Submitted:
29 March 2024
Posted:
29 March 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Gene and Genome Sequences of H. sinensis Strains and Natural C. sinensis
Transcriptome and Metatranscriptome Assemblies and Transcripts of Mating-Type Genes of H. sinensis Strains and Natural C. sinensis
Sequence Alignment Analysis
Amino Acid Property and Scale Analysis
Results
Differential Occurrence of Mating-Type Genes in H. sinensis Strains
Differential Transcription of Mating-Type Genes in the Transcriptome of H. sinensis Strains
Differential Occurrence of Mating-Type Genes in Natural and Cultivated C. sinensis Insect-Fungi Complexes
Differential Transcription of Mating-Type Genes in Natural and Cultivated C. sinensis Insect-Fungi Complexes
Transcription of Mating-Type Genes in the Metatranscriptome Assemblies of Natural C. sinensis
Transcription of Mating-Type Genes in Unassembled Metatranscriptome Sequencing Read Archives of Natural and Cultivated C. sinensis
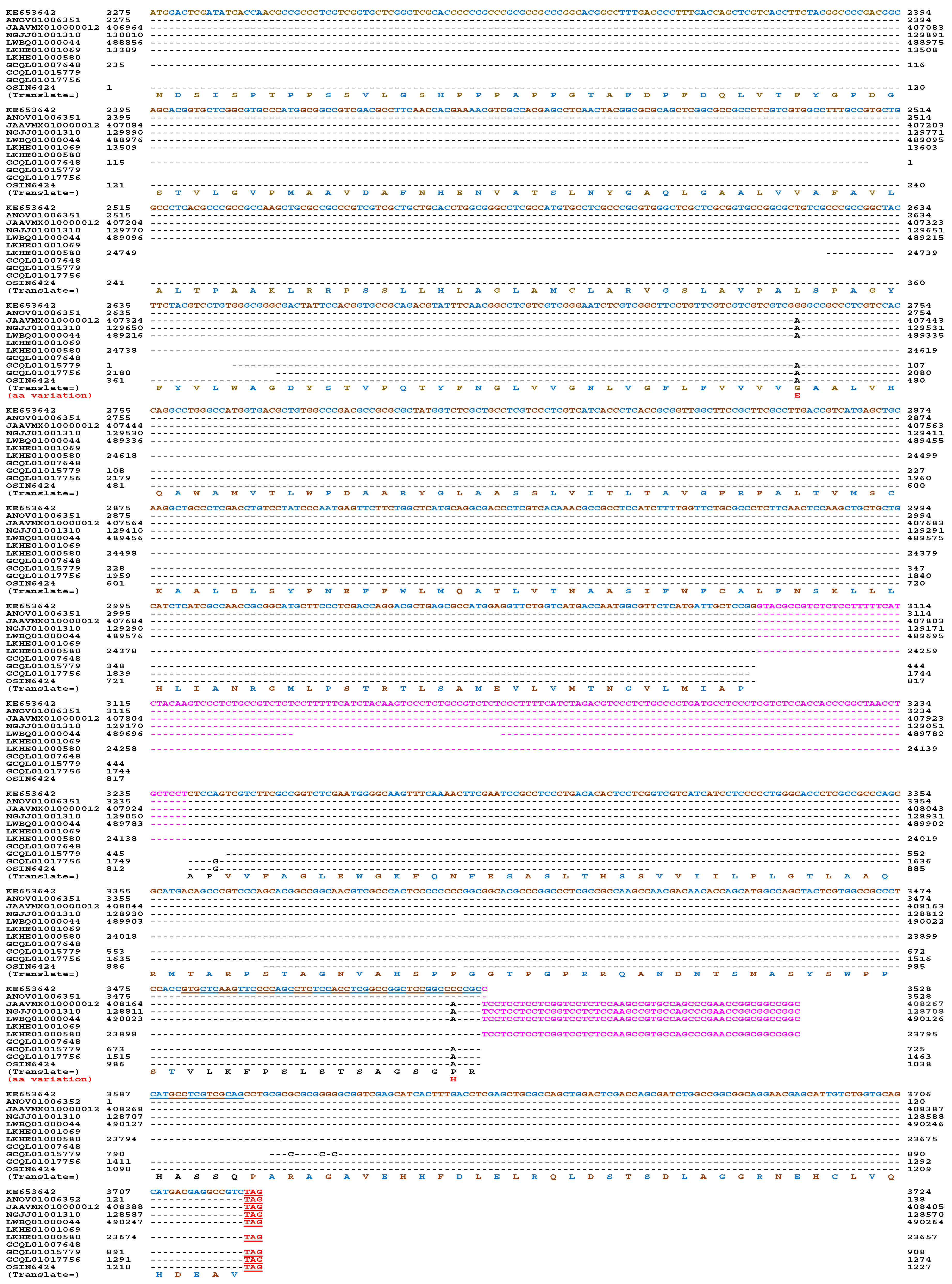
Variations in Mating Proteins
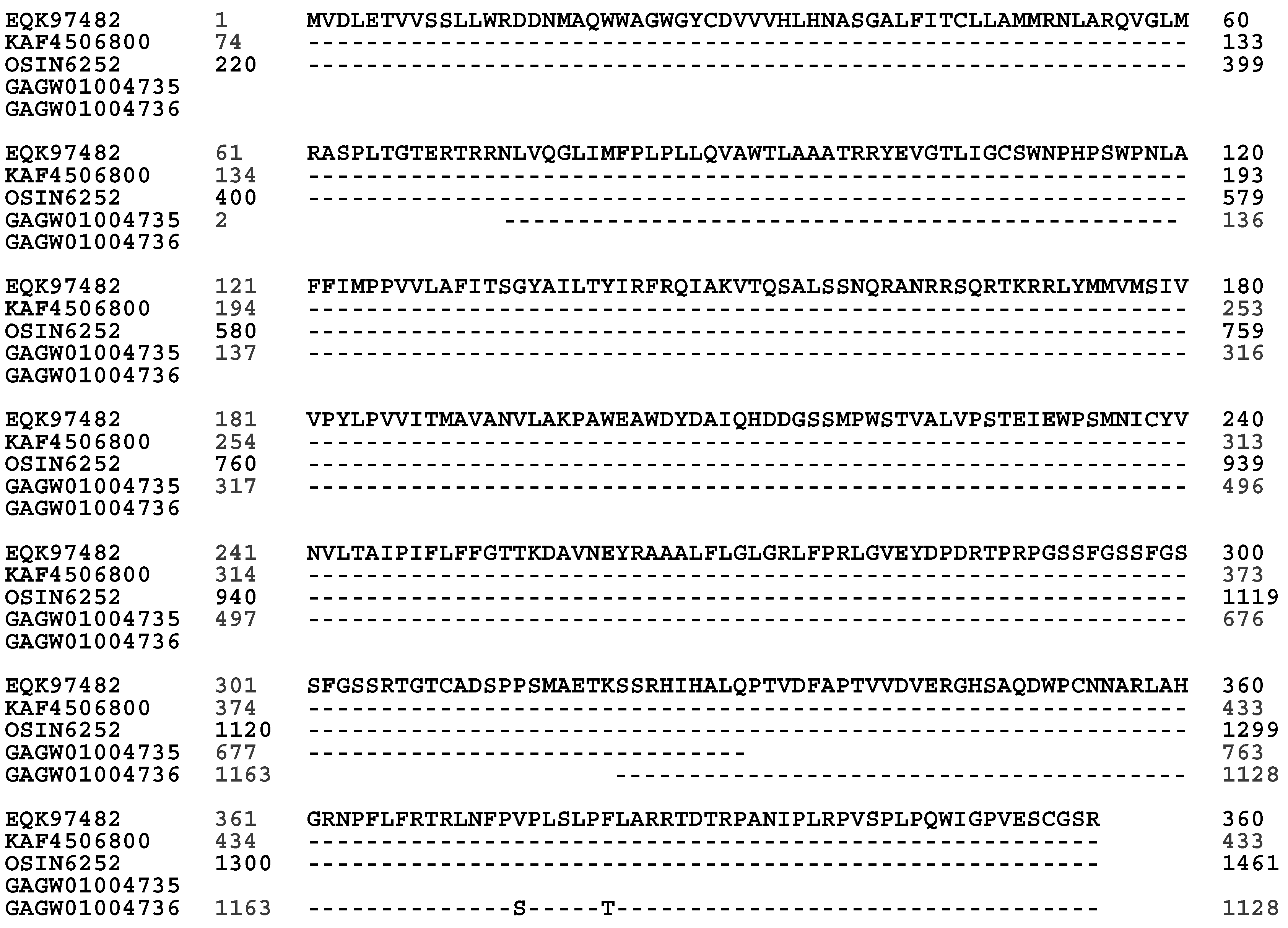
a-Factor-Like Pheromone Receptor in H. sinensis and Natural C. sinensis
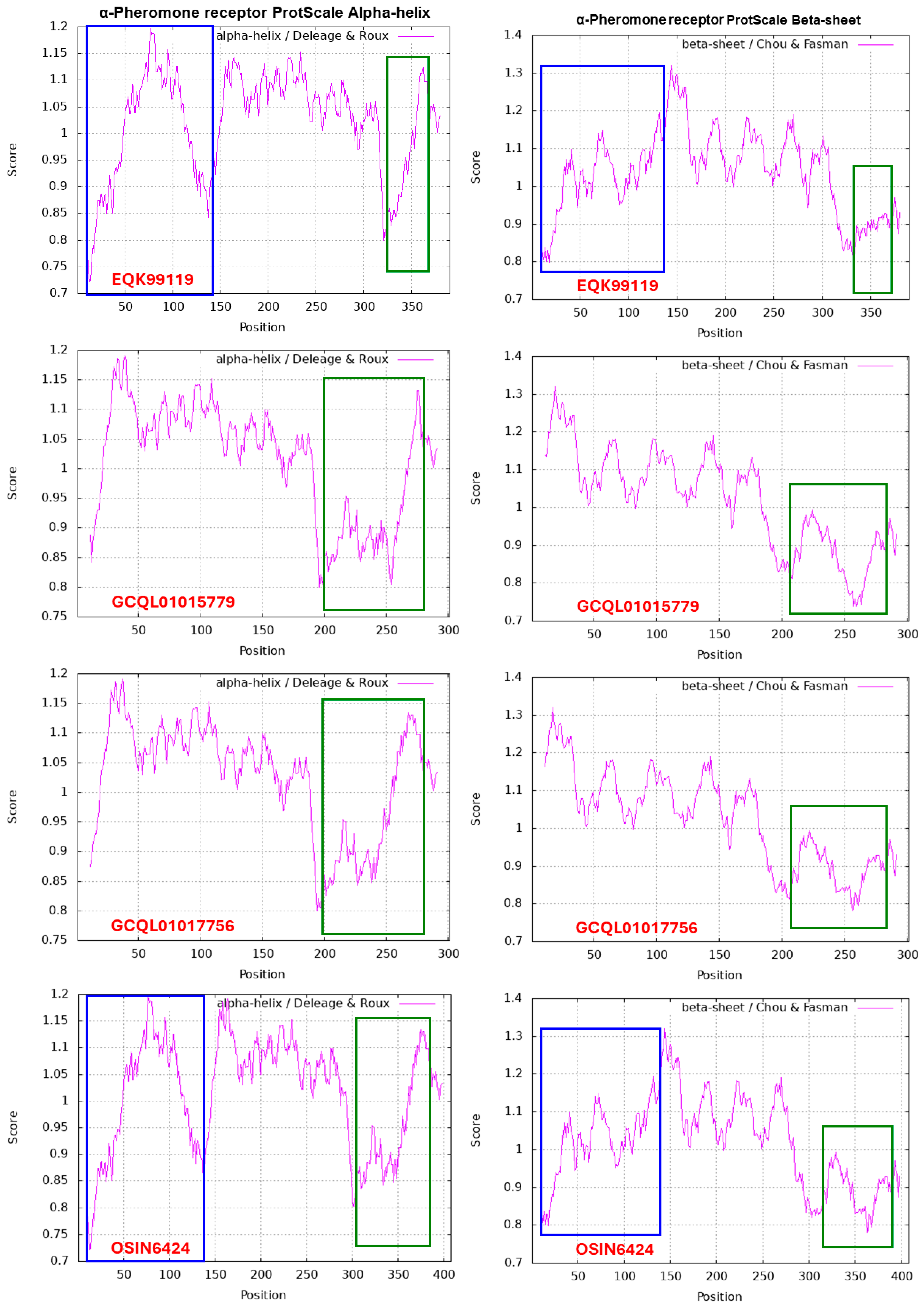
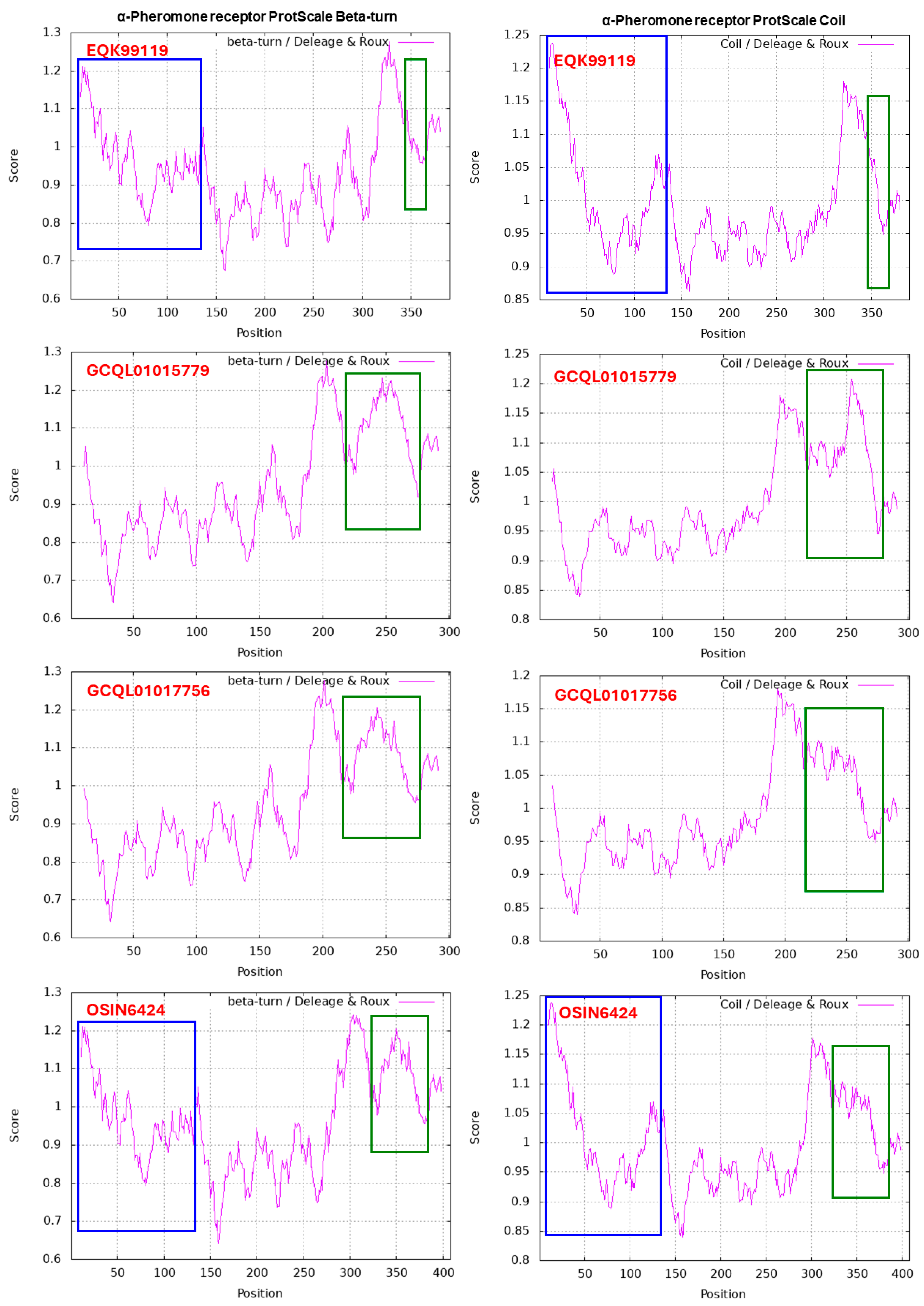
α-Factor-like Pheromone Receptor in H. sinensis and Natural C. sinensis
Other Pheromone-Related Genes in H. sinensis and Natural C. sinensis
Discussion
Reproductive Behavior of H. sinensis, Genotype #1 of O. sinensis
Sexual Reproduction Strategy during the Lifecycle of Natural C. sinensis
| Percent similarity | ||||
|---|---|---|---|---|
| ITS1-5.8S-ITS2 | ITS1 | 5.8S | ITS2 | |
| vs. Genotype #13 of O. sinensis (KT339190) | ||||
| Genotype #1 AB067721 O. sinensis | 86.3% | 100% | 94.8% | 64.2% |
| Group E AB067719 fungus | 88.2% | 71.5% | 100% | 99% |
| vs. Genotype #14 of O. sinensis (KT339178) | ||||
| Genotype #1 AB067721 O. sinensis | 87.7% | 67.9% | 94.9% | 100% |
| Group E AB067719 fungus | 89.2% | 100% | 100% | 71.5% |
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barseghyan, G.S.; Holliday, J.C.; Price, T.C.; Madison, L.M.; Wasser, S.P. Growth and cultural-morphological characteristics of vegetative mycelia of medicinal caterpillar fungus Ophiocordyceps sinensis G.H. Sung et al. (Ascomycetes) Isolates from Tibetan Plateau (P.R.China). Intl. J. Med. Mushrooms. 2011, 13, 565–581. [Google Scholar] [CrossRef]
- Bennett, R.J.; Johnson, A.D. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003, 22, 2505–2515. [Google Scholar] [CrossRef] [PubMed]
- Bushley, K.E.; Li, Y.; Wang, W.-J.; Wang, X.-L.; Jiao, L.; Spatafora, J.W.; Yao, Y.-J. Isolation of the MAT1-1 mating type idiomorph and evidence for selfing in the Chinese medicinal fungus Ophiocordyceps sinensis. Fungal Biol. 2013, 117, 599–610. [Google Scholar] [CrossRef]
