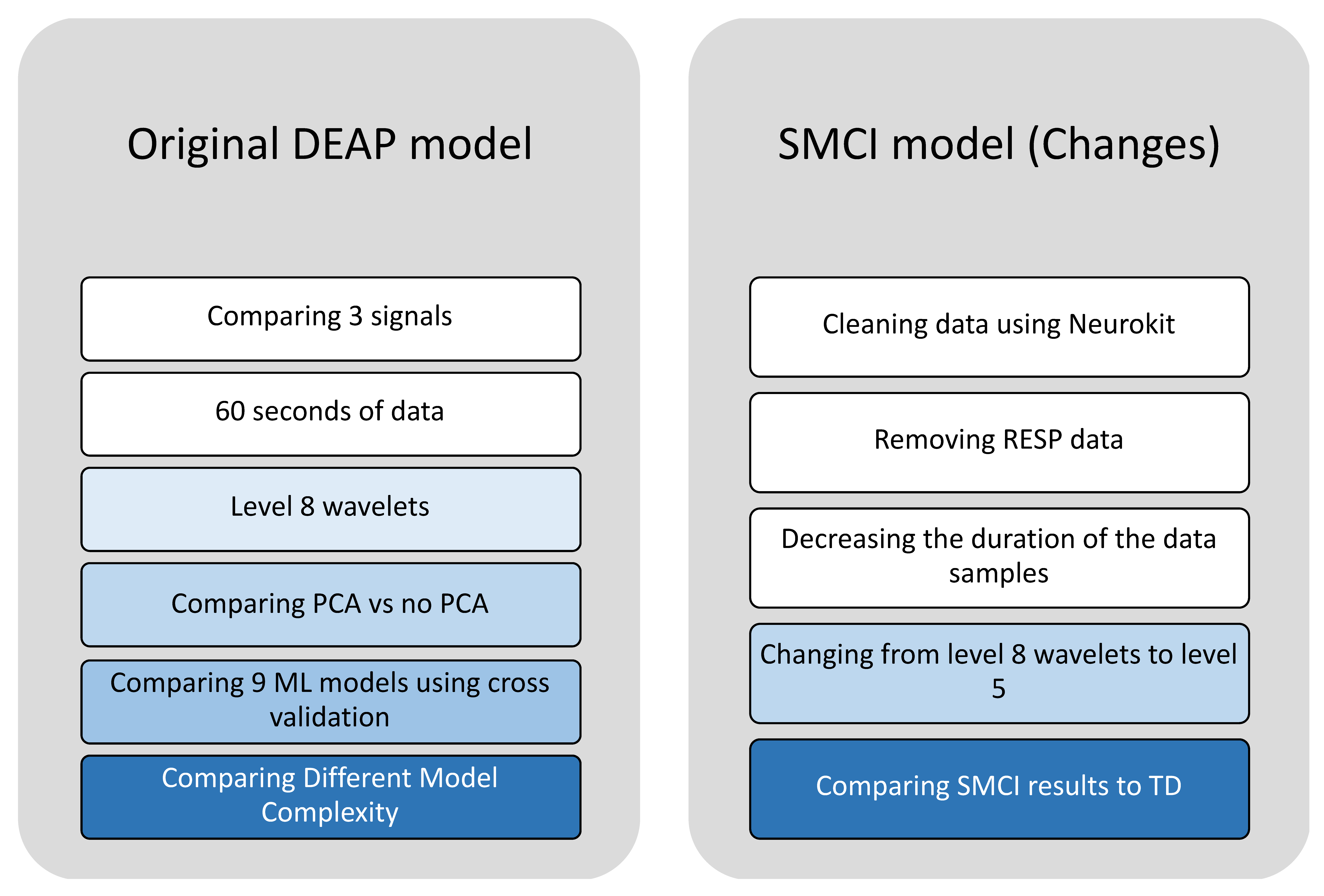
The same experimental protocol used for the DEAP dataset could not be replicated with participants who had SMCI. Changes in data collection were required and duration of trials was reduced. As a result, the DEAP model had to be modified to account for these differences. The following changes to the DEAP model were implemented (and are further discussed): removal of the RESP signal, cleaning the BDAT data was required using Neurokit, modifying the duration of the data samples (BDAT was only collected for 6 seconds, while DEAP was collected for 60 seconds), and the use of level 5 wavelets instead of the level 8 wavelets. Instead of looking at the various model complexities with different categories, the nine categories were evaluated and the emotion word from the detected physiological signals was compared to the DEAP equivalent.
