Submitted:
09 March 2025
Posted:
10 March 2025
You are already at the latest version
Abstract
The MAT1-1-1 and MAT1-2-1 proteins are essential for the sexual reproduction of Ophiocordyceps sinensis. Although Hirsutella sinensis has been postulated to be the sole-anamorph of O. sinensis and to undergo self-fertilization under homothallism or pseudohomothallism, little is known about the three-dimensional (3D) structures of the mating proteins in the natural Cordyceps sinensis insect-fungal complex, which is a valuable therapeutic agent in traditional Chinese medicine. However, the alternative splicing and differential occurrence and translation of the MAT1-1-1 and MAT1-2-1 genes have been revealed in H. sinensis, negating the self-fertilization hypothesis but rather suggesting the occurrence of self-sterility under heterothallic or hybrid outcrossing. In this study, the MAT1-1-1 and MAT1-2-1 proteins in 173 H. sinensis strains and wild-type C. sinensis isolates were clustered into 6 and 5 clades in the Bayesian clustering trees and belonged to 24 and 21 diverse AlphaFold-predicted 3D structural morphs, respectively. Over 3 quarters of the strains/isolates contained either MAT1-1-1 or MAT1-2-1 proteins but not both. The diversity of the heteromorphic 3D structures of the mating proteins suggested functional alterations of the proteins and provided additional evidence supporting the self-sterility hypothesis under heterothallism and hybridization for H. sinensis, Genotype #1 of the 17 genome-independent O. sinensis genotypes. The heteromorphic stereostructures and mutations of the MAT1-1-1 and MAT1-2-1 proteins in the wild-type C. sinensis isolates and natural C. sinensis insect-fungi complex suggest various sources of the mating proteins produced by two or more cooccurring heterospecific fungal species in natural C. sinensis that have been discovered in mycobiotic, molecular, metagenomic, and metatranscriptomic studies and may inspire future studies on the biochemistry of mating and pheromone-receptor proteins and the reproductive physiology of O. sinensis.