- Chen, C.-S.; Hseu, R.-S.; Huang, C.-T. Quality control of Cordyceps sinensis teleomorph, anamorph, and Its products. Chapter 12, in (Shoyama, Y., Ed.) Quality Control of Herbal Medicines and Related Areas. InTech, Rijeka, Croatia (2011). pp. 223–238. www.intechopen.com.
- Chen, Y.-Q.; Hu, B.; Xu, F.; Zhang, W.-M.; Zhou, H.; Qu, L.-H. Genetic variation of Cordyceps sinensis, a fruit-body-producing entomopathegenic species from different geographical regions in China. FEMS Microbiol. Lett. 2004, 230, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Chou, P.Y.; Fasman, G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas. Mol. Biol. 1978, 47, 45–148. [Google Scholar] [CrossRef]
- Dai, R.-Q.; Lan, J.-L.; Chen, W.-H.; Li, X.-M.; Chen, Q.-T.; Shen, C.-Y. Discovery of a new fungus Paecilomyces hepiali Chen & Dai. Acta Agricult. Univ. Pekin. 1989, 15, 221–224. [Google Scholar]
- Debuchy R, Turgeo BG. Mating-Type Structure, Evolution, and Function in Euascomycetes. In (eds. Kües U. & Fischer, R.) Growth, Differentiation and Sexuality. Springer (2006). pp. 293–323.
- Deleage, G.; Roux, B. An algorithm for protein secondary structure prediction based on class prediction. Protein Engineering, Design and Selection 1987, 1, 289–294. [Google Scholar] [CrossRef]
- Dong, Y.-Z.; Zhang, L.-J.; Wu, Z.-M.; Gao, L.; Yao, Y.-S.; Tan, N.-Z.; Wu, J.-Y.; Ni, L.-Q.; Zhu, J.-S. Altered proteomic polymorphism in the caterpillar body and stroma of natural Cordyceps sinensis during maturation. PLoS ONE. 2014, 9, e109083. [Google Scholar] [CrossRef]
- Du, X.-H.; Wu, D.-M.; Kang, H.; Wang, H.-C.; Xu, N.; Li, T.-T.; Chen, K.-L. Heterothallism and potential hybridization events inferred for twenty-two yellow morel species. IMA Fungus. 2020, 11, 4. [Google Scholar] [CrossRef]
- Engh, I.B. Molecular phylogeny of the Cordyceps-Tolypocladium complex. Candidate scientific thesis, Department of Biology, University of Oslo, 1999.
- Gao, L.; Li, X.-H.; Zhao, J.-Q.; Lu, J.-H.; Zhao, J.-G.; Zhu, J.-S. Maturation of Cordyceps sinensis associates with alterations of fungal expressions of multiple Ophiocordyceps sinensis mutants with transition and transversion point mutations in stroma of Cordyceps sinensis. J. Peking Univ. (Health Sci.). 2012, 44, 454–463. [Google Scholar]
- Gao, L.; Li, X.-H.; Zhao, J.-Q.; Lu, J.-H.; Zhu, J.-S. Detection of multiple Ophiocordyceps sinensis mutants in premature stroma of Cordyceps sinensis by MassARRAY SNP MALDI-TOF mass spectrum genotyping. J. Peking Univ. (Health Sci.). 2011, 43, 259–266, https://www.ncbi.nlm.nih.gov/pubmed/21503123. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server. Chapter 52 (In) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005). pp. 571–607.