Figure 2.
Modifications to the DEAP model when adapting for participants with SMCI (the colours match the process steps identified in
Figure 1).
Figure 2.
Modifications to the DEAP model when adapting for participants with SMCI (the colours match the process steps identified in
Figure 1).
3.2.1. Preprocessing
When creating the adapted SMCI model, the DEAP data was used for training and testing, however the RESP signal was removed. The Gazepoint system used for data collection of participants with SMCI did not collect respiration data. The average baseline GSR was also subtracted from all values to normalize the data and keep it within the same range as the DEAP GSR data.
The data were cleaned using the neurokit process function. This cleaning function was conducted on the pre-processed DEAP data in addition to the BDAT data. The additional cleaning phase decreased the training accuracy by less than one percent for each of the pleasure, arousal, and dominance from the original DEAP model.
Since the stimuli in the BDAT dataset were only shown for six seconds, an adaptation to the DEAP model was required to decrease the duration of the training trials in the model. However, as the duration decreased, the number of unique outputs for validation also decreased. For the DEAP data, neurokit could not process training data that was less than 11 seconds due to the limited number of peaks. Thus, the SMCI model was trained on trials of 12 seconds from the DEAP dataset, while the test set for persons with SMCI was 6 seconds (neurokit was able to process original data from SMCI participants over 6 seconds).
3.2.2. Feature Extraction
When extracting features with the adapted model and the SMCI data, the Daubechies 4 level 8 wavelet was not compatible. The highest wavelet transform that would run was a level 5 wavelet. As a result, the adapted model for SMCI used a Daubechies 4 level 5 wavelet for the PPG and GSR signals. For both PPG, and GSR, the number of features extracted with level 5 wavelets was 72 for each.
3.2.3. Feature Selection
No significant changes were made in the PCA parameters for the adapted model, and PCA was refit with a 95% explained variance threshold. Before PCA, there were 144 features and after PCA there were 16 features.
3.2.4. Classification
The nine-category classification using the adaptive SMCI model was trained and tested using the DEAP data before using the BDAT dataset. The DEAP model was compared to the SMCI model on the same DEAP training data, and the models had a maximum difference of 2% accuracy. Pleasure and dominance models were similar in accuracy to the original models (
Table 5).
3.2.5. Evaluation-BDAT Model
The SMCI model using physiological signals from participants with SMCI was combined with the EI model [
15] to identify a word that represented an emotion. This was validated against the word provided by the participant with SMCI. The accuracy of identifying an emotion from six potential outcomes was 14.81% (less than chance, 1/6 or 16%).
Data collection issues were encountered. Of the 10 participants (9 SMCI and 1 TD), 5 had valid (continuously recorded data throughout trial) GSR and HR data values for at least one image stimulus (of a potential 30 images). SMCI01 and SMCI07 each had valid data for only one image, while SMCI08 had valid data for 13 images, and SMCI09 had valid data for 9 images. The typically developing participant (TDM01) had valid data for 2 images. Therefore, 84.62% of the valid data came from two participants (SMCI08 and SMCI09). The model only predicted the correct emotions for two participants: TDM01 and SMCI08. The accuracy for predicting emotion was 50% for TDM01 and 23.08% for SMCI08.
There were 20 different IAPS image stimuli that had valid GSR and HR data values that could be used to predict emotions out of 300 IAPS image stimuli that were collected from participants with SMCI. While the protocol had been tested and checked, it became evident that flexibility in data collection is required when working with participants with disabilities. From the 20 images with usable data, six were viewed by two participants, while fourteen were viewed by only one. Of those, 2 image stimuli resulted in physiological signals that mapped to the intended emotion with 100% accuracy, while another two resulted in 50% accuracy. The other sixteen image stimuli resulted in physiological signals that could not be mapped correctly to an emotion.
By removing the participant data from those participants for which the model could not correctly predict emotions, the accuracy of the model improved from 14.81% to 33.33%. It was evident that the limited number of image stimuli that had valid data and small number of participants with valid GSR and HR data were the main factors contributing to the lower accuracy with the model.