Keywords:
1. Introduction
2. Materials and Methods
2.1. C. sinensis Isolates and Accession Numbers of the MAT1-1-1 and MAT1-2-1 Proteins
2.2. Genome, Transcriptome, and Metatranscriptome Assemblies of H. sinensis Strains and the Natural C. sinensis Insect-Fungal Complex
2.3. Statistical Clustering Analysis for the MAT1-1-1 and MAT1-2-1 Protein Sequences
2.4.AlphaFold. -Based Prediction of 3D Structures of Mating Proteins
2.5. Alignment Analysis of Protein Sequences
2.6. Amino Acid Properties and Scale Analysis
3. Results
3.1. Diversity of the MAT1-1-1 and MAT1-2-1 Proteins in H. sinensis Strains and Wild-Type C. sinensis Isolates on the Basis of the AlphaFold-Predicted 3D Structures
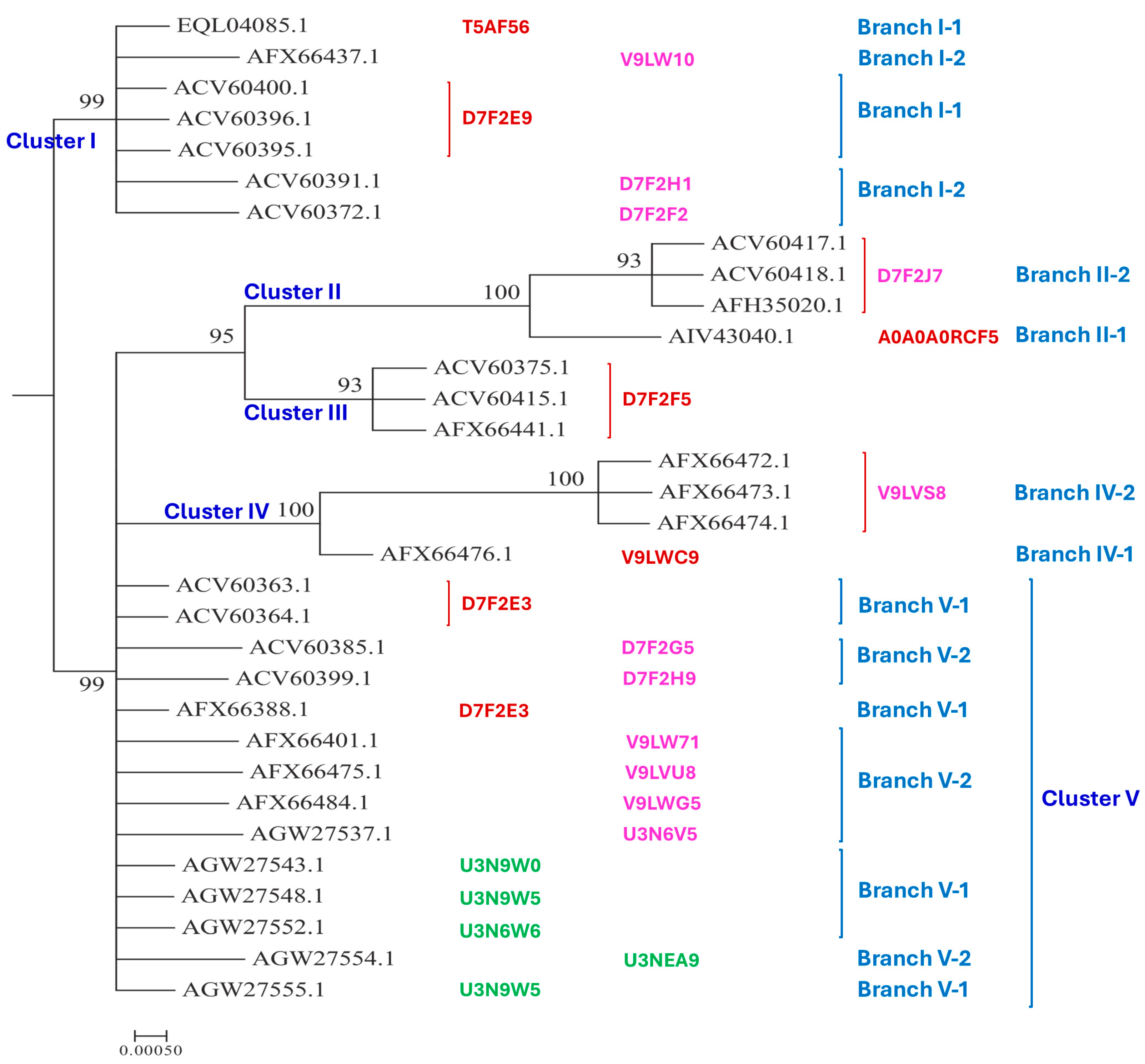
3.2. Bayesian Analysis of the MAT1-1-1 and MAT1-2-1 Proteins
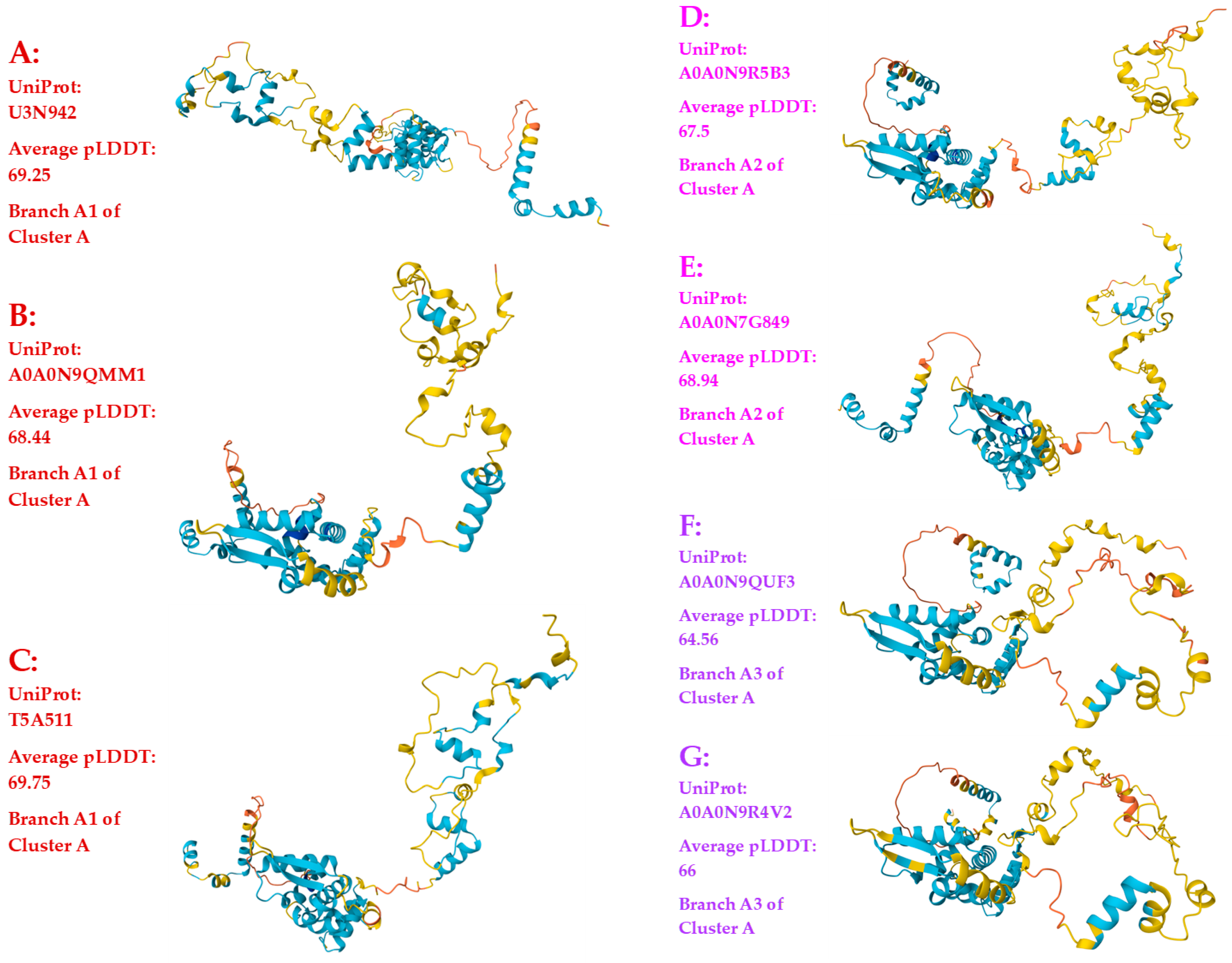
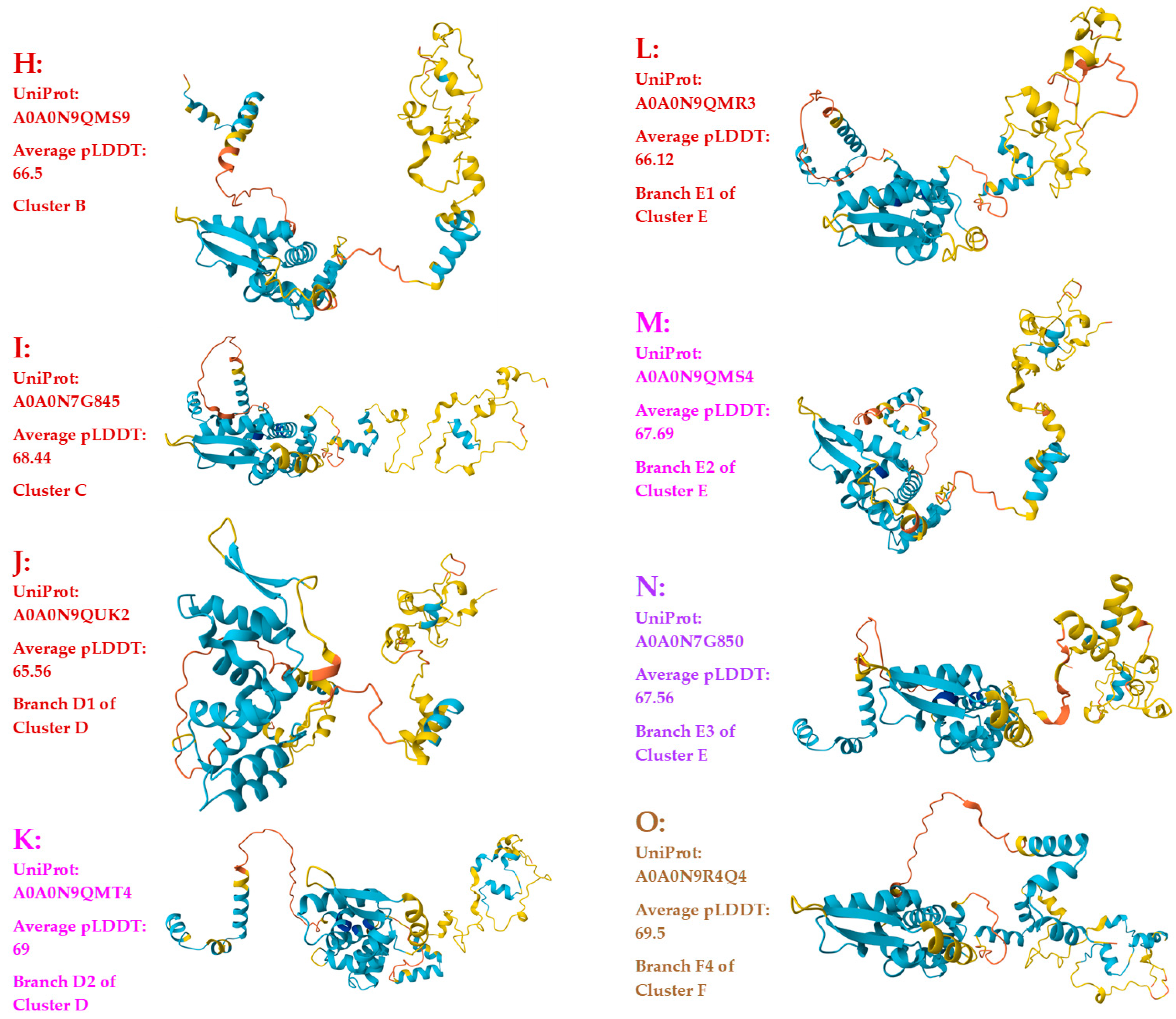
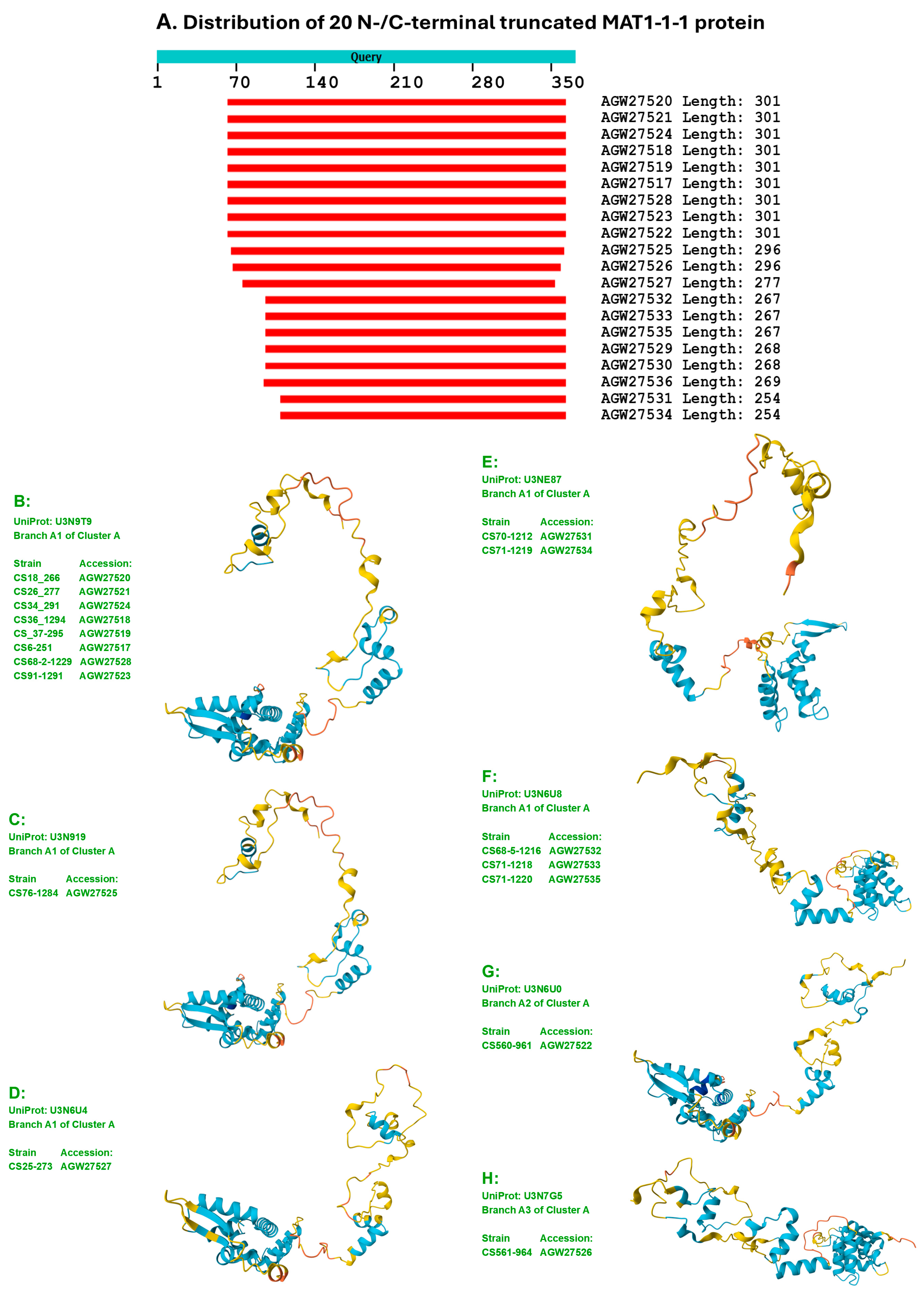
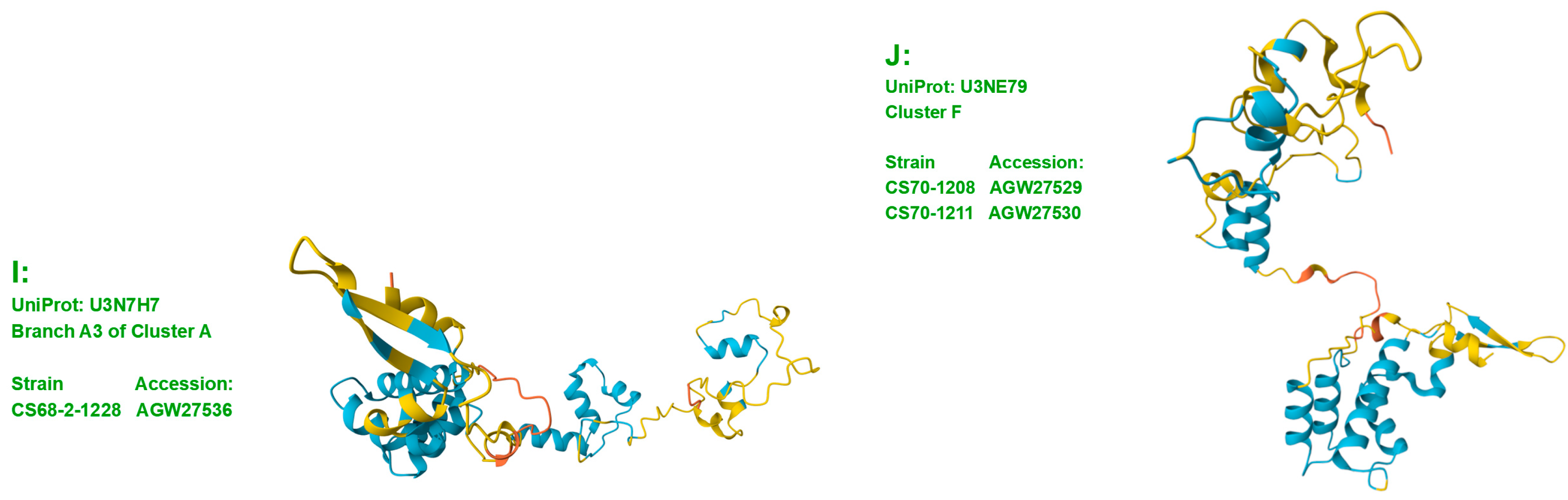
3.3. Heteromorphic AlphaFold-Predicted 3D Structures of the MAT1-1-1 Proteins
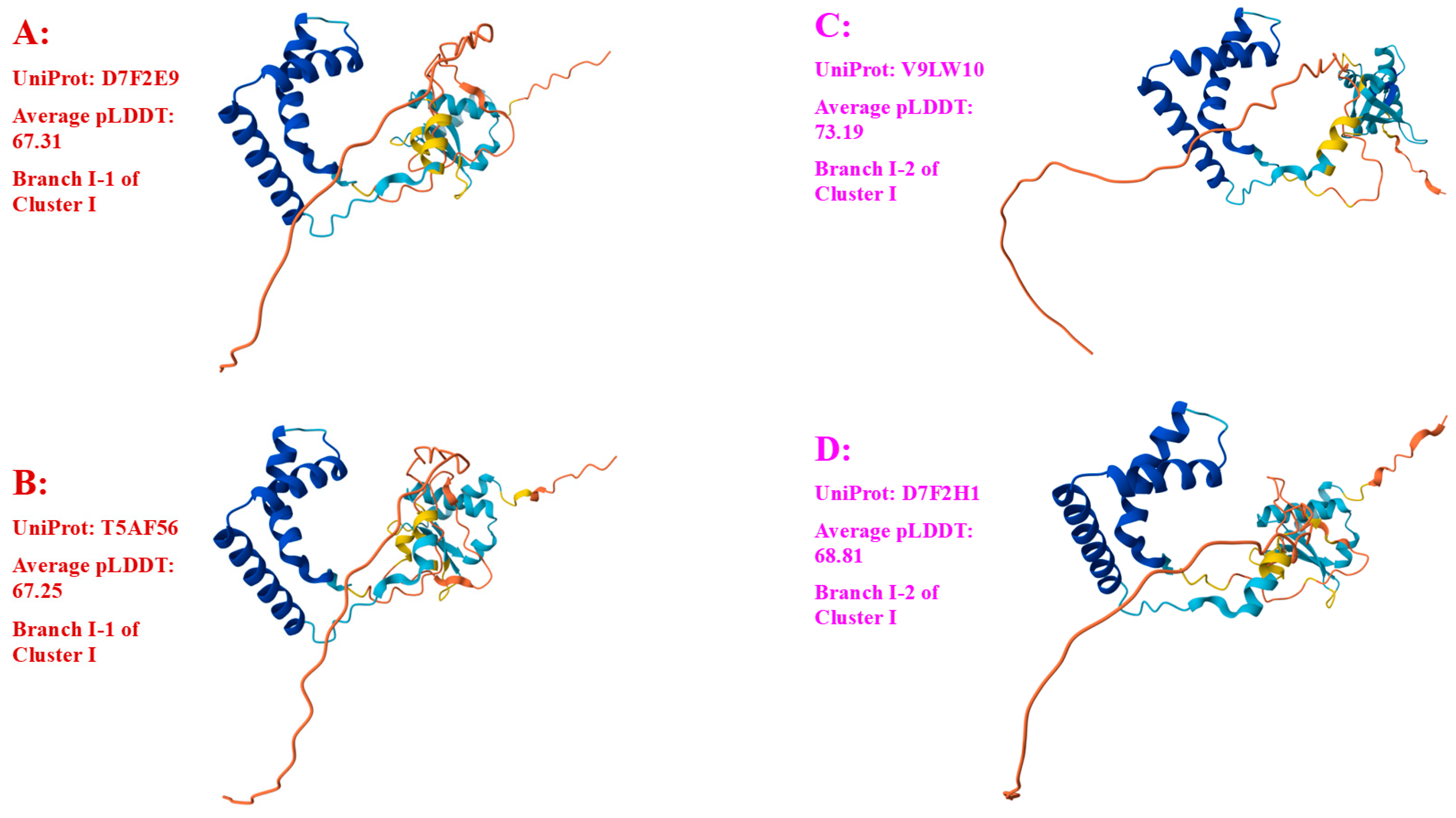
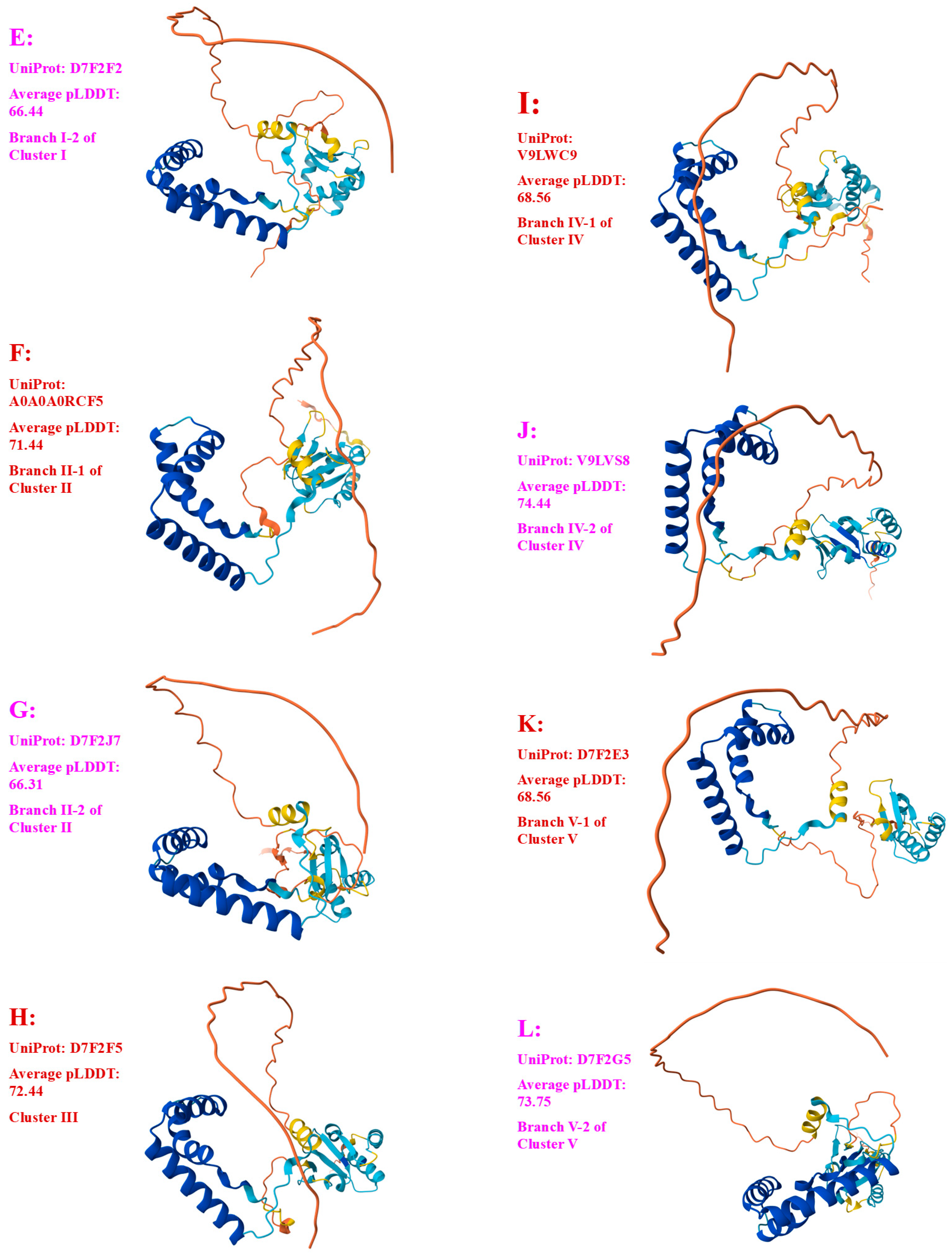
3.4. Heteromorphic AlphaFold-Predicted 3D Structures of the MAT1-2-1 Proteins
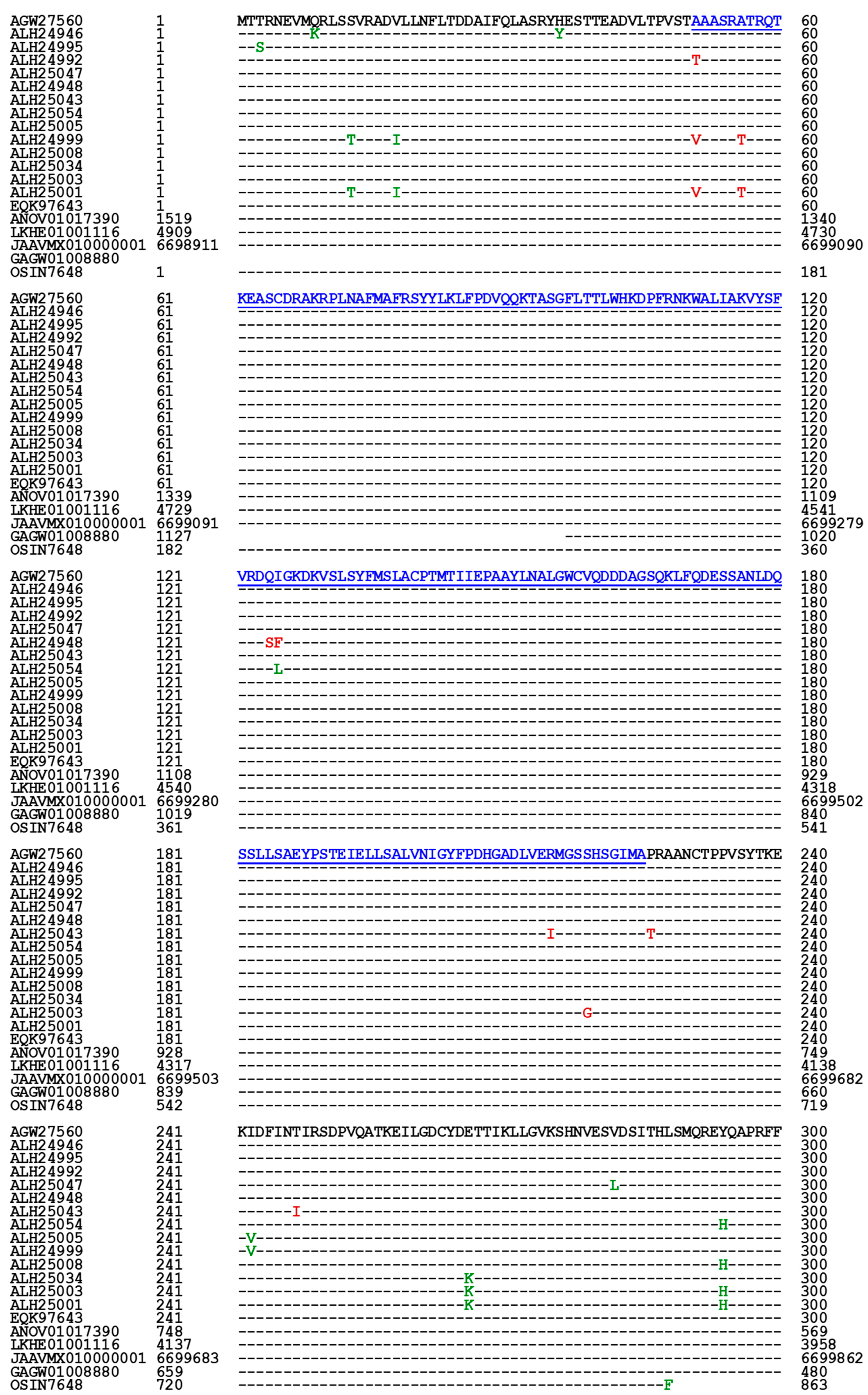
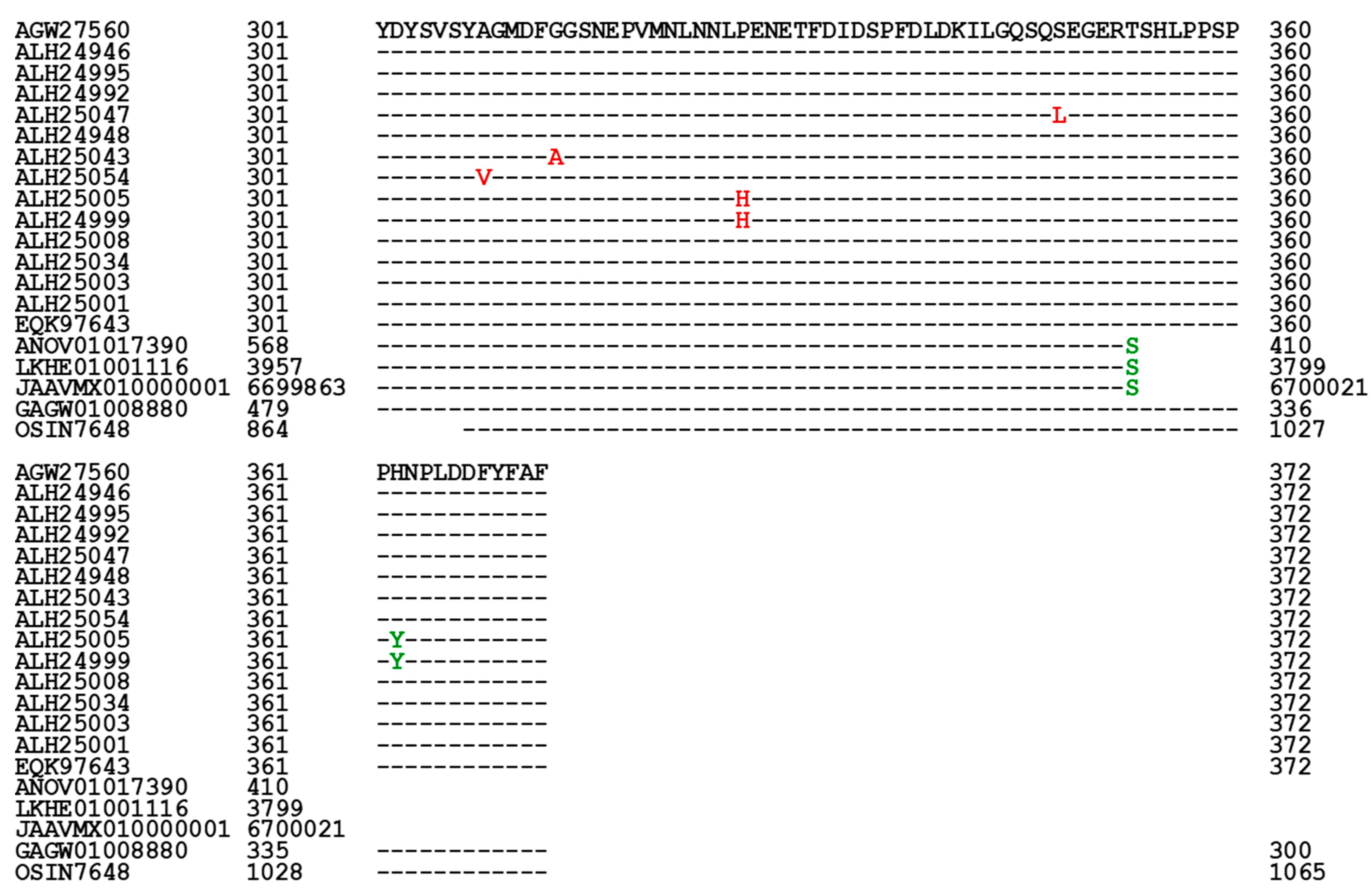
3.5. Primary Structures of the MAT1-1-1 Proteins
3.6. Primary Structures of the MAT1-2-1 Proteins
3.7. Differential Genomic Occurrence of the MAT1-1-1 and MAT1-2-1 Proteins in H. sinensis
3.8. Differential Transcriptomic and Metatranscriptomic Occurrences of the MAT1-1-1 and MAT1-2-1 Proteins in H. sinensis and the Natural C. sinensis Insect-Fumgal Complex
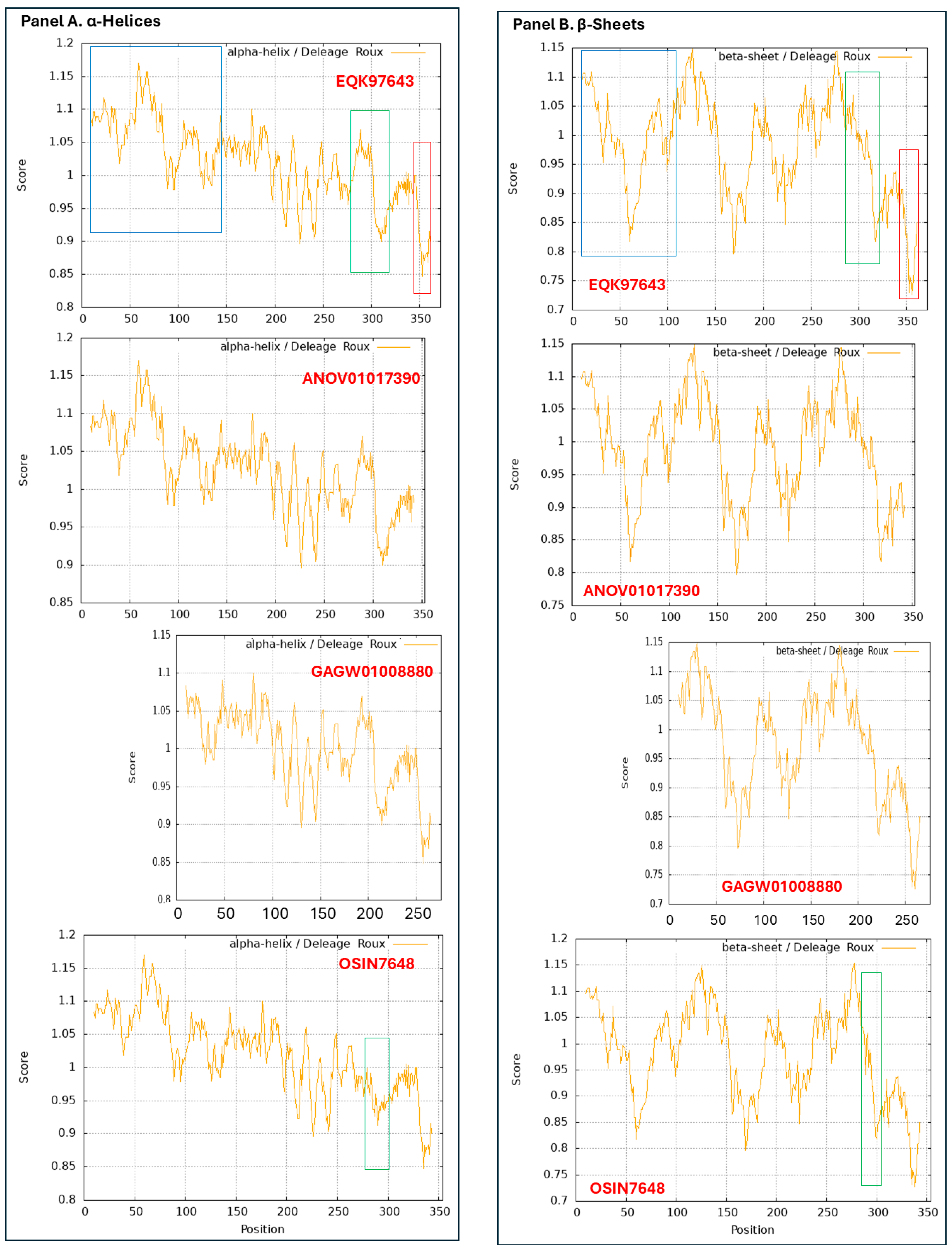
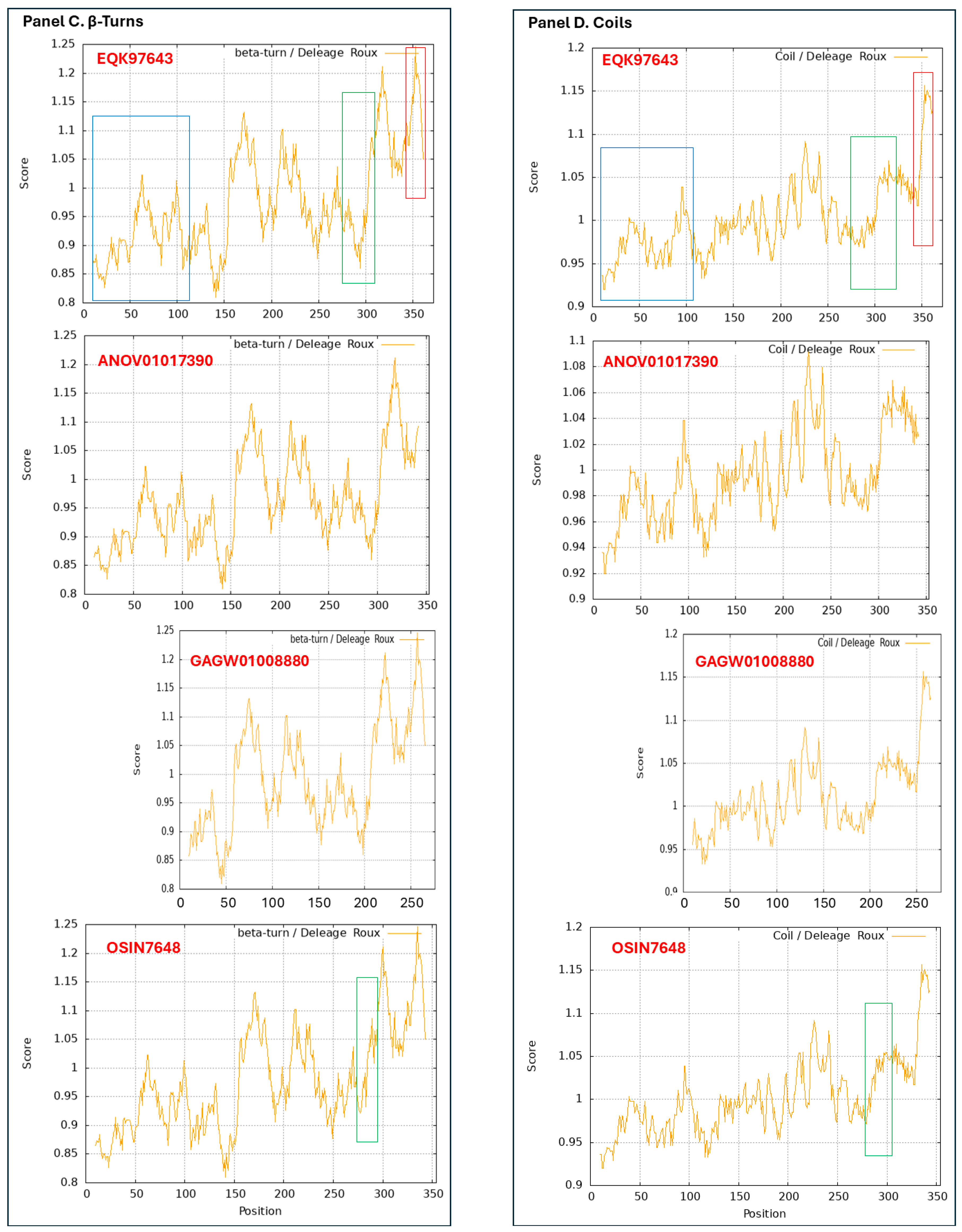
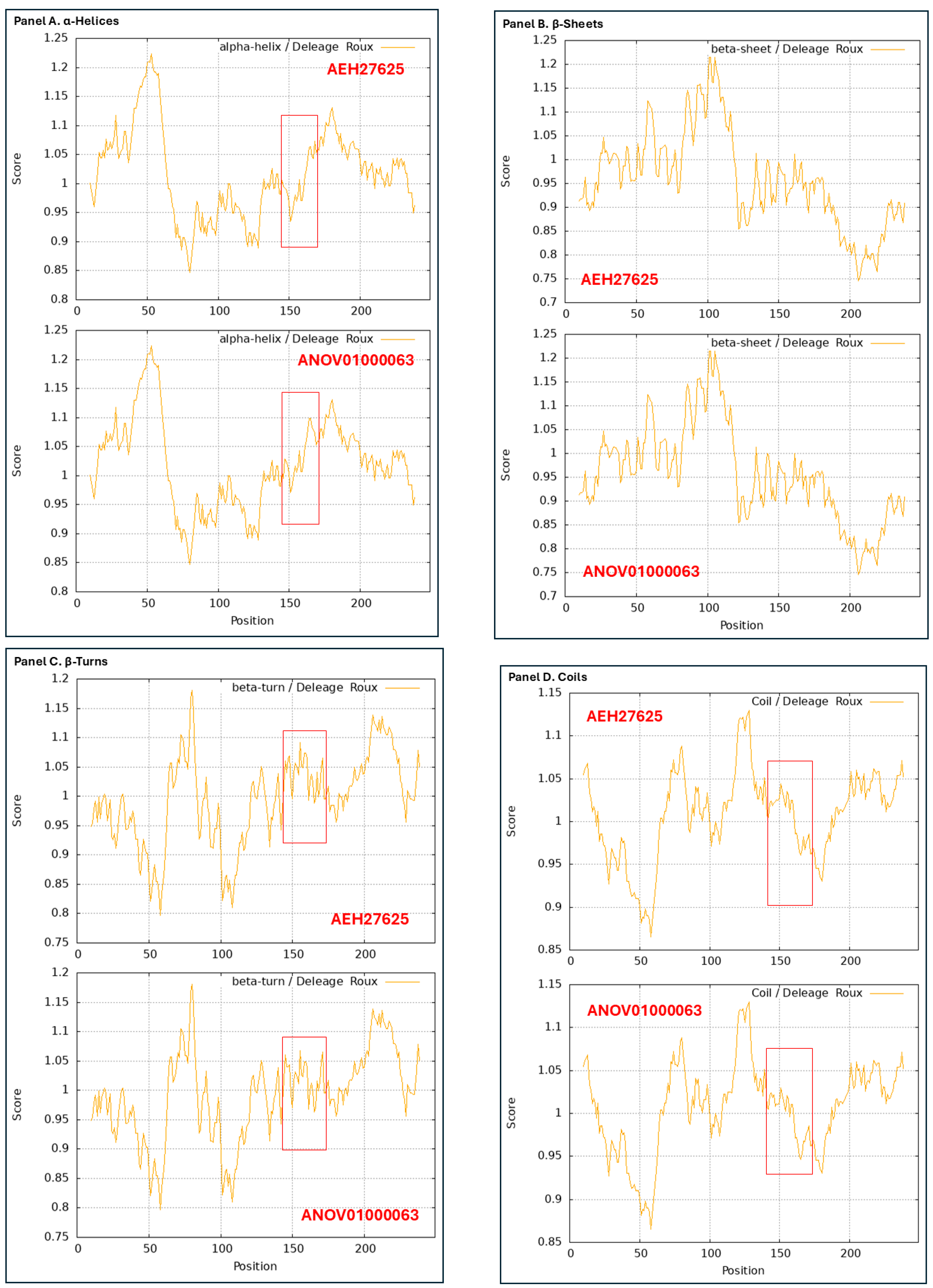
3.9. Diverse Secondary (2D) Structures of the MAT1-1-1 Proteins Encoded by the Genome of H. sinensis and the Metatranscriptome of Natural C. sinensis Insect-Fumgal Complex