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; Chen, Z.-H.; Mauceli, E.; Hacohen, N.; Gnirke, A.; Rhind, N.; di Palma, F.; Birren, B.W.; Nusbaum, C.; Lindblad-Toh, K.; Friedman, N.; Regev, A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–130. [Google Scholar] [CrossRef] [PubMed]
- Hėnault, M.; Marsit, S.; Charron, G.; Landry, C.R. The effect of hybridization on transposable element accumulation in an undomesticated fungal species. eLife. 2020, 9, e60474. [Google Scholar] [CrossRef] [PubMed]
- Holliday, J.; Cleaver, M. Medicinal value of the caterpillar fungi species of the genus Cordyceps (Fr. ) Link (Ascomycetes). A review. Int. J. Med. Mushrooms. 2008, 10, 219–234. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.-J.; Xiao, G.-H.; Zheng, P.; Xia, Y.-L.; Zhang, X.-Y.; St Leger, R.J.; Liu, X.-Z.; Wang, C.-S. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 2013, 58, 2846–2854. [Google Scholar] [CrossRef]
- Jiang, Y.; Yao, Y.-J. A review for the debating studies on the anamorph of Cordyceps sinensis. Mycosistema. 2003, 22, 161–176. [Google Scholar]
- Jin, L. -.Q.; Xu, Z.-.W.; Zhang, B.; Yi, M.; Weng, C.-.Y.; Lin, S.; Wu, H.; Qin, X.-T.; Xu, F.; Teng, Y.; Yuan, S.-J.; Liu, Z.-Q.; Zheng, Y.-G. Genome sequencing and analysis of fungus Hirsutella sinensis isolated from Ophiocordyceps sinensis. AMB Expr. 2020, 10, 105. [Google Scholar] [CrossRef]
- Jones, S.K.; Bennett, R.J. Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 2011, 48, 668–676. [Google Scholar] [CrossRef]
- Kinjo, N.; Zang, M. Morphological and phylogenetic studies on Cordyceps sinensis distributed in southwestern China. Mycoscience. 2001, 42, 567–574. [Google Scholar] [CrossRef]
- Leff, S.E.; Rosenfeld, M.G.; Evans, R.M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Ann. Rev. Biochem. 1986, 55, 1091–117. [Google Scholar] [CrossRef]
- Leung, P.-H.; Zhang, Q.-X.; Wu, J.-Y. Mycelium cultivation, chemical composition and antitumour activity of a Tolypocladium sp. fungus isolated from wild Cordyceps sinensis. J. Appl. Microbiol. 2006, 101, 275–283. [Google Scholar] [CrossRef]
- Li, C.-L. A study of Tolypocladium sinense C.L. Li. sp. nov. and cyclosporin production. Acta Mycol. Sinica. 1988, 7, 93–98. [Google Scholar]
- Li, X.; Wang, F.; Liu, Q.; Li, Q.-P.; Qian, Z.-M.; Zhang, X.-L.; Li, K.; Li, W.-J.; Dong, C.-H. Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genomics. 2019, 20, 337. [Google Scholar] [CrossRef]
- Li, X.-Z.; Li, Y.-L.; Wang, Y.-N.; Zhu, J.-S. Genetic and phylogenetic features of the repetitive ITS copies in the Hirsutella sinensis genome and the genomically independent AT-biased Ophiocordyceps sinensis genotypes. BioRxiv (the preprint server for biology) February 03, 2024. https://www.biorxiv.org/content/10.1101/2022.12.09. 03 February 5198. [Google Scholar]
- Li, X.-Z.; Li, Y.-L.; Yao, Y.-S.; Xie, W.-D.; Zhu, J.-S. Further discussion with Li et al. (2013, 2019) regarding the “ITS pseudogene hypothesis” for Ophiocordyceps sinensis. Mol. Phylogenet. Evol. 2020, 146, 106728. [Google Scholar] [CrossRef]
- Li, X.-Z.; Li, Y.-L.; Yao, Y.-S.; Zhu, J.-S. Differential expression of Hirsutella sinensis genes and intraspecific genetic variation among H. sinensis strains. Biochem. Mol. Biol. J. 2022, 8, 58. [Google Scholar] [CrossRef]
- Li, X.-Z.; Li, Y.-L.; Zhu, J.-S. Differential expression of mating-type genes in Hirsutella sinensis and natural Cordyceps sinensis Proc. 2021 Intl. Conf. Info. Technol. Biomed. Engineering, 2021, pp. 318−333. https://conferences.computer.org/icitbepub/pdfs/ICITBE2021-6JNtJiThlp10s9N5Sodvuy/009900a318/009900a318.pdf. 2021. [Google Scholar]
- Li, Y.; Hsiang, T.; Yang, R.-H.; Hu, X.-D.; Wang, K.; Wang, W.-J.; Wang, X.-L.; Jiao, L.; Yao, Y.-J. Comparison of different sequencing and assembly strategies for a repeat-rich fungal genome, Ophiocordyceps sinensis. J. Microbiol. Methods. 2016, 128, 1–6. [Google Scholar] [CrossRef]
- Li, Y.; Jiao, L.; Yao, Y.-J. Non-concerted ITS evolution in fungi, as revealed from the important medicinal fungus Ophiocordyceps sinensis. Mol. Phylogenet. Evol. 2013, 68, 373–379. [Google Scholar] [CrossRef]
- Li, Y.-L.; Gao, L.; Yao, Y.-S.; Wu, Z.-M.; Lou, Z.-Q.; Xie, W.-D.; Wu, J.-Y.; Zhu, J.-S. Altered GC- and AT-biased genotypes of Ophiocordyceps sinensis in the stromal fertile portions and ascospores of natural Cordyceps sinensis. PLoS ONE. 2023, 18, e0286865. [Google Scholar] [CrossRef]
- Li, Y.-L.; Li, X.-Z.; Yao, Y.-S.; Wu, Z.-M.; Gao, L.; Tan, N.-Z.; Lou, Z.-Q.; Xie, W.-D.; Wu, J.-Y.; Zhu, J.-S. Differential cooccurrence of multiple genotypes of Ophiocordyceps sinensis in the stromata, stromal fertile portion (ascocarps) and ascospores of natural Cordyceps sinensis. PLoS ONE. 2023, 18, e0270776. [Google Scholar] [CrossRef]
- Li, Y.-L.; Li, X.-Z.; Yao, Y.-S.; Xie, W.-D.; Zhu, J.-S. Molecular identification of Ophiocordyceps sinensis genotypes and the indiscriminate use of the Latin name for the multiple genotypes and the natural insect-fungi complex. Am J BioMed Sci. 2022, 14, 115–135. [Google Scholar] [CrossRef]
- Li, Y.-L.; Yao, Y.-S.; Xie, W.-D.; Zhu, J.-S. Molecular heterogeneity of natural Cordyceps sinensis with multiple Ophiocordyceps sinensis fungi challenges the anamorph-teleomorph connection hypotheses and implementation of the Amsterdam Declaration of IMA. Am. J. Biomed. Sci. 2016, 8, 123–159. [Google Scholar] [CrossRef]
- Li, Y.-L.; Yao, Y.-S.; Zhang, Z.-H.; Xu, H.-F.; Liu, X.; Ma, S.-L.; Wu, Z.-M.; Zhu, J.-S. Synergy of fungal complexes isolated from the intestines of Hepialus lagii larvae in increasing infection potency. J. Fungal Res. 2016, 14, 96–112. [Google Scholar]
- Liang, Z.-Q.; Han, Y.-F.; Liang, J.-D.; Dong, X.; Du, W. Issues of concern in the studies of Ophiocordyceps sinensis. Microbiol. Chin. 2010, 37, 1692–1697. [Google Scholar]
- Liu, J.; Guo, L.-N.; Li, Z.-W.; Zhou, Z.; Li, Z.; Li, Q.; Xiaochen Bo, X.-C.; Wang, S.-Q.; Wang, J.-L.; Ma, S.-C.; Zheng, J.; Yang, Y. Genomic analyses reveal evolutionary and geologic context for the plateau fungus Ophiocordyceps sinensis. Clin. Med. 2020, 15, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Q.; Lin, S.; Baker, P.J.; Wu, L.-F.; Wang, X.-R.; Wu, H.; Xu, F.; Wang, H.-Y.; Brathwaite, M.E.; Zheng, Y.-G. Transcriptome sequencing and analysis of the entomopathogenic fungus Hirsutella sinensis isolated from Ophiocordyceps sinensis. BMC Genomics. 2015, 16, 106–123. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-L.; St Leger, R.J. Chapter Seven - Insect Immunity to Entomopathogenic Fungi, Editors: Lovett B, St. Leger RJ. Advanc Genet, Academic Press, 2016. Vol. 94; pp. 251–85.
- Mao, X.-M.; Zhao, S.-M.; Cao, L.; Yan, X.; Han, R.-C. The morphology observation of Ophiocordyceps sinensis from different origins. J. Environ. Entomol. 2013, 35, 343–353. [Google Scholar]
- Meng, Q.; Yu, H.-Y.; Zhang, H.; Zhu, W.; Wang, M.-L.; Zhang, J.-H.; Zhou, G.-L.; Li, X.; Qin, Q.-L.; Hu, S.-N.; Zou, Z. Transcriptomic insight into the immune defenses in the ghost moth, Hepialus xiaojinensis, during an Ophiocordyceps sinensis fungal infection. Insect Biochem. Mol. Biol. 2015, 64, 1–15. [Google Scholar] [CrossRef]
- Nakamura, N.; Tanaka, C.; Takeuchi-Kaneko, Y. Transmission of antibiotic-resistance markers by hyphal fusion suggests partial presence of parasexuality in the root endophytic fungus Glutinomyces brunneus. Mycol. Progress 2019, 18, 453–462. [Google Scholar] [CrossRef]
- Peters, C.; Elofsson, A. Why is the biological hydrophobicity scale more accurate than earlier experimental hydrophobicity scales? Proteins. 2014, 82, 2190–2198. [Google Scholar] [CrossRef]
- Pfennig, K.S. Facultative Mate Choice Drives Adaptive Hybridization. Sci. 2007, 318, 965–967. [Google Scholar] [CrossRef]
- Qiu W-F, Chen Q-T, Qian C-R, Shen C-Y, Xu J-T, Wang L-Y, Zhang W-J. “Study on Cordyceps sinensis, a new species Paecilomyces hepiali” certificate of identification for the achievement in science and technology. In Archive of Institution of Chinese Materia Medica, Chinese Academy of Traditional Chinese Medicine (Unpublished; January 8, 1987) Docs #000021, #000044, and #000045.