4. Discussion
The focus of this study was to develop a model that could predict emotion from physiological signals of children with SMCI. Based on the DEAP model, data from typically developing individuals could be used to identify emotions with results greater than chance. However, when the model was applied to data collected from children with SMCI, the results were less convincing. Only in one case, SMCI08, the emotional outcomes were similar in accuracy to the DEAP dataset.
When training the model, different training durations were evaluated. Based on the standard IAPS protocol, images are shown for six seconds, which was implemented in this study for participants with SMCI. However, Verduyn et al. [
21] found that different emotions have different durations. It is perceivable that different time windows may allow for better identification of different emotions. Since this study was focused on the basic six emotions, it is possible that the duration did not allow for accurate identification. Verduyn et al. [
21] also found that women feel emotion for a longer time period than men. Since the DEAP and SMCI models were not person-specific, nuances in emotional response were not evaluated and may be considered for future work.
Another aspect that could contribute to accuracy when detecting emotions is that different event importance also causes different durations of feeling an emotion. Verduyn et al. [
21] found that different training durations were better or worse depending on the duration of the signal evaluated. A song as used in DEAP may elicit an emotion for longer than the IAPS image used in the BDAT dataset. This suggests that the stimulus used in the training of the model should be the same as the stimulus used for testing. Additionally, when training a model for everyday use, more environmentally suitable stimuli are required.
Kim and Andre [
22] found that a person dependent model is more accurate than an independent model when identifying emotions (using a pseudoinverse linear determinant analysis). Person dependent models could rely on data collected over longer timeframes that are updated based on prior experiences. Given the diversity of the population of persons with disability, it is more appropriate to consider a person specific model that focuses on one individual and updates the model as it learns from changes in their physiological signals and emotions over time.
To the best of our knowledge, no researchers have attempted to model all 9 levels of emotion response from the PAD scores, and most researchers do not attempt to model dominance. The choice to include all the levels for pleasure, arousal, and dominance was undertaken as a first step in the design of a system that can provide information about emotional expression by an individual who has severe motor and communication impairment. The expansion of the research to include all aspects of emotion, rather than only positive and negative, identifies the importance of a pursuit towards more useable and applicable measures of emotion.
5. Conclusions
This study demonstrates the feasibility of using a machine learning model to identify emotions in children with severe motor and communication impairments (SMCI) by analyzing physiological signals. By adapting a model developed for typically developing participants from the DEAP dataset, we created a version suitable for SMCI participants using the BDAT dataset. Key adaptations, such as removing the respiratory signal and training the model on shorter duration segments, allowed comparable accuracy levels to be identified. The model performed better than chance suggesting potential for future refinement and application in healthcare settings.
Future work should focus on expanding the dataset for SMCI participants, integrating additional signals like ECG for improved accuracy, and developing personalized models to account for individual variations. Incorporating the dominance dimension and recognizing a broader range of emotions could further enhance the model's utility. This research highlights the potential of machine learning to improve understanding and support of the emotional well-being of children with complex communication needs.
Author Contributions
“Conceptualization, C.V, and T.C.D.; methodology, T.C.D..; software, C.V. and K.P.; validation, C.V.; formal analysis, C.V.; data curation, C.V; writing—original draft preparation, C.V. and K.P.; writing—review and editing, T.C.D; supervision, T.C.D; project administration, C.V. and T.C.D.; funding acquisition, T.C.D. All authors have read and agreed to the published version of the manuscript.”
Funding
This work was supported by a Discovery Grant from the Natural Science and Engineering Research Council of Canada [NSERC RGPIN 2016-04669 and RGPIN 2023-05354] and a CREATE grant [NSERC-READI].
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Queen’s University Health Sciences and Affiliated Teaching Hospitals Research Ethics Board (HSREB) on August 24, 2022 MECH-76-22.
Informed Consent Statement
“Informed consent was obtained from the parents of participants involved in the study while participants provided informed assent.
Data Availability Statement
All data from the development of the DEAP model is available online [
16]. Permission was obtained to collect physiological data from parents and participants for the BDAT dataset, but not to share publicly.
Acknowledgments
We’d like to acknowledge the support and data collection assistance of Mackenzie Collins and Sydney van Engelen.