3.10. Diverse 2D Structures of the MAT1-2-1 Proteins Encoded by the Genomes, Transcriptomes, and Metatranscriptomes of H. sinensis and the Natural C. sinensis Insect-Fumgal Complex
4. Discussion
4.1. Protein 3D Structure Analysis via the AI-Based AlphaFold Prediction System in Combination with Statistical Bayesian Clustering Technology to Stratify 3D Structure Models
4.2. Heteromorphic 3D Structures of the MAT1-1-1 and MAT1-2-1 Proteins in H. sinensis Strains and Wild-Type C. sinensis Isolates
4.3. Differential Occurrences of MAT1-1-1 and MAT1-2-1 Proteins with Heteromorphic Sereostructures in H. sinensis Strains and C. sinensis Isolates
4.4. Heteromorphic 3D Structures of the Mating Proteins and Sexual Reproductive Behavior of H. sinensis, Genotype #1 of O. sinensis
4.5. Heteromorphic 3D Structures of the Mating Proteins and Sexual Reproduction Strategy During the Lifecycle of the Natural C. sinensis
- (1)
-
Evidence for the differential cooccurrence of multiple genotypes of O. sinensis in the compartments of the natural C. sinensis insect-fungi complex:
- (1-b)
- Differential occurrence of AT-biased Genotype #4 or #5 of O. sinensis without the cooccurrence of GC-biased H. sinensis in natural C. sinensis samples collected from different production areas in geographically remote locations [Engh 1999; Kinjo and Zang 2001; Stensrud et al. 2005, 2007; Mao et al. 2013].
- (1-c)
- Multiple cooccurring GC- and AT-biased genotypes of O. sinensis have been observed differentially in different combinations in the stroma, caterpillar body, ascocarps and ascospores of natural C. sinensis [Xiao et al. 2009; Zhu et al. 2010; Li et al. 2013, 2022, 2023c, 2023d; Mao et al. 2013]. The abundances of the O. sinensis genotypes underwent dynamic alterations in an asynchronous, disproportional manner in the caterpillar bodies and stromata of C. sinensis specimens during maturation, with a consistent predominance