- Ren, Y.; Wan, D.-G.; Lu, X.-M.; Guo, J.-L. The study of scientific name discussion for TCM Cordyceps. LisShenzhen Med. Material Medica Res. 2013, 24, 2211–2212. [Google Scholar]
- Samarasinghe, H.; You, M.; Jenkinson, T.S.; Xu, J.-P.; James, T.Y. Hybridization Facilitates Adaptive Evolution in Two Major Fungal Pathogens. Genes. 2020, 11, 101. [Google Scholar] [CrossRef]
- Seervai, R.N.H.; Jones, S.K.; Hirakawa, M.P.; Porman, A.M.; Bennett, R.J. Parasexuality and ploidy change in Candida tropicalis. Eukaryotic Cell. 2013, 12, 1629–1640. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, R.K.; Bennett, R.J. Fungal meiosis and parasexual reproduction--lessons from pathogenic yeast. Current Opinion Microbiol. 2009, 12, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Shu, R.-H.; Zhang, J.-H.; Meng, Q.; Zhang, H.; Zhou, G.-L.; Li, M.-M.; Wu, P.-P.; Zhao, Y.-N.; Chen, C.; Qin, Q. A new high-quality draft genome assembly of the Chinese cordyceps Ophiocordyceps sinensis. Genome Biol. Evol. 2020, 12, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Simm S, Einloft J, Mirus O, Schleiff E. 50 years of amino acid hydrophobicity scales: revisiting the capacity for peptide classification. Biol. Res. 2016, 49, 31. [Google Scholar] [CrossRef]
- Steensels, J.; Gallone, B.; Verstrepen, K.J. Interspecific hybridization as a driver of fungal evolution and Adaptation. Nat. Rev. Microbiol. 2021, 19, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Stensrud Ø, Hywel-Jones NL, Schumacher T. Towards a phylogenetic classification of Cordyceps: ITS nrDNA sequence data confirm divergent lineages and paraphyly. Mycol. Res. 2005, 109, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Stensrud Ø, Schumacher T, Shalchian-Tabrizi K, Svegardenib IB, Kauserud H. Accelerated nrDNA evolution and profound AT bias in the medicinal fungus Cordyceps sinensis. Mycol. Res. 2007, 111, 409–415. [Google Scholar] [CrossRef]
- Stone, R. Improbable partners aim to bring biotechnology to a Himalayan kingdom. Sci. 2010, 327, 940–941. [Google Scholar] [CrossRef]
- Sung, G.-H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Turgeon, B.G.; Yoder, O.C. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 2000, 31, 1–5. [Google Scholar]
- Wang, J.-B.; St Leger, R.J.; Wang, C.-S. Advances in Genomics of Entomopathogenic Fungi. Advan. Genetics 2016, 94, 1–39. [Google Scholar] [CrossRef]
- Wang, Y.; Stata, M.; Wang, W.; Stajich, J.E.; White, M.M.; Moncalvo, J.M. Comparative genomics reveals the core gene toolbox for the fungus-insect symbiosis. mBio. 2018, 9, e00636–e18. [Google Scholar] [CrossRef]
- Wang, Y.-B.; Wang, Y.; Fan, Q.; Duan, D.-E.; Zhang, G.-D.; Dai, R.-Q.; Dai, Y.-D.; Zeng, W.-B.; Chen, Z.-H.; Li, D.-D.; Tang, D.-X.; Xu, Z.-H.; Sun, T.; Nguyen, T.-T.; Tran, N.-L.; Dao, V.-M.; Zhang, C.-M.; Huang, L.-D.; Liu, Y.-J.; Zhang, X.-M.; Yang, D.-R.; Sanjuan, T.; Liu, X.-Z.; Yang, Z.-L.; Yu, H. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Diversity. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Wei, J.-C.; Wei, X.-L.; Zheng, W.-F.; Guo, W.; Liu, R.-D. Species identification and component detection of Ophiocordyceps sinensis cultivated by modern industry. Mycosystema. 2016, 35, 404–410. [Google Scholar]
- Wei, X.-L.; Yin, X.-C.; Guo, Y.-L.; Shen, N.-Y.; Wei, J.-C. Analyses of molecular systematics on Cordyceps sinensis and its related taxa. Mycosystema. 2006, 25, 192–202. [Google Scholar]
- Wilson, A.M.; Wilken, P.M.; van der Nest, M.A.; Steenkamp, E.T.; Wingfield, M.J.; Wingfield, B.D. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus. 2015, 6, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.-H.; Yang, D.-R.; Jiang, J.-J.; Zhang, Q.-J.; Liu, Y.; Liu, Y.-L.; Zhang, Y.; Zhang, H.-B.; Shi, C.; Tong, Y.; Kim, C.-H.; Chen, H.; Peng, Y.-Q.; Yu, Y.; Zhang, W.; Eichler, E.E.; Gao, L.-Z. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 2017, 7, 1806. [Google Scholar] [CrossRef]
- Xia, F.; Liu, Y.; Shen, G.-L.; Guo, L.-X.; Zhou, X.-W. Investigation and analysis of microbiological communities in natural Ophiocordyceps sinensis. Can. J. Microbiol. 2015, 61, 104–111. [Google Scholar] [CrossRef]
- Xiang, L.; Li, Y.; Zhu, Y.; Luo, H.; Li, C.; Xu, X.; Sun, C.; Song, J.-Y.; Shi, L.-H.; He, L.; Sun, W.; Chen, S.-L. Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics. 2014, 103, 154–159. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, J.-P.; Zhu, P.; Cheng, K.-D.; He, H.-X.; Zhu, H.-X.; Wang, Q. Non-support of species complex hypothesis of Cordyceps sinensis by targeted rDNA-ITS sequence analysis. Mycosystema. 2009, 28, 724–730. [Google Scholar]
- Yang, J.-L.; Xiao, W.; He, H.-H.; Zhu, H.-X.; Wang, S.-F.; Cheng, K.-D.; Zhu, P. Molecular phylogenetic analysis of Paecilomyces hepiali and Cordyceps sinensis. Acta Pharmaceut. Sinica. 2008, 43, 421–426. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Tong, X.-X.; He, C.-Y.; Bai, J.; Wang, F.; Guo, J.-L. Comparison of endogenetic microbial community diversity between wild Cordyceps sinensis, artificial C. sinensis and habitat soil. Chin. J. Chin. Materia Medica. 2021, 46, 3106–3115. [Google Scholar]
- Yao, Y.-S.; Gao, L.; Li, Y.-L.; Ma, S.-L.; Wu, Z.-M.; Tan, N.-Z.; Wu, J.-Y.; Ni, L.-Q.; Zhu, J.-S. Amplicon density-weighted algorithms for analyzing dissimilarity and dynamic alterations of RAPD polymorphisms of Cordyceps sinensis. Beijing Da Xue Xue Bao Yi Xue Bao. 2014, 46, 618–628, (http://xuebao.bjmu.edu.cn/EN/Y2014/V46/I4/618). [Google Scholar]
- Yao, Y.-S.; Zhou, Y.-J.; Gao, L.; Lu, J.-H.; Zhu, J.-S. Dynamic alterations of the differential fungal expressions of Ophiocordyceps sinensis and its mutant genotypes in stroma and caterpillar during maturation of natural Cordyceps sinensis. J. Fungal Res. 2011, 9, 37–49. [Google Scholar]
- Zhang, S.; Zhang, Y.-J. Molecular evolution of three protein-coding genes in the Chinese caterpillar fungus Ophiocordyceps sinensis. Microbiol. China. 2015, 42, 1549–1560. [Google Scholar]
- Zhang, S.; Zhang, Y.-J.; Liu, X.-Z.; Wen, H.-A.; Wang, M.; Liu, D.-S. Cloning and analysis of the MAT1-2-1 gene from the traditional Chinese medicinal fungus Ophiocordyceps sinensis. Fungal Biol. 2011, 115, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang S, Zhang Y-J, Shrestha B, Xu J-P, Wang C-S, Liu X-Z. Ophiocordyceps sinensis and Cordyceps militaris: research advances, issues and perspectives. Mycosystema. 2013, 32, 577−597.
- Zhang, S.-W.; Cen, K.; Liu, Y.; Zhou, X.-W.; Wang, C.-S. Metatranscriptomics analysis of the fruiting caterpillar fungus collected from the Qinghai-Tibetan plateau. Sci. Sinica Vitae. 2018, 48, 562–570. [Google Scholar]
- Zhang Y-J, Li E-W, Wang C-S, Li Y-L, Liu X-Z. Ophiocordyceps sinensis, the flagship fungus of China: terminology, life strategy and ecology. Mycol. 2012, 3, 2–10, https://www.tandfonline.com/doi/full/10.1080/21501203.2011.654354.
- Zhang Y-J, Sun B-D, Zhang S, Wàngmŭ, Liu X-Z, Gong W-F. Mycobiotal investigation of natural Ophiocordyceps sinensis based on culture-dependent investigation. Mycosistema. 2010, 29, 518–527. [Google Scholar]
- Zhang Y-J, Xu L-L, Zhang S, Liu X-Z, An Z-Q, Wàngmŭ, et al. Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: implications for its evolution and conservation. BMC Evol. Biol. 2009, 9, 290. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Zhang, S.; Li, Y.-L.; Ma, S.-L.; Wang, C.-S.; Xiang, M.-C.; Liu, X.; An, Z.-Q.; Xu, J.-P.; Liu, X.-Z. Phylogeography and evolution of a fungal–insect association on the Tibetan Plateau. Mol. Ecol. 2014, 23, 5337–5355. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-N.; Zhang, J.-H.; Meng, Q.; Zhang, H.; Zhou, G.-L.; Li, M.-M.; Wu, P.-P.; Shu, R.-H.; Gao, X.-X.; Guo, L.; Tong, Y.; Cheng, L.-Q.; Guo, L.; Chen, C.; Qin, Q. Transcriptomic analysis of the orchestrated molecular mechanisms underlying fruiting body initiation in Chinese cordyceps. Gene. 2020, 763, 145061. [Google Scholar] [CrossRef]
- Zheng, P.; Wang, C.-S. Sexuality Control and Sex Evolution in Fungi. Sci Sinica Vitae. 2013, 43, 1090–1097. [Google Scholar]
- Zhong, X.; Gu, L.; Wang, H.-Z.; Lian, D.-H.; Zheng, Y.-M.; Zhou, S.; Zhou, W.; Gu, J.; Zhang, G.; Liu, X. Profile of Ophiocordyceps sinensis transcriptome and differentially expressed genes in three different mycelia, sclerotium and fruiting body developmental stages. Fungal Biol. 2018, 122, 943–951. [Google Scholar] [CrossRef]
- Zhu, J.-S.; Gao, L.; Li, X.-H.; Yao, Y.-S.; Zhou, Y.-J.; Zhao, J.-Q.; Zhou, Y.-J. Maturational alterations of oppositely orientated rDNA and differential proliferations of CG:AT-biased genotypes of Cordyceps sinensis fungi and Paecilomyces hepiali in natural C. sinensis. Am. J. Biomed. Sci. 2010, 2, 217–238. [Google Scholar] [CrossRef]
- Zhu, J.-S.; Guo, Y.-L.; Yao, Y.-S.; Zhou, Y.-J.; Lu, J.-H.; Qi, Y.; Chen, W.; Zheng, T.-Y.; Zhang, L.; Wu, Z.-M.; Zhang, L.-J.; Liu, X.-J.; Yin, W.-T. Maturation of Cordyceps sinensis associates with co-existence of Hirsutella sinensis and Paecilomyces hepiali DNA and dynamic changes in fungal competitive proliferation predominance and chemical profiles. J. Fungal Res. 2007, 5, 214–224. [Google Scholar]
- Zhu, J.-S.; Halpern, G.M.; Jones, K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis: Part I. J. Altern. Complem. Med. 1998, 4, 289–303. [Google Scholar] [CrossRef]
- Zhu, J.-S.; Halpern, G.M.; Jones, K. The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis: Part II. J. Altern. Complem. Med. 1998, 4, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-S.; Li, C.-L.; Tan, N.-Z.; Berger, J.L.; Prolla, T.A. Combined use of whole-gene expression profiling technology and mouse lifespan test in anti-aging herbal product study. Proc. TCM Products Innovation and Industrial Development Summit, Hangzhou, China (Nov 27, 2011). pp. 443–448. https://xueshu.baidu.com/usercenter/paper/show?paperid=08341c17fa58c8f85584b92572b90f75&site=xueshu_se.