Conflicts of Interest
Declare conflicts of interest or state “The authors declare no conflicts of interest.”
References
- Cohen, E., et al., Children with medical complexity: an emerging population for clinical and research initiatives. Pediatrics, 2011. 127(3): p. 529-38.
- Cohen, E., et al., Patterns and costs of health care use of children with medical complexity. Pediatrics, 2012. 130(6): p. e1463-70.
- Berry, J.G., What Children with Medical Complexity, Their Families, and Healthcare Providers Deserve from an Ideal Healthcare System. 2015, Lucile Packard's Foundation for Children's Health.
- Pollard, E.L. and P.D. Lee, Child Well-being: A Systematic Review of the Literature. Social Indicators Research, 2003. 61(1): p. 59-78.
- Noyek, S., et al., Direct assessment of emotional well-being from children with severe motor and communication impairment: a systematic review. Disabil Rehabil Assist Technol, 2022. 17(5): p. 501-514.
- Nations, U., Convention on the Rights of the Child. 1990: New York.
- Broekens, J. and W.-P. Brinkman, AffectButton: A method for reliable and valid affective self-report. International Journal of Human-Computer Studies, 2013. 71(6): p. 641-667.
- Choi, K.-H., et al., Is heart rate variability (HRV) an adequate tool for evaluating human emotions? – A focus on the use of the International Affective Picture System (IAPS). Psychiatry Research, 2017. 251: p. 192-196.
- Zied, G., et al., Emotion recognition from physiological signals using fusion of wavelet based features. 2015.
- Goshvarpour, A. and A. Goshvarpour, Evaluation of Novel Entropy-Based Complex Wavelet Sub-bands Measures of PPG in an Emotion Recognition System. Journal of Medical and Biological Engineering, 2020. 40(3): p. 451-461.
- Goshvarpour, A., A. Abbasi, and A. Goshvarpour, An accurate emotion recognition system using ECG and GSR signals and matching pursuit method. Biomedical Journal, 2017. 40(6): p. 355-368.
- Katsis, C.D., et al., Toward Emotion Recognition in Car-Racing Drivers: A Biosignal Processing Approach. IEEE Transactions on Systems, Man, and Cybernetics - Part A: Systems and Humans, 2008. 38(3): p. 502-512.
- Domínguez-Jiménez, J.A., et al., A machine learning model for emotion recognition from physiological signals. Biomedical Signal Processing and Control, 2020. 55: p. 101646.
- Yoo, G. and S. Hong. Emotion Evaluation Analysis and System Design of Biosignal. in 2016 International Conference on Platform Technology and Service (PlatCon). 2016.
- C. Vowles, M.C.C., T.C. Davies, Assessing Basic Emotion via Machine Learning: Comparative Analysis of Number of Basic Emotions and Algorithms, in Annual International Conference IEEE Engineering in Medicine and Biology Society. 2024, EMBC: Florida, USA.
- Koelstra, S., et al., DEAP: A Database for Emotion Analysis ;Using Physiological Signals. IEEE Transactions on Affective Computing, 2012. 3(1): p. 18-31.
- Lang, P.J., Bradley, M. M., & Cuthbert, B. N. , International affective picture system (IAPS): Affective ratings of pictures and instruction manual. 2008: Gainesville, FL: University of Florida.
- developers, s.-l. User Guide. 2024 [cited 2024; Available from: https://scikit-learn.org/stable/user_guide.html.
- Makowski, D., Pham, T., Lau, Z. J., Brammer, J. C., Lespinasse, F., Pham, H., Schölzel, C., & Chen, S. A. , NeuroKit2: A Python toolbox for neurophysiological signal processing. Behavior Research Methods, 2021 53(4): p. 1689–1696.
- Vowles, C. and C. Davies, Emotion Based on the PAD Theory of Emotion: A Comparison of DEAP and ANEW Datasets. Pre-print 2023.
- Verduyn, P., et al., Predicting the duration of emotional experience: two experience sampling studies. (1528-3542 (Print)).
- Kim, J. and E. Andre, Emotion Recognition Based on Physiological Changes in Music Listening. IEEE transactions on pattern analysis and machine intelligence, 2009. 30: p. 2067-83.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).