of the AT-biased genotypes of O. sinensis, not the GC-biased H. sinensis, in the stromata, indicating that the sequences of O. sinensis genotypes were present in independent genomes of different fungi [Xiao et al. 2009; Zhu et al. 2010; Gao et al. 2011, 2012; Hu et al. 2013; Li et al. 2013, 2016a, 2020, 2022, 2023c; Zhu & Li 2017; Jin et al. 2020; Liu et al. 2020; Shu et al. 2020].
- (1-d)
- The GC-biased Genotypes #1 and #2 of O. sinensis cooccur in the stromata of natural C. sinensis. The abundance of the GC-biased genotypes was dynamically altered during C. sinensis maturation [Zhu et al. 2010; Gao et al. 2012].
- (1-e)
- The cooccurrence of GC-biased genomically independent Genotypes #1 and #7 of O. sinensis was detected in the same specimen of natural C. sinensis [Chen et al. 2011].
- (1-f)
- A species contradiction between the anamorphic inoculants (GC-biased Genotype #1 H. sinensis strains) and the sole-teleomorph of AT-biased Genotype #4 of O. sinensis was detected in the fruiting body of cultivated C. sinensis in a product-oriented industrial setting [Wei et al. 2016].
- (1-g)
- Discovery of Genotypes #13 and #14 of O. sinensis in semi-ejected and fully ejected multicellular heterokaryotic ascospores, respectively, collected from the same C. sinensis samples [Li et al. 2023d].
- (1-h)
- The genetic heterogeneity of ascospores and SFP, the reproductive cells and organs of natural C. sinensis, involves multiple GC- and AT-biased O. sinensis genotypes in different combinations [Li et al. 2013, 2022, 2023c, 2023d].
- (2)
-
Evidence for the differential cooccurrence of heterospecific fungal species in different compartments of the natural C. sinensis insect-fungi complex:
- (2-a)
- Mycobiota findings for differential cooccurrence of >90 fungal species of at least 37 fungal genera in the caterpillar bodies and stromata of natural C. sinensis [Zhang et al. 2010, 2018; Xia et al. 2015; Guo et al. 2017; Zhong et al. 2018; Kang et al. 2024].