- Zhu, J.-S.; Li, Y.-L. A Precious Transitional Chinese Medicine, Cordyceps sinensis: Multiple heterogeneous Ophiocordyceps sinensis in the insect-fungi complex. Lambert Academic Publishing, Saarbrüchen. Germany (.
- Zhu, J.-S.; Li, Y.-L.; Yao, Y.-S.; Xie, W.-D. Molecular evidence not supporting the use of Hirsutella sinensis strain EFCC7287 as the taxonomic standard for multiple Ophiocordyceps sinensis fungi. Intl. J. Mol. Biol. 2018, 3, 114–115. [Google Scholar]
- Zhu, J.-S.; Li, Y.-L.; Yao, Y.-S.; Xie, W.-D. The multiple genotypes of Ophiocordyceps sinensis and the “ITS pseudogene” hypothesis. Mol. Phylogenet. Evol. 2019, 139, 106322. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-S.; Wu, J.-Y. Genetic heterogeneity of natural Cordyceps sinensis with co-existence of multiple fungi. Chin. J. Cell Biol. 2015, 37, 284–298. [Google Scholar]
- Zhu, J.-S.; Zhao, J.-G.; Gao, L.; Li, X.-H.; Zhao, J.-Q.; Lu, J.-H. Dynamically altered expressions of at least 6 Ophiocordyceps sinensis mutants in the stroma of Cordyceps sinensis. J. Fungal Res. 2012, 10, 100–112. [Google Scholar]
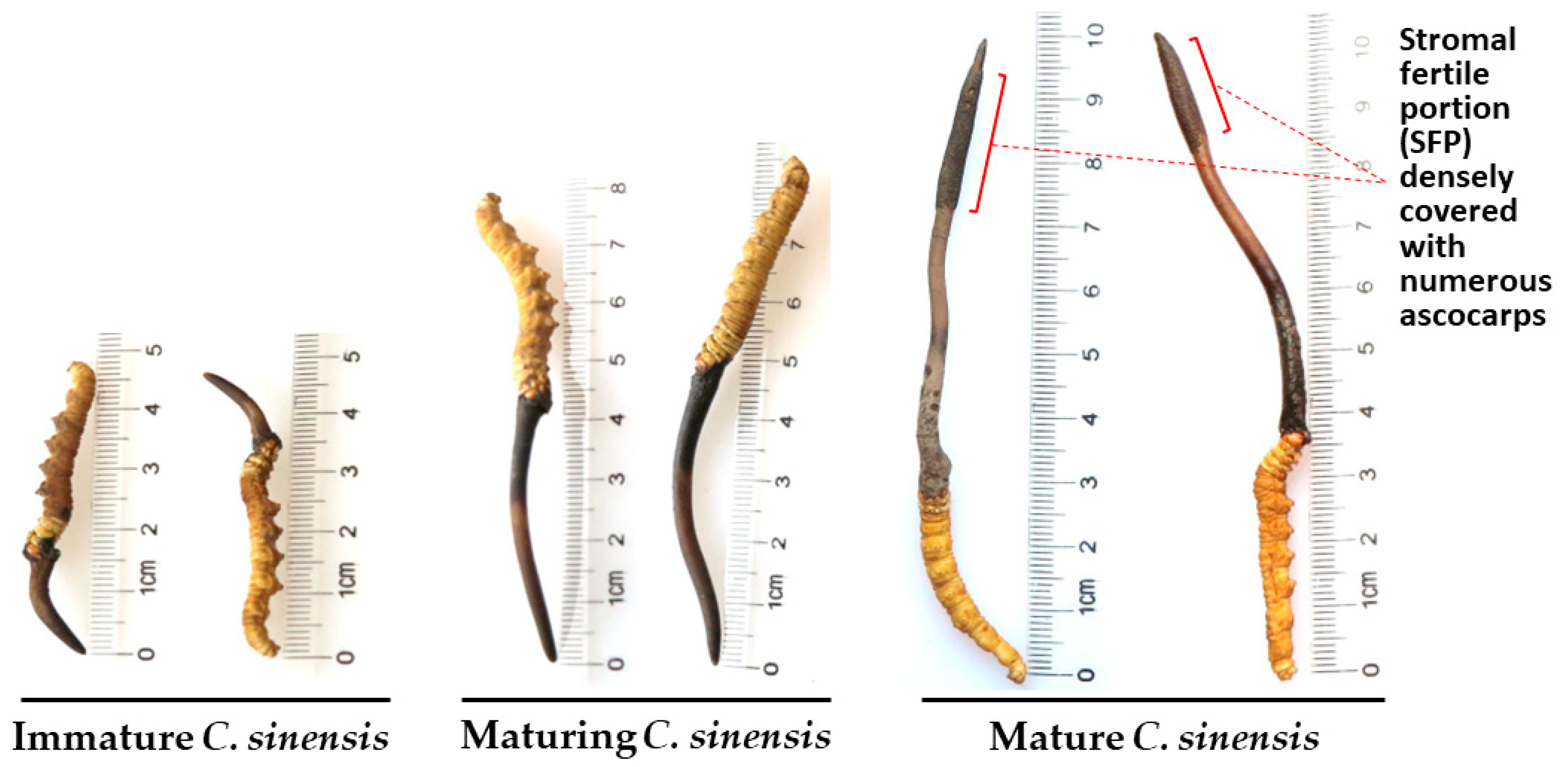
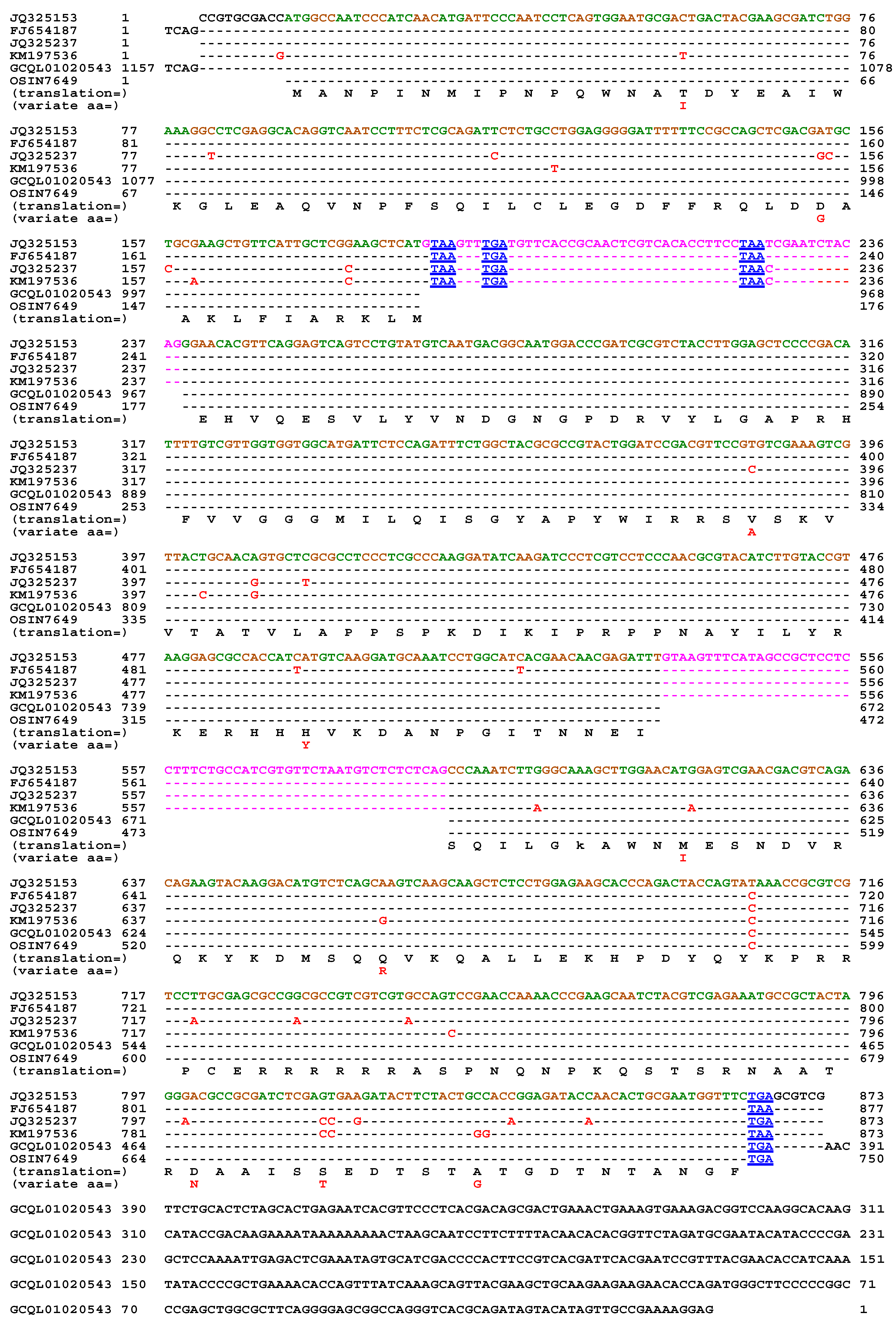
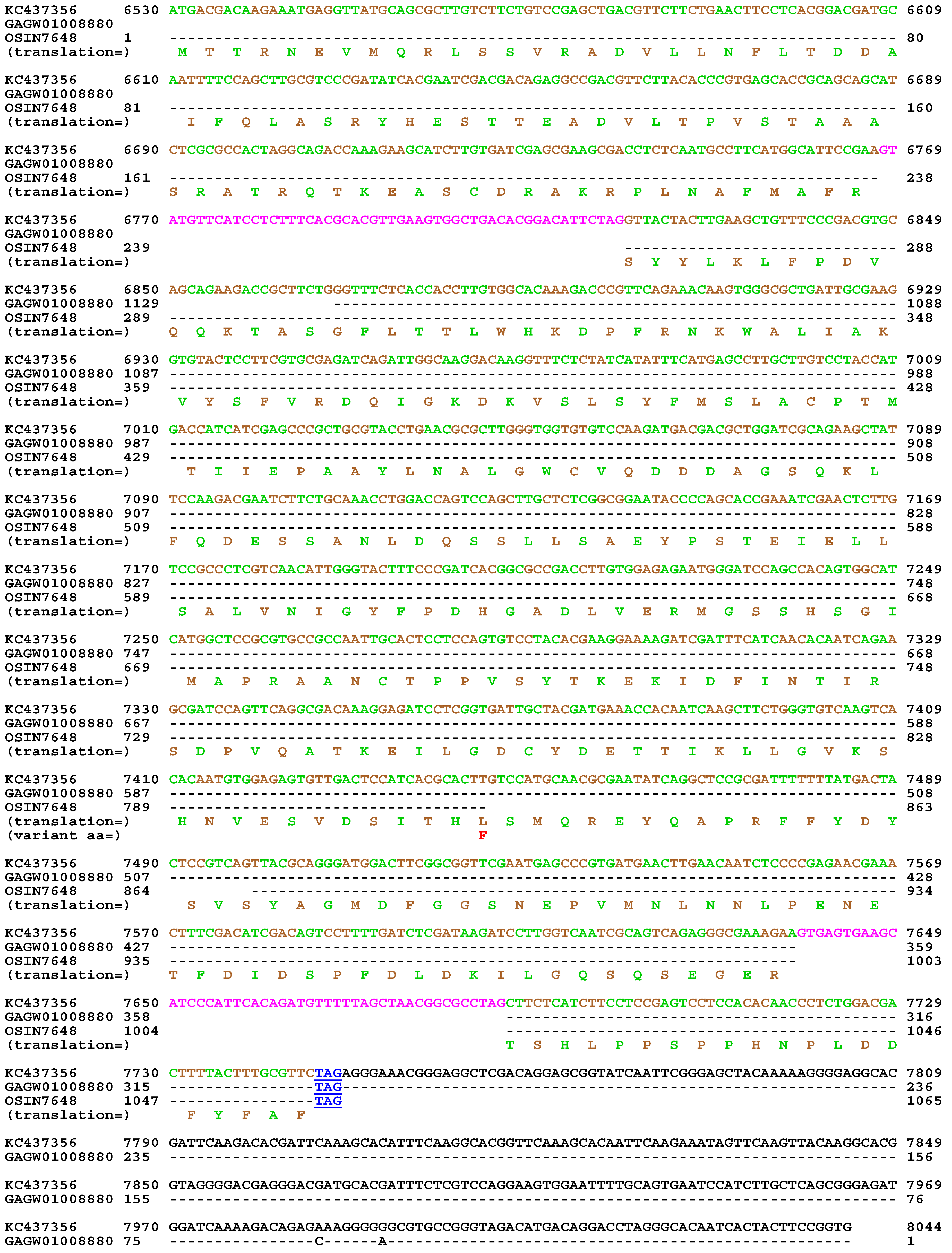
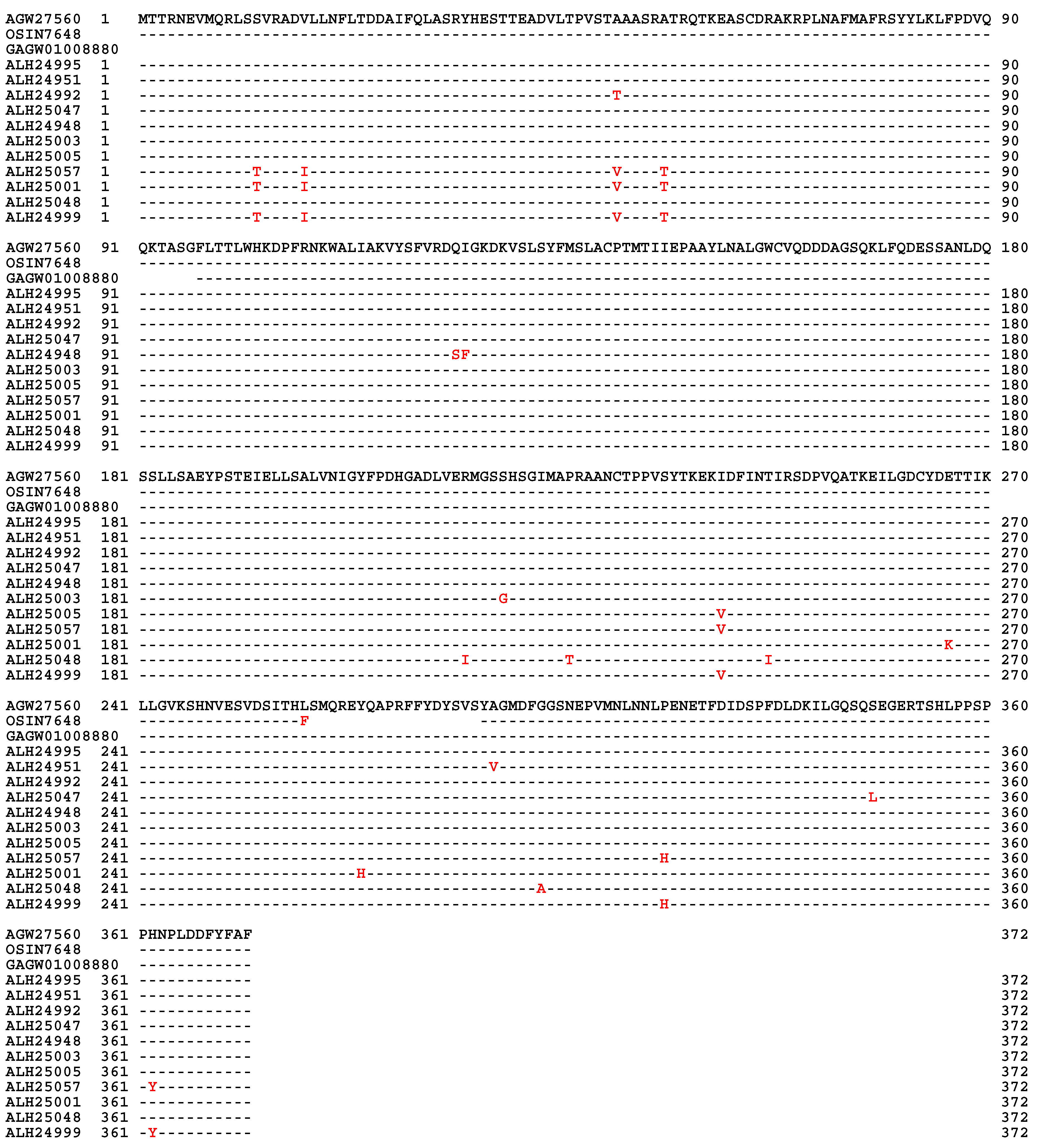
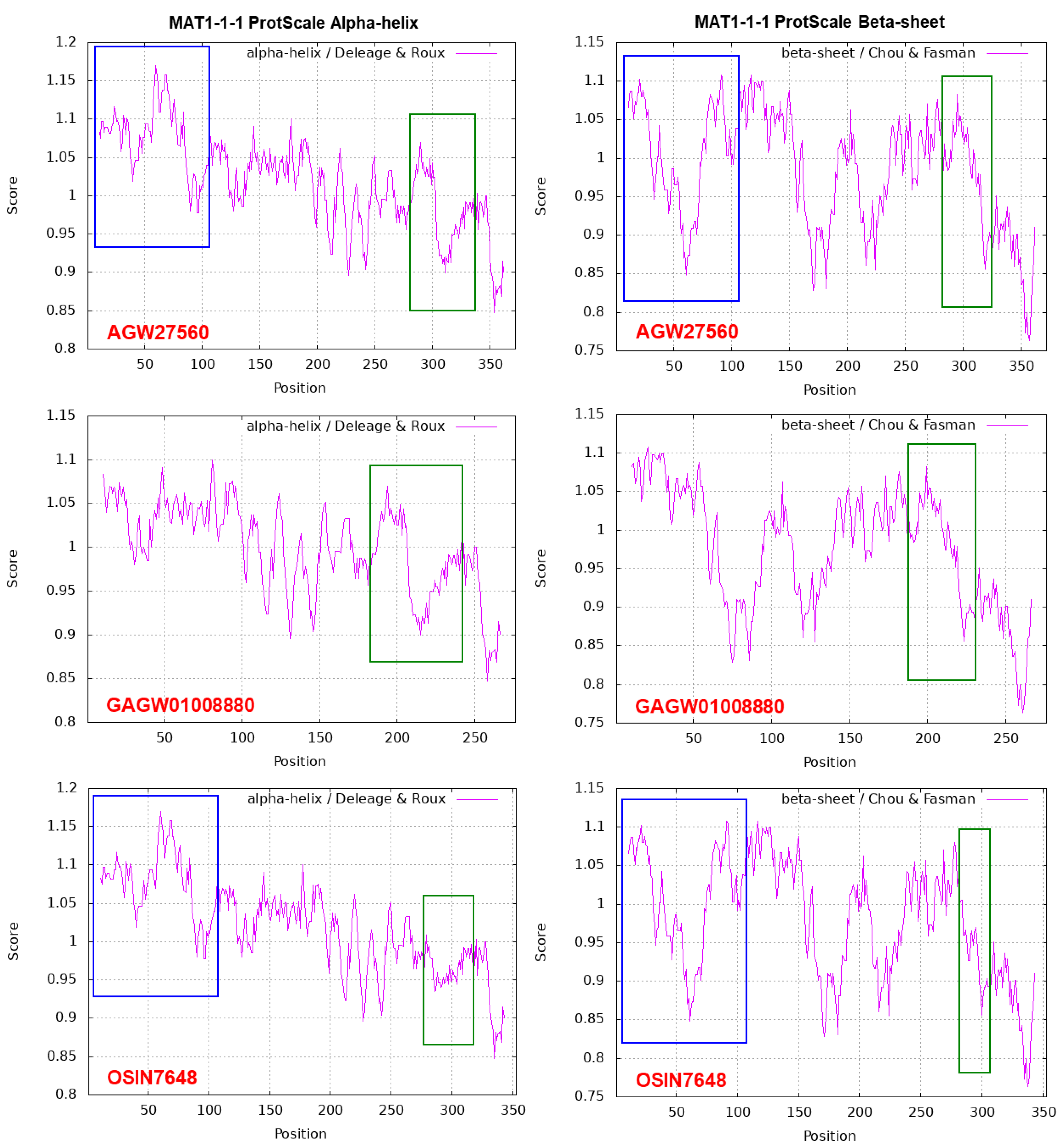
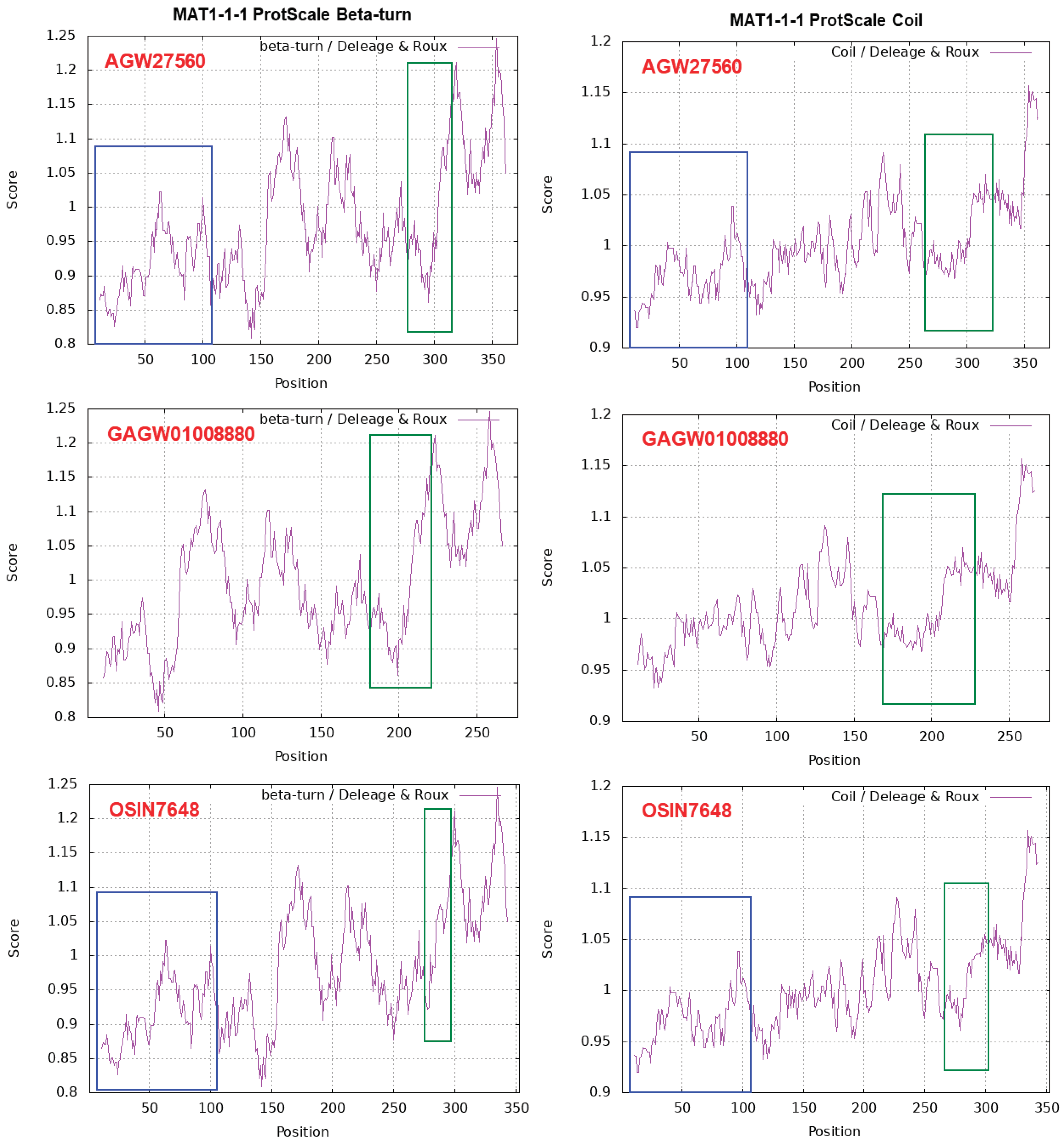
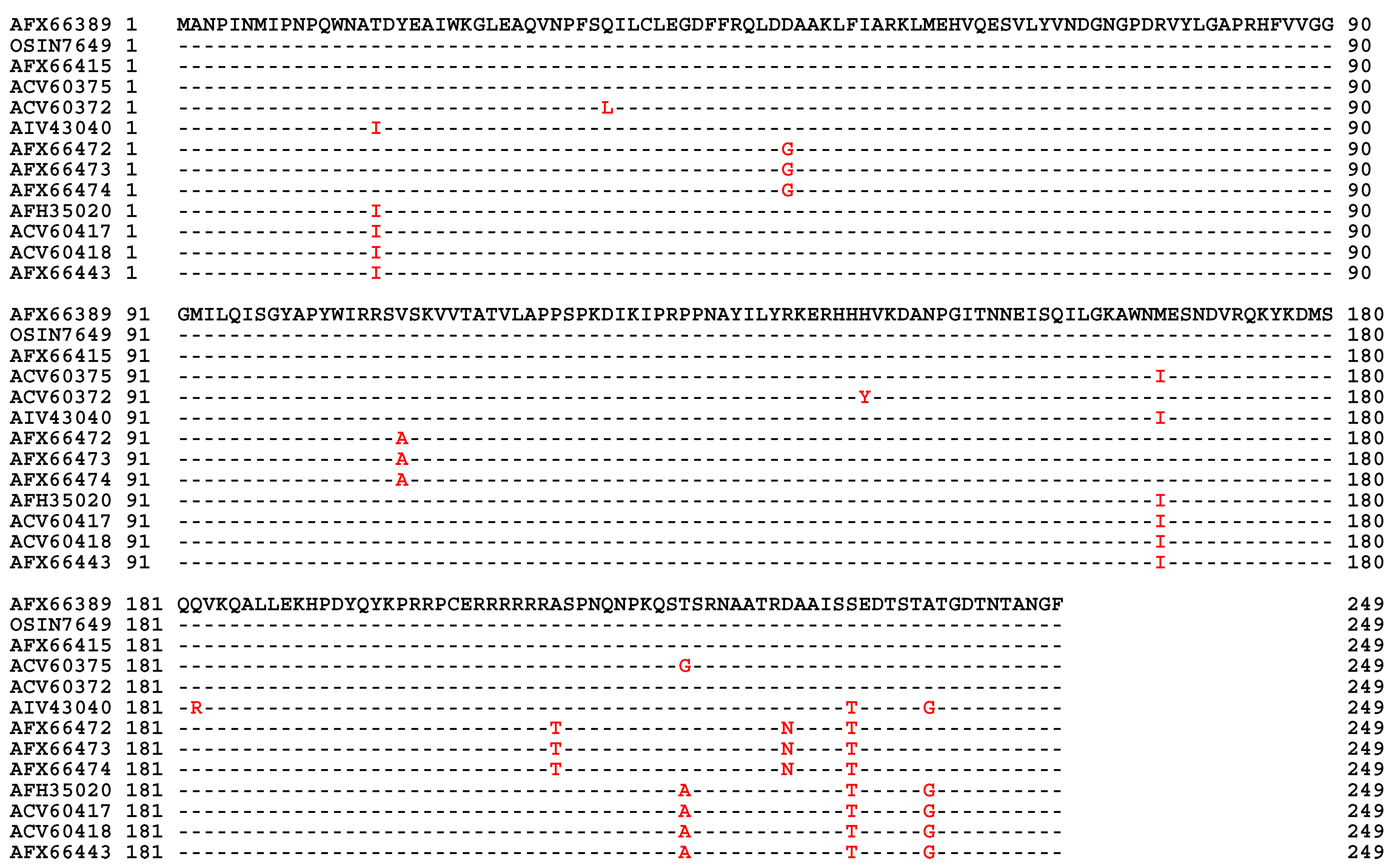
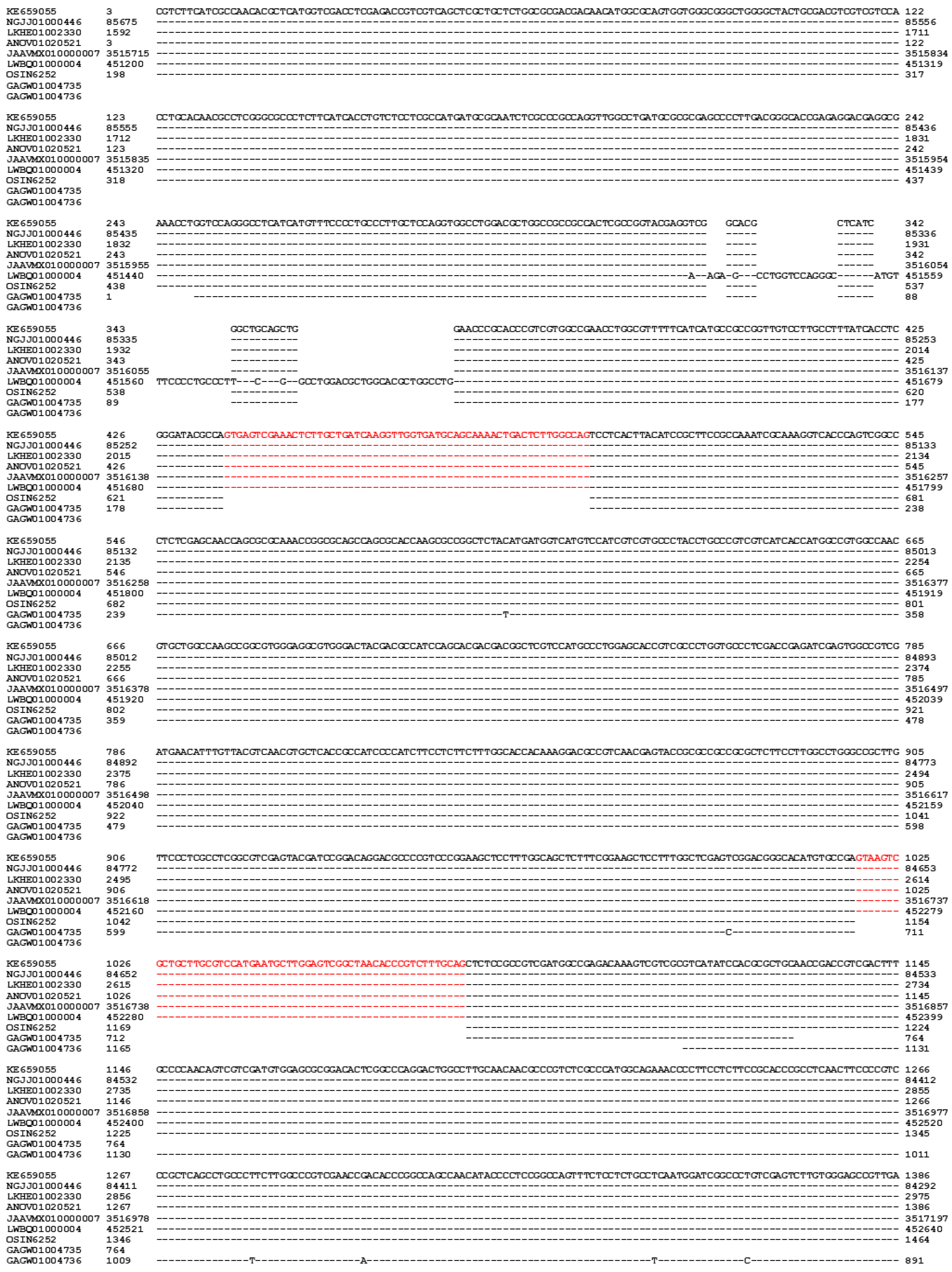

| 22 strains | 66 strains | 149 strains | ||||
|---|---|---|---|---|---|---|
| containing only the MAT1-1-1 gene | containing only the MAT1-2-1 gene | containing both the MAT1-1-1 and MAT1-2-1 genes | ||||
| CS09-143 | CC1406-203 | XZ-CD-41 | 1229 | NP10_1 | SC09_47 | XZ09_32 |
| CS09-229 | CS2 | XZ-CD-59 | Co18 | NP10_2 | SC09_57 | XZ09_46 |
| GS03 | GS09_337 | XZ-CD-64 | CS6-251 | QH01 | SC09_65 | XZ09_48 |
| IOZ07 | QH02 | XZ-ML-191 | CS18-266 | QH03 | SC09_77 | XZ09_59 |
| QH07_188 | QH05 | XZ-LZ05-6 | CS25-273 | QH04 | SC09_87 | XZ09_71 |
| QH07_197 | QH09_11 | XZ-LZ06_1 | CS26-277 | QH06 | SC09_97 | XZ09_80 |
| XZ05_3 | QH09_187 | XZ-LZ06_7 | CS34-291 | QH07 | SC09_107 | XZ09_95 |
| XZ05_6 | QH-LJ-214 | XZ-LZ06_21 | CS36-1294 | QH08 | SC09_117 | XZ09_100 |