- (2-b)
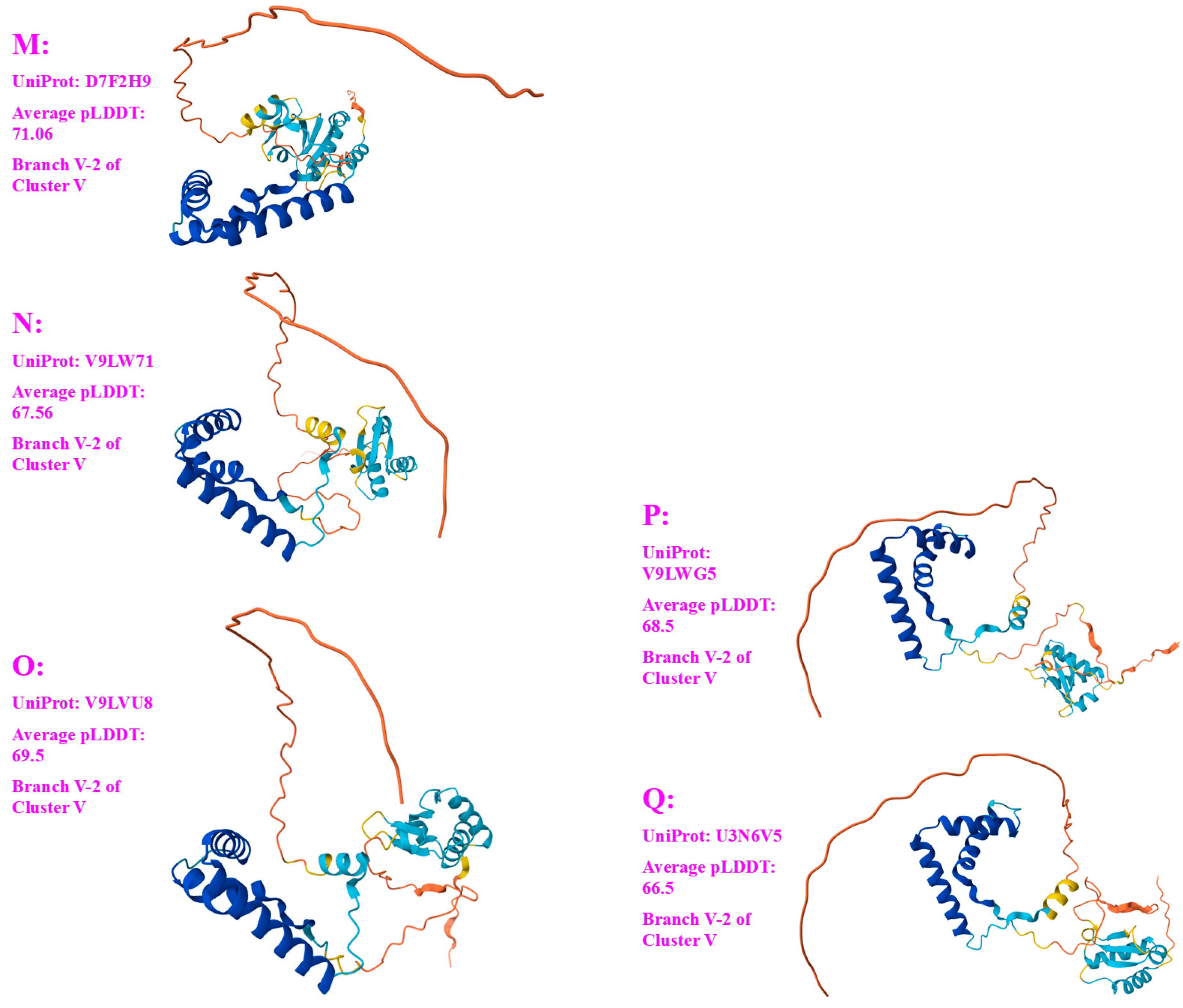
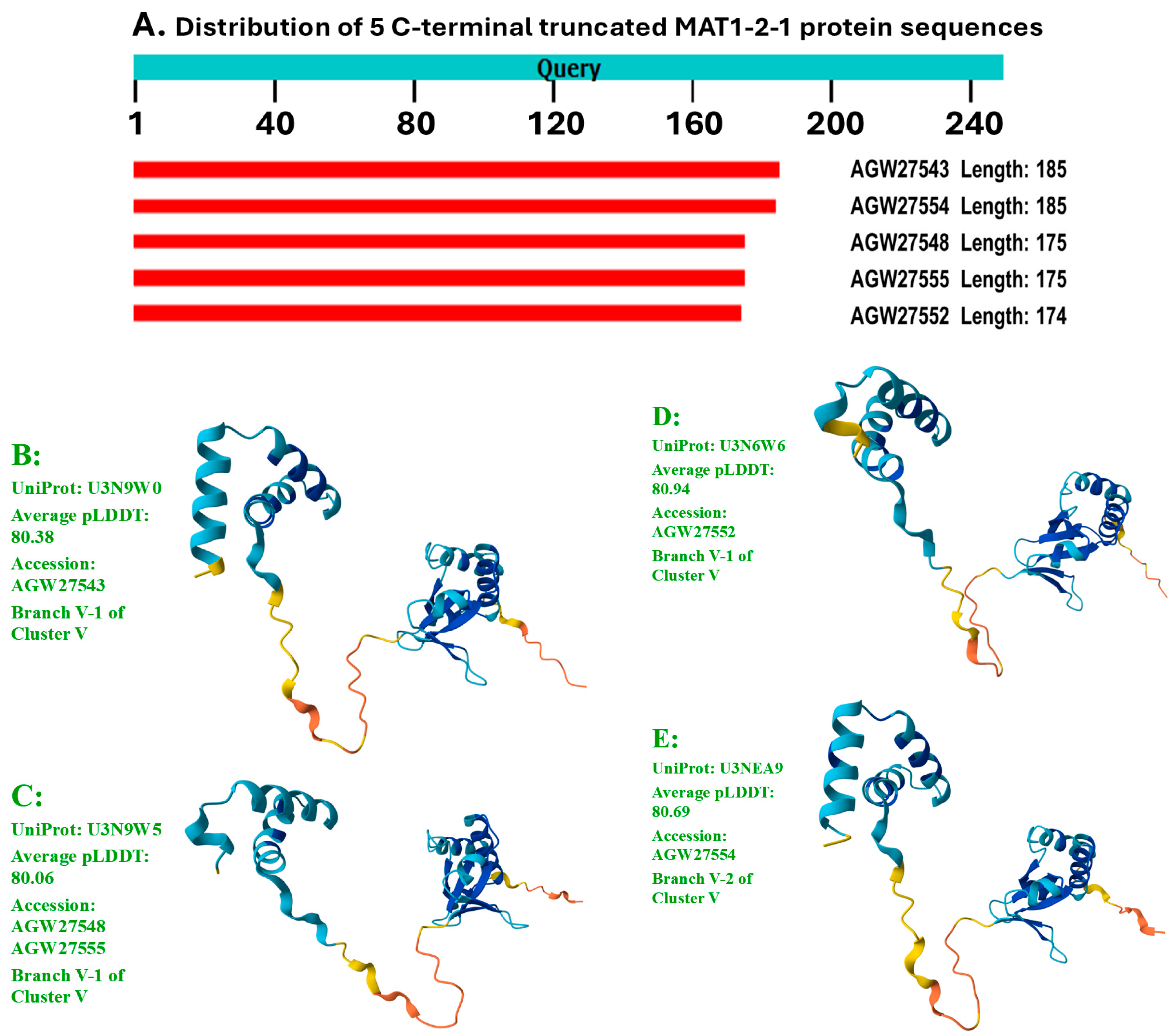
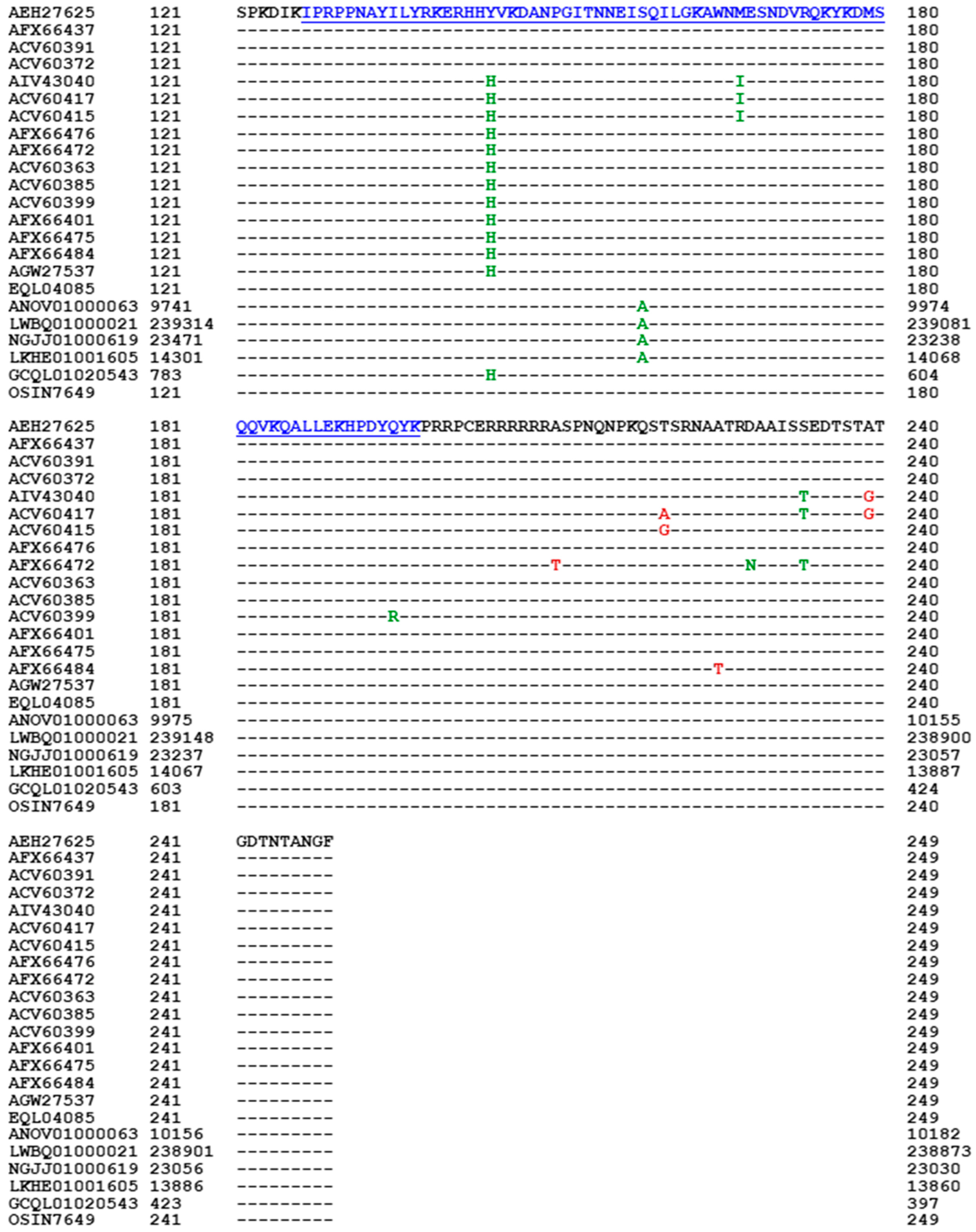
- A good number of C. sinensis isolates contained mutant MAT1-1-1 and MAT1-2-1 proteins, especially those proteins with C- and/or N-terminal truncations that belong to 9 and 4 diverse 3D structural morphs (cf. Figure 4 and Figure 6), respectively. The mutant proteins were either clustered into a separate Bayesian clade or clustered within the main clustering branches in the Bayesian trees (cf. Figure 1 and Figure 2). The MAT1-1-1 and MAT1-2-1 proteins encoded by metatranscriptome assemblies of natural C. sinensis also exhibited either large-segment truncation or sequence variations similar to those observed in wild-type C. sinensis isolates (cf. Figure 7, Figure 8, Figure 9 and Figure 10). Some of the mutant proteins might be produced by heterospecific fungi in impure wild-type C. sinensis isolates and in natural C. sinensis insect-fungi complexes.
- (2-c)
- Discoveries of the formation of the heterospecific Cordyceps-Tolypocladium complex in natural C. sinensis [Engh 1999; Stensrud et al. 2005, 2007] and the dual anamorphs of O. sinensis, involving psychrophilic H. sinensis and mesophilic Tolypocladium sinensis [Li 1988; Chen et al. 2004; Leung et al. 2006; Barseghyan et al. 2011].
- (2-d)
- A close association of psychrophilic H. sinensis and mesophilic S. hepiali (≡ P. hepiali) was found in the caterpillar body, stroma, ascospores, and stromal fertile portion, which was densely covered with numerous ascocarps of natural C. sinensis, and in the wild-type C. sinensis complexes, which appeared to be difficult to purify [Dai et al. 1989; Jiang & Yao 2003; Chen et al. 2004; Zhu et al. 2007, 2010; Yang et al. 2008; Li et al. 2016b, 2023d; Zhu & Li 2017].
- (2-e)
- Although Genotypes #13−14 are among the 17 genotypes of O. sinensis, these 2 GC-biased genotypes feature precise reciprocal cross substitutions of large DNA segments among 2 heterospecific parental fungi, namely, H. sinensis and an AB067719-type fungus. The taxonomic position of the AB067719-type fungus is undetermined to date, and more than 900 heterospecific fungal sequences, which are highly homologous to AB067719, have been uploaded to GenBank [Li et al. 2023d]. Chromosomal intertwining and genetic material recombination may occur after plasmogamy and karyogamy of the heterospecific parental fungi under sexual reproduction hybridization or parasexuality, which is characterized by the prevalence of heterokaryosis and results in concerted chromosome loss for transferring-substituting genetic materials without conventional meiosis [Bennett & Johnson 2003; Sherwood & Bennett 2009; Bushley et al. 2013; Seervai et al. 2013; Nakamura et al. 2019; Mishra et al. 2021; Kück et al. 2022; Li et al. 2023d].
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O'Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Žemgulytė A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Žídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024; 630(8016): 493-500. [CrossRef]
- Barseghyan GS, Holliday JC, Price TC, Madison LM, Wasser SP. Growth and cultural-morphological characteristics of vegetative mycelia of medicinal caterpillar fungus Ophiocordyceps sinensis G.H. Sung et al. (Ascomycetes) Isolates from Tibetan Plateau (P. R. China). Intl. J. Med. Mushrooms. 2011; 13(6): 565−581. [CrossRef]
- Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003; 22(10): 2505−2515. [CrossRef]
- Bushley KE, Li Y, Wang W-J, Wang X-L, Jiao L, Spatafora JW, Yao Y-J. Isolation of the MAT1-1 mating type idiomorph and evidence for selfing in the Chinese medicinal fungus Ophiocordyceps sinensis. Fungal Biol. 2013; 117(9): 599−610. [CrossRef]
- Chen C-S, Hseu R-S, Huang C-T. Quality control of Cordyceps sinensis teleomorph, anamorph, and Its products. Chapter 12, in (Shoyama, Y., Ed.) Quality Control of Herbal Medicines and Related Areas. InTech, Rijeka, Croatia (2011). pp. 223–238. www.intechopen.com.
- Chen Y-Q, Hu B, Xu F, Zhang W, Zhou H, Qu L-H. Genetic variation of Cordyceps sinensis, a fruit-body-producing entomopathogenic species from different geographical regions in China. FEMS Microbiol Lett. 2004; 230: 153–158. [CrossRef]
- China Ministry of Agriculture and Rural Affairs. Announcement (No. 15 of 2021) of National Forestry and Grassland Administration: List of National Key Protected Wild Plants. September 7, 2021.
- Dai R-Q, Lan J-L, Chen W-H, Li X-M, Chen Q-T, Shen C-Y. Discovery of a new fungus Paecilomyces hepiali Chen & Dai. Acta Agricult. Univ. Pekin. 1989; 15(2): 221−224.
- David A, Islam S, Tankhilevich E, Sternberg MJE. The AlphaFold Database of Protein Structures: A Biologist’s Guide. J Mol Biol. 2022; 434(2): 167336. [CrossRef]
- Debuchy R, Turgeo BG. Mating-Type Structure, Evolution, and Function in Euascomycetes. In (eds. Kües U. & Fischer, R.) Growth, Differentiation and Sexuality. Springer (2006). pp. 293–323.
- Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Engineering, Design and Selection 1987; 1: 289–294. [CrossRef]
- Du X-H, Wu D-M, Kang H, Wang H-C, Xu N, Li T-T, Chen K-L. Heterothallism and potential hybridization events inferred for twenty-two yellow morel species. IMA Fungus. 2020; 11: 4. [CrossRef]
- Engh IB. Molecular phylogeny of the Cordyceps-Tolypocladium complex. Candidate scientific thesis, Department of Biology, University of Oslo, 1999.
- Gao L, Li X-H, Zhao J-Q, Lu J-H, Zhao J-G, Zhu J-S. Maturation of Cordyceps sinensis associates with alterations of fungal expressions of multiple Ophiocordyceps sinensis mutants with transition and transversion point mutations in stroma of Cordyceps sinensis. Beijing Da Xue Xue Bao. 2012; 44(3): 454–463. [CrossRef]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. Chapter 52 (In) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005). pp. 571–607.
- Guo M-Y, Liu Y, Gao Y-H, Jin T, Zhang H-B, Zhou X-W. Identification and bioactive potential of endogenetic fungi isolated from medicinal caterpillar fungus Ophiocordyceps sinensis from Tibetan Plateau. Int J Agric Biol. 2017; 19: 307‒313. [CrossRef]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, and 82 other authors. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011; 2: 105−112. [CrossRef]
- Hėnault M, Marsit S, Charron G, Landry CR. The effect of hybridization on transposable element accumulation in an undomesticated fungal species. eLife. 2020; 9: e60474. [CrossRef]
- Holliday J, Cleaver M. Medicinal value of the caterpillar fungi species of the genus Cordyceps (Fr.) Link (Ascomycetes). A review. Int. J. Med. Mushrooms. 2008; 10: 219–234. [CrossRef]
- Hu X, Zhang Y-J, Xiao G-H, Zheng P, Xia Y-L, Zhang X-Y, St Leger RJ, Liu X-Z, Wang C-S. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 2013; 58: 2846–2854. [CrossRef]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformat. 2001; 17: 754−755. [CrossRef]
- Jacobsen S, Wittig M, Pöggeler S. Interaction Between Mating-Type Proteins From the Homothallic Fungus Sordaria macrospora. Curr. Genet. 2002; 41: 150–158. [CrossRef]
- Jiang Y, Yao Y-J. A review for the debating studies on the anamorph of Cordyceps sinensis. Mycosistema. 2003; 22(1): 161–176.
- Jin L-Q, Xu Z-W, Zhang B, Yi M, Weng C-Y, Lin S, Wu H, Qin X-T, Xu F, Teng Y, Yuan S-J, Liu Z-Q, Zheng Y-G. Genome sequencing and analysis of fungus Hirsutella sinensis isolated from Ophiocordyceps sinensis. AMB Expr. 2020; 10: 105. [CrossRef]
- Jones SK, Bennett RJ. Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 2011; 48(7): 668–676. [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature 2021; 596: 583–589. [CrossRef]
- Kang Q, Zhang J, Chen F, Dong C, Qin Q, Li X, Wang H, Zhang H and Meng Q. Unveiling mycoviral diversity in Ophiocordyceps sinensis through transcriptome analyses. Front. Microbiol. 2024; 15: 1493365. [CrossRef]
- Kinjo N, Zang M. Morphological and phylogenetic studies on Cordyceps sinensis distributed in southwestern China. Mycoscience. 2001; 42: 567–574. [CrossRef]
- Kück U, Bennett RJ, Wang L and Dyer PS. Editorial: Sexual and Parasexual Reproduction of Human Fungal Pathogens. Front. Cell. Infect. Microbiol. 2022; 12: 934267. [CrossRef]
- Leung P-H, Zhang Q-X, Wu J-Y. Mycelium cultivation, chemical composition and antitumour activity of a Tolypocladium sp. fungus isolated from wild Cordyceps sinensis. J. Appl. Microbiol. 2006; 101: 275−283. [CrossRef]
- Li C-L. A study of Tolypocladium sinense C.L. Li. sp. nov. and cyclosporin production. Acta Mycol. Sinica. 1988; 7: 93−98.
- Li M-M, Zhang J-H, Qin Q-L, Zhang H, Li X, Wang H-T, Meng Q. Transcriptome and Metabolome Analyses of Thitarodes xiaojinensis in Response to Ophiocordyceps sinensis Infection. Microorganisms 2023a; 11: 2361. [CrossRef]
- Li X, Wang F, Liu Q, Li Q-P, Qian Z-M, Zhang X-L, Li K, Li W-J, Dong C-H. Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genomics. 2019; 20: 337. [CrossRef]
- Li X-Z, Li Y-L, Wang Y-N, Zhu J-S. Translations of mutant repetitive genomic sequences in Hirsutella sinensis and changes in secondary structures and functional specifications of the encoded proteins. International J Mol Sci 2024a; 25(20), 11178. [CrossRef]
- Li X-Z, Li Y-L, Yao Y-S, Xie W-D, Zhu J-S. Further discussion with Li et al. (2013, 2019) regarding the “ITS pseudogene hypothesis” for Ophiocordyceps sinensis. Mol. Phylogenet. Evol. 2020; 146: 106728. [CrossRef]
- Li X-Z, Li Y-L, Zhu J-S. Differential transcription of mating-type genes during sexual reproduction of natural Cordyceps sinensis. Chin. J. Chin. Materia Medica. 2023b; 48(10): 2829–2840. [CrossRef]
- Li X-Z, Xiao M-J, Li Y-L, Gao L, Zhu J-S. Mutations and differential transcription of mating-type and pheromone receptor genes in Hirsutella sinensis and the natural Cordyceps sinensis insect‒fungi complex. Biol. (Basel) 2024b; 13(8): 632. [CrossRef]
- Li Y, Hsiang T, Yang R-H, Hu X-D, Wang K, Wang W-J, Wang X-L, Jiao L, Yao Y-J. Comparison of different sequencing and assembly strategies for a repeat-rich fungal genome, Ophiocordyceps sinensis. J. Microbiol. Methods. 2016a; 128: 1−6. [CrossRef]
- Li Y, Jiao L, Yao Y-J. Non-concerted ITS evolution in fungi, as revealed from the important medicinal fungus Ophiocordyceps sinensis. Mol. Phylogenet. Evol. 2013; 68: 373−379. [CrossRef]
- Li Y-L, Gao L, Yao Y-S, Wu Z-M, Lou Z-Q, Xie W-D, Wu J-Y, Zhu J-S. Altered GC- and AT-biased genotypes of Ophiocordyceps sinensis in the stromal fertile portions and ascospores of natural Cordyceps sinensis. PLoS ONE. 2023c; 18(6): e0286865. [CrossRef]
- Li Y-L, Li X-Z, Yao Y-S, Wu Z-M, Gao L, Tan N-Z, Lou Z-Q, Xie W-D, Wu J-Y, Zhu J-S. Differential cooccurrence of multiple genotypes of Ophiocordyceps sinensis in the stromata, stromal fertile portion (ascocarps) and ascospores of natural Cordyceps sinensis. PLoS ONE. 2023d; 18(3): e0270776. [CrossRef]
- Li Y-L, Li X-Z, Yao Y-S, Xie W-D, Zhu J-S. Molecular identification of Ophiocordyceps sinensis genotypes and the indiscriminate use of the Latin name for the multiple genotypes and the natural insect-fungi complex. Am J BioMed Sci. 2022b; 14(3): 115–135. [CrossRef]
- Li Y-L, Yao Y-S, Zhang Z-H, Xu H-F, Liu X, Ma S-L, Wu Z-M, Zhu J-S. Synergy of fungal complexes isolated from the intestines of Hepialus lagii larvae in increasing infection potency. J. Fungal Res. 2016b; 14: 96−112.
- Liu J, Guo L-N, Li Z-W, Zhou Z, Li Z, Li Q, Bo X-C, Wang S-Q, Wang J-L, Ma S-C, Zheng J, Yang Y. Genomic analyses reveal evolutionary and geologic context for the plateau fungus Ophiocordyceps sinensis. Clin. Med. 2020; 15: 107‒119. [CrossRef]
- Liu Z-Q, Lin S, Baker PJ, Wu L-F, Wang X-R, Wu H, Xu F, Wang H-Y, Brathwaite ME, Zheng Y-G. Transcriptome sequencing and analysis of the entomopathogenic fungus Hirsutella sinensis isolated from Ophiocordyceps sinensis. BMC Genomics. 2015; 16: 106‒123. [CrossRef]
- Lu H-L, St Leger RJ. Chapter Seven - Insect Immunity to Entomopathogenic Fungi, Editors: Lovett B, St. Leger RJ. Advanc Genet, Academic Press, 2016. Vol. 94; pp. 251–85.
- Mariani V, Biasini M, Barbato A, Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013; 29(21): 2722‒2728. [CrossRef]
- Mao X-M, Zhao S-M, Cao L, Yan X, Han R-C. The morphology observation of Ophiocordyceps sinensis from different origins. J. Environ. Entomol. 2013; 35(3): 343‒353.
- Meng Q, Yu H-Y, Zhang H, Zhu W, Wang M-L, Zhang J-H, Zhou G-L, Li X, Qin Q-L, Hu S-N, Zou Z. Transcriptomic insight into the immune defenses in the ghost moth, Hepialus xiaojinensis, during an Ophiocordyceps sinensis fungal infection. Insect Biochem. Mol. Biol. 2015; 64: 1−15. [CrossRef]
- Mishra A, Forche A and Anderson MZ. Parasexuality of Candida Species. Front. Cell. Infect. Microbiol. 2021; 11: 796929. [CrossRef]
- Monzon V, Haft DH, Bateman A. Folding the unfoldable: using AlphaFold to explore spurious proteins. Bioinformatics Advances, 2022; 1(2): vbab043. [CrossRef]
- Nakamura N, Tanaka C, Takeuchi-Kaneko Y. Transmission of antibiotic-resistance markers by hyphal fusion suggests partial presence of parasexuality in the root endophytic fungus Glutinomyces brunneus. Mycol. Progress 2019; 18: 453–462. [CrossRef]
- Pfennig KS. Facultative Mate Choice Drives Adaptive Hybridization. Sci. 2007; 318: 965–967. [CrossRef]
- Peters C, Elofsson A. Why is the biological hydrophobicity scale more accurate than earlier experimental hydrophobicity scales? Proteins. 2014; 82(9): 2190–2198. [CrossRef]
- Rams B, Kück U. The Penicillium chrysogenum tom1 gene a major target of transcription factor MAT1-1-1 encodes a nuclear protein involved in sporulation. Front. Fungal Bio. 2022; 3: 937023. [CrossRef]
- Ren Y, Wan D-G, Lu X-M, Guo J-L. The study of scientific name discussion for TCM Cordyceps. LisShenzhen Med. Material Medica Res. 2013; 24(9): 2211−2212.
- Rettie SA, Campbell KV, Bera AK, Kang A, Kozlov S, De La Cruz J, Adebomi V, Zhou G, DiMaio F, Ovchinnikov S, Bhardwaj G. Cyclic peptide structure prediction and design using AlphaFold. bioRxiv [Preprint]. 2023 Feb 26:2023.02.25.529956. [CrossRef]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space, Systematic Biology, 2012; 61(3): 539–542. [CrossRef]
- Samarasinghe H, You M, Jenkinson TS, Xu J-P, James TY. Hybridization Facilitates Adaptive Evolution in Two Major Fungal Pathogens. Genes. 2020; 11: 101. [CrossRef]
- Seervai RNH, Jones SK, Hirakawa MP, Porman AM, Bennett RJ. Parasexuality and ploidy change in Candida tropicalis. Eukaryotic Cell. 2013; 12(12): 1629–1640. [CrossRef]
- Sherwood RK, Bennett RJ. Fungal meiosis and parasexual reproduction--lessons from pathogenic yeast. Current Opinion Microbiol. 2009; 12(6): 599–607. [CrossRef]
- Shu R-H, Zhang J-H, Meng Q, Zhang H, Zhou G-L, Li M-M, Wu P-P, Zhao Y-N, Chen C, Qin Q-L. A new high-quality draft genome assembly of the Chinese cordyceps Ophiocordyceps sinensis. Genome Biol. Evol. 2020; 12(7): 1074−1079. [CrossRef]
- Simm S, Einloft J, Mirus O, Schleiff E. 50 years of amino acid hydrophobicity scales: revisiting the capacity for peptide classification. Biol. Res. 2016; 49(1): 31. [CrossRef]
- Steensels J, Gallone B, Verstrepen KJ. Interspecific hybridization as a driver of fungal evolution and Adaptation. Nat. Rev. Microbiol. 2021; 19: 485–500.