| XZ07_H2 | QH-LJ-236 | XZ-LZ06_61 | CS37-295 | QH09_20L | SC09_128 | XZ09_106 |
| XZ07_108 | QH-QL-206 | XZ-LZ06_108 | CS68-2-1228 | QH09_33L | SC09_147 | XZ09_113 |
| XZ07_154 | QH-QL-207 | XZ-LZ07_30 | CS68-2-1229 | QH09_37 | SC09_157 | XZ09_118 |
| XZ07_166 | QH-YS-188 | XZ-LZ07_64 | CS68-5-1216 | QH09_46 | SC09_167 | XZ10_7 |
| XZ07_176 | QH-YS-189 | XZ-LZ07_H1 | CS70-1208 | QH09_56 | SC09_180 | XZ10_15 |
| XZ07_180 | QH-YS-196 | XZ-LZ07_H2 | CS70-1211 | QH09_66 | SC09_190 | XZ10_17 |
| XZ08_4 | QH-YS-197A | XZ-NQ_74 | CS70-1212 | QH09_78 | SC09_200 | XZ10_23 |
| XZ08_10 | QH-YS-197B | XZ-NQ_80 | CS71-1218 | QH09_93 | SC10_4 | XZ12_1 |
| XZ08_26 | QH-YS-199 | XZ-NQ_84 | CS71-1219 | QH09_122 | SC10_18 | XZ12_16 |
| XZ08_59 | SC-2 | XZ-NQ_86 | CS71-1220 | QH09_131 | SC10_21 | XZ12_33 |
| XZ08_A1 | SC-3 | XZ-NQ_92 | CS76-1284 | QH09_151 | TB01 | XZ12_43 |
| XZ08_B1 | SC-4 | XZ-NQ_139 | CS91-1291 | QH09_164 | TB02 | YN01 |
| YN07_6 | SC-5 | XZ-NQ_154 | CS560-961 | QH09_173 | TB03 | YN02 |
| YN07_8 | SC-7 | XZ-NQ_155 | CS561-964 | QH09_201 | TB04 | YN03 |
| SCK05-4-3 | XZ-NQ_156 | GS01 | QH09_210 | TB05 | YN09_3 | |
| XZ07_11 | XZ-NQ_166 | GS02 | QH10_1 | TB06 | YN09_6 | |
| XZ07_46 | XZ-NQ_176 | GS04 | QH10_4 | TB07 | YN09_22 | |
| XZ08_33 | XZ-NQ_180 | GS05 | QH10_7 | TB08 | YN09_51 | |
| XZ08_38 | XZ-SN_44 | GS09_111 | SC01 | XZ05_2 | YN09_61 | |
| XZ-CD-A1 | YN-1 | GS09_121 | SC02 | XZ05_7 | YN09_64 | |
| XZ-CD-B1 | YN-4 | GS09_131 | SC03 | XZ05_8 | YN09_72 | |
| XZ-CD-4 | YN-5 | GS09_143 | SC04 | XZ05_12 | YN09_81 | |
| XZ-CD-10 | YN-6 | GS09_201 | SC05 | XZ06_124 | YN09_85 | |
| XZ-CD-26 | YN-8 | GS09_225 | SC06 | XZ06_152 | YN09_89 | |
| XZ-CD-30 | ZJB12195 | GS09_229 | SC07 | XZ06_260 | YN09_96 | |
| GS09_281 | SC08 | XZ07_133 | YN09_101 | |||
| GS09_311 | SC09_1 | XZ08_24 | YN09_140 | |||
| GS10_1 | SC09_21 | XZ08_56 | ||||
| GS10_10 | SC09_36 | XZ09_4 | ||||
| ID10_1 | SC09_37 | XZ09_15 | ||||
| Chemical-physical property | α-Helix | β-Sheet | β-Turn | Coil | |||
|---|---|---|---|---|---|---|---|
| Aspartic acid | Asp | D | Acidic | 0.924 | 0.540 | 1.197 | 1.197 |
| Glutamic acid | Glu | E | Acidic | 1.504 | 0.370 | 1.149 | 0.761 |
| Alanine | Ala | A | Aliphatic | 1.489 | 0.830 | 0.788 | 0.824 |