- Stensrud Ø, Hywel-Jones NL, Schumacher T. Towards a phylogenetic classification of Cordyceps: ITS nrDNA sequence data confirm divergent lineages and paraphyly. Mycol. Res. 2005; 109: 41−56. [CrossRef]
- Stensrud Ø, Schumacher T, Shalchian-Tabrizi K, Svegardenib IB, Kauserud H. Accelerated nrDNA evolution and profound AT bias in the medicinal fungus Cordyceps sinensis. Mycol. Res. 2007; 111: 409–415. [CrossRef]
- Stone, R. Stone R. Improbable partners aim to bring biotechnology to a Himalayan kingdom. Sci. 2010; 327: 940–941. [CrossRef]
- Sung G-H, Hywel-Jones NL, Sung J-M, Luangsa-ard JJ, Shrestha B, Spatafora JW. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007; 57: 5−59. [CrossRef]
- Tunyasuvunakool, K., Adler, J., Wu, Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, Kleywegt GJ, Bateman A, Evans R, Pritzel A, Figurnov M, Ronneberger O, Bates R, Kohl SAA, Potapenko A, Ballard AJ, Romera-Paredes B, Nikolov S, Jain R, Clancy E, Reiman D, Petersen S, Senior AW, Kavukcuoglu K, Birney E, Kohli P, Jumper J, Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature 2021; 596: 590–596. [CrossRef]
- Turgeon BG, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 2000; 31: 1‒5. [CrossRef]
- Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, Tsenkov M, Nair S, Mirdita M, Yeo J, Kovalevskiy O, Tunyasuvunakool K, Laydon A, Žídek A, Tomlinson H, Hariharan D, Abrahamson J, Green T, Jumper J, Birney E, Steinegger M, Hassabis D, Velankar S. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024; 52(D1): D368–D375. [CrossRef]
- Wang Y, Stata M, Wang W, Stajich JE, White MM, Moncalvo JM. Comparative genomics reveals the core gene toolbox for the fungus-insect symbiosis. mBio. 2018; 9: e00636-18. [CrossRef]
- Wang Y-B, Wang Y, Fan Q, Duan D-E, Zhang G-D,· Dai R-Q, Dai Y-D, Zeng W-B, Chen Z-H, Li D-D, Tang D-X, Xu Z-H, Sun T, Nguyen T-T, Tran N-L, Dao V-M, Zhang C-M, Huang L-D, Liu Y-J, Zhang X-M, Yang D-R, Sanjuan T, Liu X-Z, Yang Z-L, Yu H. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers 2020; 103: 1. [CrossRef]
- Wei J-C, Wei X-L, Zheng W-F, Guo W, Liu R-D. Species identification and component detection of Ophiocordyceps sinensis cultivated by modern industry. Mycosystema. 2016; 35(4): 404‒410.
- Wei X-L, Yin X-C, Guo Y-L, Shen N-Y, Wei, J.-C. Analyses of molecular systematics on Cordyceps sinensis and its related taxa. Mycosystema. 2006; 25(2): 192–202.
- Wilson AM, Wilken PM, van der Nest MA, Steenkamp ET, Wingfield MJ, Wingfield BD. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus. 2015; 6(1): 207–214. [CrossRef]
- Wroblewski K, Kmiecik S. Integrating AlphaFold pLDDT Scores into CABS-flex for enhanced protein flexibility simulations. Comput Struct Biotechnol J. 2024; 30(23): 4350–4356. [CrossRef]
- Xia E-H, Yang D-R, Jiang J-J, Zhang Q-J, Liu Y, Liu Y-L, Zhang Y, Zhang H-B, Shi C, Tong Y, Kim C-H, Chen H, Peng Y-Q, Yu Y, Zhang W, Eichler EE, Gao L-Z. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 2017; 7: 1806. [CrossRef]
- Xia F, Liu Y, Shen G-L, Guo L-X, Zhou X-W. Investigation and analysis of microbiological communities in natural Ophiocordyceps sinensis. Can. J. Microbiol. 2015; 61: 104‒111. [CrossRef]
- Xiang L, Li Y, Zhu Y, Luo H, Li C, Xu X, Sun C, Song J-Y, Shi L-H, He L, Sun W, Chen S-L. Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics. 2014; 103: 154−159. [CrossRef]
- Xiao W, Yang J-P, Zhu P, Cheng K-D, He H-X, Zhu H-X, Wang Q. Non-support of species complex hypothesis of Cordyceps sinensis by targeted rDNA-ITS sequence analysis. Mycosystema. 2009; 28(6): 724–730.
- Xu T, Xu Q, Li J-Y. Toward the appropriate interpretation of Alphafold2. Front Artif Intell. 2023; 6: 1149748. [CrossRef]
- Yang J-L, Xiao W, He H-X, Zhu H-X, Wang S-F, Cheng -KD, Zhu P. Molecular phylogenetic analysis of Paecilomyces hepiali and Cordyceps sinensis. Acta Pharmaceut. Sinica 2008; 43(4): 421‒426.
- Yang J-Y, Tong X-X, He C-Y, Bai J, Wang F, Guo J-L. Comparison of endogenetic microbial community diversity between wild Cordyceps sinensis, artificial C. sinensis and habitat soil. Chin. J. Chin. Materia Medica. 2021; 46(12): 3106‒3115.
- Yao Y-S, Zhu J-S. Indiscriminate use of the Latin name for natural Cordyceps sinensis and Ophiocordyceps sinensis fungi. Chin. J. Chin. Mater. Med. 2016; 41(7): 1316–1366.
- Zhang S; Zhang Y-J. Molecular evolution of three protein-coding genes in the Chinese caterpillar fungus Ophiocordyceps sinensis. Microbiol. China. 2015; 42(8): 1549−1560.
- Zhang S, Zhang Y-J, Liu X-Z, Wen H-A, Wang M, Liu D-S. Cloning and analysis of the MAT1-2-1 gene from the traditional Chinese medicinal fungus Ophiocordyceps sinensis. Fungal Biol. 2011; 115: 708−714.
- Zhang S, Zhang Y-J, Shrestha B, Xu J-P, Wang C-S, Liu X-Z. Ophiocordyceps sinensis and Cordyceps militaris: research advances, issues and perspectives. Mycosystema. 2013; 32: 577−597.
- Zhang S-W, Cen K, Liu Y, Zhou X-W, Wang C-S. Metatranscriptomics analysis of the fruiting caterpillar fungus collected from the Qinghai-Tibetan plateau. Sci. Sinica Vitae. 2018; 48(5): 562−570.
- Zhang Y-J, Li E-W, Wang C-S, Li Y-L, Liu X-Z. Ophiocordyceps sinensis, the flagship fungus of China: terminology, life strategy and ecology. Mycol. 2012; 3(1): 2–10.
- Zhang Y-J, Sun B-D, Zhang S, Wàngmŭ, Liu X-Z, Gong W-F. Mycobiotal investigation of natural Ophiocordyceps sinensis based on culture-dependent investigation. Mycosistema. 2010; 29(4): 518–527.
- Zhang Y-J, Xu L-L, Zhang S, Liu X-Z, An Z-Q, Wàngmŭ, Guo Y-L. Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: implications for its evolution and conservation. BMC Evol. Biol. 2009; 9: 290. [CrossRef]
- Zhang Y-J, Zhang S, Li Y-L, Ma S-L, Wang C-S, Xiang M-C, Liu X, An Z-Q, Xu J-P, Liu X-Z. Phylogeography and evolution of a fungal–insect association on the Tibetan Plateau. Mol. Ecol. 2014; 23: 5337−5355. [CrossRef]
- Zhao Y-N, Zhang J-H, Meng Q, Zhang H, Zhou G-L, Li M-M, Wu P-P, Shu R-H, Gao X-X, Guo L, Tong Y, Cheng L-Q, Guo L, Chen C, Qin Q. Transcriptomic analysis of the orchestrated molecular mechanisms underlying fruiting body initiation in Chinese cordyceps. Gene. 2020; 763: 145061. [CrossRef]
- Zheng, P.; Wang, C.-S. Sexuality Control and Sex Evolution in Fungi. Sci. Sin. Vitae 2013, 43, 1090–1097.
- Zhong X, Gu L, Wang H-Z, Lian D-H, Zheng Y-M, Zhou S, Zhou W, Gu J, Zhang G, Liu X. Profile of Ophiocordyceps sinensis transcriptome and differentially expressed genes in three different mycelia, sclerotium and fruiting body developmental stages. Fungal Biol. 2018; 122: 943‒951. [CrossRef]
- Zhu J-S, Gao L, Li X-H, Yao Y-S, Zhou Y-J, Zhao J-Q, Zhou Y-J. Maturational alterations of oppositely orientated rDNA and differential proliferations of CG:AT-biased genotypes of Cordyceps sinensis fungi and Paecilomyces hepiali in natural C. sinensis. Am. J. Biomed. Sci. 2010; 2(3): 217–238. [CrossRef]
- Zhu J-S, Guo Y-L, Yao Y-S, Zhou Y-J, Lu J-H, Qi Y, Chen W, Zheng T-Y, Zhang L, Wu Z-M, Zhang L-J, Liu X-J, Yin W-T. Maturation of Cordyceps sinensis associates with co-existence of Hirsutella sinensis and Paecilomyces hepiali DNA and dynamic changes in fungal competitive proliferation predominance and chemical profiles. J. Fungal Res. 2007; 5(4): 214–224.
- Zhu J-S, Gray GM. Renaturative catalytic blotting of enzyme proteins. Chapter 17. in Dunbar BS (ed.), Protein blotting: A practical approach (IRL series). Oxford University Press, Oxford, 1994, pp. 221–238. [CrossRef]
- Zhu J-S, Halpern GM, Jones K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis: Part I. J. Altern. Complem. Med. 1998a; 4(3): 289–303. [CrossRef]
- Zhu J-S, Halpern GM, Jones K. The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis: Part II. J. Altern. Complem. Med. 1998b; 4(4): 429–457. [CrossRef]
- Zhu J-S, Li C-L, Tan N-Z, Berger JL, Prolla TA. Combined use of whole-gene expression profiling technology and mouse lifespan test in anti-aging herbal product study. Proc. 2011 New TCM Products Innovation and Industrial Development Summit, Hangzhou, China (Nov 27, 2011). pp. 443–448. https://xueshu.baidu.com/usercenter/paper/show?paperid=08341c17fa58c8f85584b92572b90f75&site=xueshu_se.
- Zhu J-S, Li Y-L. A Precious Transitional Chinese Medicine, Cordyceps sinensis: Multiple heterogeneous Ophiocordyceps sinensis in the insect-fungi complex. Lambert Academic Publishing, Saarbrüchen. Germany, 2017.
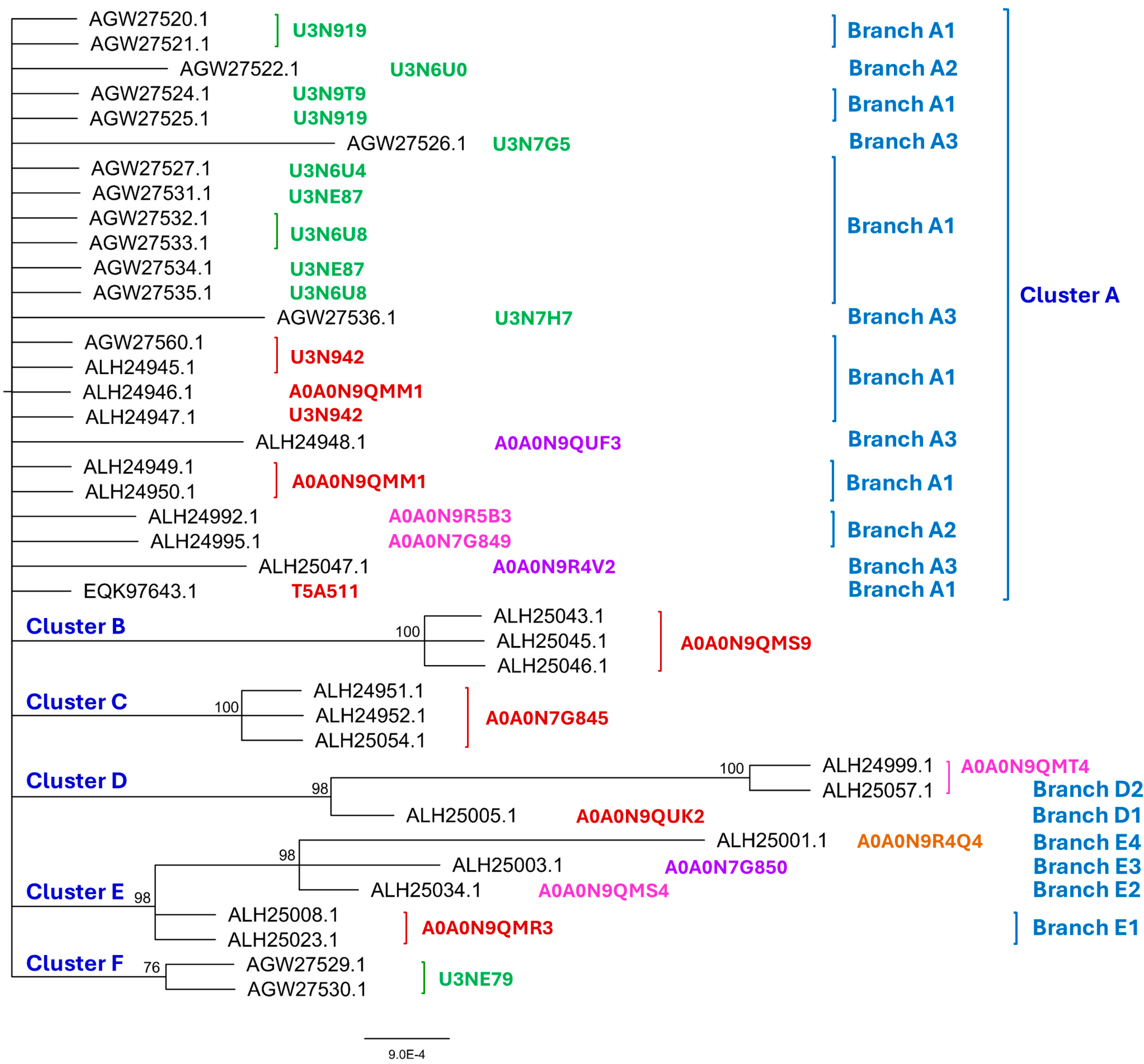

| AlphaFold UniProt code (Bayesian cluster/branch*) | Strain/isolate number (GenBank accession number) |
| U3N942 (A1) | CS68-2-1229 (AGW27560)(AGW27528),GS09_111 (ALH24945),GS09_131 (ALH24947), ID10_1 (ALH24954), IOZ07 (KAF4512729), NP10_1 (ALH24955), NP10_2 (ALH24956), QH07_188 (ALH24957), QH07_197 (ALH24958), QH09_37 (ALH24968), QH09_46 (ALH24969), QH09_56 (ALH24970), QH09_66 (ALH24971), QH09_78 (ALH24972), QH09_93 (ALH24973), QH09_122 (ALH24959), QH09_131 (ALH24960), QH09_151 (ALH24961), QH09_20L (ALH24965), QH09_33L (ALH24967), QH10_1 (ALH24974), QH10_4 (ALH24975), QH10_7 (ALH24976), SC09_21 (ALH24987), SC09_36 (ALH24988), SC09_37 (ALH24989), SC09_47 (ALH24990), SC09_57 (ALH24991), SC09_77 (ALH24993), SC09_107 (ALH24978), SC09_117 (ALH24979), SC09_128 (ALH24980), SC09_147 (ALH24981), SC09_157 (ALH24982), SC09_167 (ALH24983), SC09_180 (ALH24984), SC09_190 (ALH24985), SC09_200 (ALH24986), SC10_18 (ALH24996), SC10_21 (ALH24997), SC10_4 (ALH24998), XZ05_3 (ALH25002), XZ05_7 (ALH25004), XZ05_12 (ALH25000), XZ06_124 (ALH25006), XZ06_152 (ALH25007), XZ07_108 (ALH25009), XZ07_133 (ALH25010), XZ07_154 (ALH25011), XZ07_166 (ALH25012), XZ07_176 (ALH25013), XZ07_180 (ALH25014), XZ08_4 (ALH25018), XZ08_10 (ALH25015), XZ08_24 (ALH25016), XZ08_26 (ALH25017), XZ08_56 (ALH25019), XZ08_59 (ALH25020), XZ08_A1 (ALH25021), XZ08_B1 (ALH25022), XZ09_4 (ALH25029), XZ09_46 (ALH25030), XZ09_48 (ALH25031), XZ09_59 (ALH25032), XZ09_71 (ALH25033), XZ09_80 (ALH25055), XZ09_106 (ALH25024), XZ09_113 (ALH25025), XZ09_118 (ALH25026), XZ09_15 (ALH25027), XZ09_32 (ALH25028), XZ10_7 (ALH25038), XZ10_15 (ALH25035), XZ10_17 (ALH25036), XZ10_23 (ALH25037), XZ12_1 (ALH25056), XZ12_33 (ALH25058), XZ12_43 (ALH25059), YN07_6 (ALH25039), YN07_8 (ALH25040), YN09_3 (ALH25044), YN09_72 (ALH25049), YN09_81 (ALH25050), YN09_85 (ALH25051), YN09_89 (ALH25052), YN09_96 (ALH25053),YN09_101 (ALH25041), YN09_140 (ALH25042) |
| A0A0N9QMM1 (A1) | GS09_121 (ALH24946), GS09_201 (ALH24949), GS09_225 (ALH24950), SC09_1 (ALH24977) |
| T5A511 (A1) | Co18 (EQK97643) (KE657544 410←1519 and ANOV01017390 410←1519) |
| A0A0N9R5B3 (A2) | SC09_65 (ALH24992) |
| A0A0N7G849 (A2) | SC09_97 (ALH24995) |
| A0A0N9QUF3 (A3) | GS09_143 (ALH24948) |
| A0A0N9R4V2 (A3) | YN09_61 (ALH25047) |
| A0A0N9QMS9 (B) | YN09_6 (ALH25046), YN09_22 (ALH25043), YN09_51 (ALH25045), YN09_64 (ALH25048) |
| A0A0N7G845 (C) | GS09_229 (ALH24951), GS09_281 (ALH24952), GS09_311 (ALH25054), GS10_1 (ALH24953), QH09_164 (ALH24962), QH09_173 (ALH24963), QH09_201 (ALH24964), QH09_210 (ALH24966), SC09_87 (ALH24994) |
| A0A0N9QUK2 (D1) | XZ05_8 (ALH25005) |
| A0A0N9QMT4 (D2) | XZ07_H2 (ALH24999), XZ12_16 (ALH25057) |
| A0A0N9QMR3 (E1) | XZ06_260 (ALH25008), XZ09_100 (ALH25023) |
| A0A0N9QMS4 (E2) | XZ09_95 (ALH25034) |
| A0A0N7G850 (E3) | XZ05_6 (ALH25003) |
| A0A0N9R4Q4 (E4) | XZ05_2 (ALH25001) |
| AlphaFold UniProt code (Bayesian cluster/branch**) | Strain/isolate number (GenBank accession number) |
| D7F2E9 (I-1) | CS2 (AEH27625)(ACV60400), CS26-277 (AGW27541), CS36-1294 (AGW27538), CS37-295 (AGW27539), SC-2 (ACV60395), SC-4 (ACV60396), SC-5 (ACV60398), SC-7 (ACV60397), SC09-37 (AFH35019), SC09_47 (AFX66423), SC09_57 (AFX66424), SC09_77 (AFX66426), SC09_97 (AFX66428), XZ05_7 (AFX66442), XZ05_12 (AFX66444), XZ06_152 (AFX66445),XZ07_11 (AFX66447), XZ07_46 (AFX66448), XZ09_106 (AFX66464), XZ09_15 (AFX66455), YN09_101 (AFX66482), YN09_72 (AFX66477), XZ09_113 (AFX66465), XZ-LZ06-1 (ACV60369), XZ-LZ06-7 (ACV60370), XZ-LZ06-21 (ACV60371), XZ-LZ06-108 (ACV60373), XZ-LZ07-30 (ACV60377), XZ-LZ07-108 (ACV60379), XZ-ML-191 (ACV60376), YN-1 (ACV60390), YN-5 (ACV60392), YN-6 (ACV60393), YN-8 (ACV60394), YN09_81 (AFX66478), YN09_85 (AFX66479), YN09_89 (AFX66480) |
| T5AF56 (I-1) | Co18 (EQL04085) (ANOV01000063 9329→10182) |
| V9LW10 (I-2) | SC09_200 (AFX66437) |
| D7F2H1 (I-2) | YN-4 (ACV60391) |
| D7F2F2 (I-2) | XZ-LZ06-61 (ACV60372) |
| A0A0A0RCF5 (II-1) | XZ12_16 (AIV43040) |
| D7F2J7 (II-2) | XZ05_8 (AFX66443), XZ06-124 (AFH35020), XZ-LZ07-H2 (ACV60418), XZ-LZ07-H1 (ACV60417) |
| D7F2F5 (III) | XZ05_2 (AFX66441), XZ06_260 (AFX66446), XZ09_80 (AFX66461), XZ09_95 (AFX66462), XZ09_100 (AFX66463), XZ-LZ05-6 (ACV60415), XZ-SN-44 (ACV60375), |
| V9LWC9 (IV-1) | YN09_64 (AFX66476) |
| V9LVS8 (IV-2) | YN09_6 (AFX66472), YN09_22 (AFX66473), YN09_51 (AFX66474) |
| D7F2E3 (V-1) | GS09_111 (AFX66388), CS560-961 (AGW275424), QH09-93 (AFH35018), XZ-NQ-154 (ACV60363), XZ-NQ-155 (ACV6036) |
| D7F2G5 (V-2) | QH-YS-199 (ACV60385) |
| D7F2H9 (V-2) | SC-3 (ACV60399) |
| V9LW71 (V-2) | QH09_11 (AFX66401) |
| V9LVU8 (V-2) | YN09_61 (AFX66475) |
| V9LWG5 (V-2) | ID10_1 (AFX66484) |
| U3N6V5 (V-2) | CS6-251 (AGW27537) |
| ‡ |
NP10_1 (AFX66485), NP10_2 (AFX66486), YN09_3 (AFX66471), YN09_96 (AFX66481), YN09_140 (AFX66483) |
| Accession number | % similarity to AGW27560 | Amino acid residue substitution | Bayesian cluster | 3D structure model | AlphaFold UniProt code | |||
| conservative | nonconservative | Branch | Cluster | |||||
| AGW27560 ALH24946 EQK97643 | 100% 99.4% 100% |
Q-to-K, H-to-Y | A1 | A | A B C |
U3N942 A0A0N9QMM1 T5A511 |
||
| ALH24992 ALH24995 |
99.7% | A-to-T | A2 | A | D E |
A0A0N9R5B3 A0A0N7G849 | ||
| T-to-S | ||||||||
| ALH25047 ALH24948 | 99.4% | S-to-L | A3 | A | F G |
A0A0N9R4V2 A0A0N9QUF3 | ||
| ALH25043 | 98.9% |
R-to-I, P-to-T, T-to-I, G-to-A, |
B | H | A0A0N9QMS9 | |||
| ALH25054 | 99.4% | I-to-L | A-to-V | C | I | A0A0N7G845 | ||
| ALH25005 | 99.2% | H-to-Y | P-to-H | D1 | D | J | A0A0N9QUK2 | |
| ALH24999 | 98.1% | S-to-T, I-to-V, H-to-Y | A-to-V, A-to-T | D2 | D | K | A0A0N9QMT4 | |
| ALH25008 | 99.7% | Y-to-H | E1 | E | L | A0A0N9QMR3 | ||
| ALH25034 | 99.4% | E-to-K, Y-to-H | E2 | E | M | A0A0N9QMS4 | ||
| ALH25003 | 99.2% | E-to-K, Y-to-H | S-to-G | E3 | E | N | A0A0N7G850 | |
| ALH25001 | 98.4% | S-to-T, V-to-I, E-to-K, Y-to-H | A-to-V, A-to-T | E4 | E | O | A0A0N9R4Q4 | |
| Accession number | % similarity to AEH27625 | Amino acid residue substitution | Bayesian cluster | 3D structure model | AlphaFold UniProt code | |||
| conservative | nonconservative | Branch | Cluster | |||||
| AEH27625 EQL04085 | 100% 100% |
I-1 | I | A B |
D7F2E9 T5AF56 |
|||
| AFX66437 ACV60391 ACV60372 | 99.6% | D-to-N | I-2 | I | C D E |
V9LW10 D7F2H1 D7F2F2 |
||
| G-to-R Q-to-L |
||||||||
| AIV43040 | 97.6% | Y-to-H, M-to-I, Q-to-R, S-to-T | T-to-I, A-to-G | II-1 | II | F | A0A0A0RCF5 | |
| ACV60417 | 97.6% | Y-to-H, M-to-I, S-to-T | T-to-I, T-to-A, A-to-G | II-2 | II | G | D7F2J7 | |
| ACV60415 | 98.8% | Y-to-H, M-to-I | T-to-G | III | H | D7F2F5 | ||
| AFX66476 | 98.8% | Y-to-H | D-to-G, V-to-A | IV-1 | IV | I | V9LWC9 | |
| AFX66472 | 97.6% | Y-to-H, D-to-N, S-to-T | D-to-G, V-to-A, A-to-T | IV-2 | IV | J | V9LVS8 | |
| ACV60363 | 99.6% | Y-to-H | V-1 | V | K | D7F2E3 | ||
| ACV60385 | 99.2% | I-to-V, Y-to-H | V-2 | V | L | D7F2G5 | ||
| ACV60399 | 99.2% | Y-to-H, Q-to-R | V-2 | V | M | D7F2H9 | ||
| AFX66401 | 99.2% | Y-to-H | D-to-I | V-2 | V | N | V9LW71 | |
| AFX66475 | 99.2% | Y-to-H | I-to-T | V-2 | V | O | V9LVU8 | |
| AFX66484 | 99.2% | Y-to-H | A-to-T | V-2 | V | P | V9LWG5 | |
| AGW27537 | 99.2% | V-to-I, Y-to-H | V-2 | V | Q | U3N6V5 | ||
| H. sinensis strain | Genome assembly segment | Percentage similarity | |
| MAT1-1-1 (vs. EQK97643) |
MAT1-2-1 (vs. AEH27625) |
||
| Co18 | ANOV01017390 (410←1519) | 99.7% | |
| ANOV01000063 (9329→10182) | 99.6% | ||
| 1229 | LKHE01001116 (3799←4909) | 99.7% | |
| LKHE01001605 (13860←14713) | 99.6% | ||
| IOZ07 | JAAVMX010000001 (6698911→6700021) | 99.7% | |
| JAAVMX000000000 | ― | ||
| ZJB12195 | LWBQ00000000 | ― | |
| LWBQ01000021 (238873←239726) | 99.6% | ||
| CC1406-20395 | NGJJ00000000 | ― | |
| NGJJ01000619 (23030←23883) | 99.6% | ||
|
H. sinensis strain or natural C. sinensis |
Transcriptome or metatranscriptome assembly segment | Percentage similarity | |
| MAT1-1-1 (vs. EQK97643) |
MAT1-2-1 (vs. AEH27625) |
||
| H. sinensis strain L0106 | GCQL00000000 | ― | |
| GCQL01020543 (397←1143) | 99.6% | ||
| Mature natural C. sinensis (Collected at Deqin, Yunnan) |
OSIN7648 (1→1065) | 94.9% | |
| OSIN7649 (1→397) | 100% | ||
| Natural C. sinensis * (Collected at Kangding, Sichuan) |
GAGW01008880 (300←1127) | 100% | |
| GAGW00000000 | ― | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