| Isoleucine | Ile | I | Aliphatic | 1.003 | 1.600 | 0.240 | 0.886 |
| Leucine | Leu | L | Aliphatic | 1.236 | 1.300 | 0.670 | 0.810 |
| Valine | Val | V | Aliphatic | 0.990 | 1.700 | 0.387 | 0.772 |
| Phenylalanine | Phe | F | Aromatic | 1.195 | 1.380 | 0.624 | 0.797 |
| Tryptophan | Trp | W | Aromatic | 1.090 | 1.370 | 0.546 | 0.941 |
| Tyrosine | Tyr | Y | Aromatic | 0.787 | 1.470 | 0.795 | 1.109 |
| Arginine | Arg | R | Basic | 1.224 | 0.930 | 0.912 | 0.893 |
| Histidine | His | H | Basic | 1.003 | 0.870 | 0.970 | 1.068 |
| Lysine | Lys | K | Basic | 1.172 | 0.740 | 1.302 | 0.897 |
| Asparagine | Asn | N | with polar neutral side chains | 0.772 | 0.890 | 1.572 | 1.167 |
| Cysteine | Cys | C | with polar neutral side chains | 0.966 | 1.190 | 0.965 | 0.953 |
| Glutamine | Gln | Q | with polar neutral side chains | 1.164 | 1.100 | 0.997 | 0.947 |
| Methionine | Met | M | with polar neutral side chains | 1.363 | 1.050 | 0.436 | 0.810 |
| Serine | Ser | S | with polar neutral side chains | 0.739 | 0.750 | 1.316 | 1.130 |
| Threonine | Thr | T | with polar neutral side chains | 0.785 | 1.190 | 0.739 | 1.148 |
| Glycine | Gly | G | Unique amino acids | 0.510 | 0.750 | 1.860 | 1.251 |
| Proline | Pro | P | Unique amino acids | 0.492 | 0.550 | 1.415 | 1.540 |
| MAT1-1-1 | MAT1-1-2 | MAT1-1-3 | MAT1-2-1 | |
|---|---|---|---|---|
| (vs. KC437356 of Strain CS68-2-1229) | (vs. JQ325153 of Strain GS09_121) | |||
| % similarity of the genome sequences | ||||
| H. sinensis Strain 1229 | 100% | 100% | 100% | 99.9% |
| H. sinensis Strain Co18 | 99.9% | 100% | 100% | 99.7% |
| H. sinensis Strain IOZ07 | 100% | 100% | 100% | ― |
| H. sinensis Strain CC1406-203 | ― | ― | ― | 99.9% |
| H. sinensis Strain ZJB12195 | ― | ― | ― | 99.9% |
|
% similarity of the coding sequences of the genes (excluding introns) and the transcriptome sequences |
||||
| H. sinensis Strain L0106 | ― | ― | ― | 99.0% |
| H. sinensis Strain 1229 | 100% | 100% | 100% | 100% |
| Natural C. sinensis (Deqin, Yunnan) | 100% | ― | ― | 100% |
| Natural C. sinensis (Kangding, Sichuan) | 100% | ― | ― | ― |
| % Amino acids of mating-type protein | |||||
|---|---|---|---|---|---|
| Hydrophobic | Acidic | Basic | Neutral | ||
| MAT1-1-1 protein | |||||
| AGW27560 | H. sinensis Strain CS68-2-1229 | 41.4% | 13.2% | 11.0% | 34.4% |
| GAGW01008880 | Natural C. sinensis | 41.3% | 14.1% | 9.8% | 34.8% |
| OSIN7648 | Natural C. sinensis | 41.9% | 13.1% | 11.1% | 33.7% |
| MAT1-2-1 protein | |||||
| AFX66389 | H. sinensis Strain GS09_121 | 41.2% | 10.0% | 16.5% | 33.3% |
| OSIN7649 | Natural C. sinensis | 41.2% | 10.0% | 16.5% | 33.3% |
| Percent similarity | ||
|---|---|---|
| a-Factor-like pheromone receptor | α-Factor-like pheromone receptor | |
| Between the sequences of pheromone receptor genes and the H. sinensis genomes | ||
| H. sinensis Strain 1229 | 100% | 99.8% |
| H. sinensis Strain CC1406-203 | 100% | 99.8% |
| H. sinensis Strain Co18 | 100% | 100% |
| H. sinensis Strain IOZ07 | 100% | 99.9% |
| H. sinensis Strain ZJB12195 | 95.2% | 97.5% |
| Between the coding sequences of the pheromone receptor genes (excluding introns) and the transcriptome sequences of H. sinensis and natural C. sinensis | ||
| H. sinensis Strain L0106 | ― | 99.3% |
| Natural C. sinensis (Deqin, Yunnan) | 100% | 99.7% |
| Natural C. sinensis (Kangding, Sichuan) | 98.5‒100% | ― |
| Between the pheromone receptor protein sequences translated from the gene sequences and the transcript sequences of H. sinensis and natural C. sinensis | ||
| H. sinensis Strain L0106 | ― | 86.4‒100% |
| Natural C. sinensis (Deqin, Yunnan) | 100% | 82.9% |
| Natural C. sinensis (Kangding, Sichuan) | 97.8‒100% | ― |
| % Amino acids of pheromone receptor protein | |||||
|---|---|---|---|---|---|
| Hydrophobic | Acidic | Basic | Neutral | ||
| a-pheromone receptor protein | |||||
| EQK97482 | H. sinensis Strain Co18 | 50.7% | 6.5% | 11.6% | 31.4% |
| GAGW01004735-GAGW01004736 | Natural C. sinensis | 50.4% | 6.2% | 11.2% | 31.2% |
| OSIN6252 | Natural C. sinensis | 50.7% | 6.5% | 11.6% | 31.4% |
| α-pheromone receptor protein | |||||
| EQK99119 | H. sinensis Strain Co18 | 55.1% | 5.6% | 7.4% | 31.8% |
| GCQL01017756 | H. sinensis Strain L0106 | 50.8% | 6.0% | 9.0% | 34.2% |
| GCQL01015779 | H. sinensis Strain L0106 | 51.8% | 6.0% | 9.0% | 33.2% |
| OSIN6424 | Natural C. sinensis | 52.7% | 5.8% | 8.6% | 32.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





