2. Genomic Signatures of Micrornas in Cervical Cancer
Non-coding RNA, as the name suggests, refers to RNA that cannot be translated into a protein. It can be divided into two major categories: housekeeping non-coding RNA and regulatory non-coding RNA. The latter includes short-chain siRNA, microRNA, piwiRNA and lncRNA. Briefly, microRNAs are a large class of single-chain, highly conserved, non-coding, short-stranded RNAs composed of 19-24 nucleotides, widely found in many eukaryotes and viruses. 50% of microRNAs are localized to chromosome regions prone to structural changes. MicroRNA directly targets mRNA and exerts posttranscriptional regulation in classical and noncanonical manners [
11]. MicroRNAs bind to the target gene 3′-UTRs through the seed sequence. When the microRNA is fully complementary to the target gene, it directly causes cleavage of the mRNA; when it is incompletely complementary to the target gene, it represses the translation of the gene [
11]. Previous studies have shown that microRNAs are involved in several biological processes, such as cell growth, development, differentiation, apoptosis, and cellular homeostasis. Dysfunction of microRNA has been reported to play a role in tumorigenesis and tumor progression by promoting uncontrolled proliferation, increasing survival rate, inhibiting differentiation, and promoting cancer cell metastasis, including in cervical cancer [
12]. This evidence allows us to propose systems biology-based cooperating microRNAs as gene therapy in cancer.
According to sequence bioinformatics analysis by different websites, including miRTarBase [
13], TargetScan [
14], miRanda [
15], miRbase [
16], MirWalk [
17], TriplexRNA [
18], miRSearch [
19], Eclipsebio [
20], miR2disease [
21], DIANA-miRPath [
22], GeneCodis [
23], miRecords [
24], and TransmiR v2.0 [
25]; a microRNA may have binding sites on hundreds of mRNAs, and a mRNAs 3′-UTR may be bound by tens of microRNAs. By posttranscriptional regulatory element sequencing, it is possible to identify the target sequences of microRNAs and RNA binding proteins in mRNAs [
26]. One major mechanism that underlies the roles of microRNAs in cancer development could be deregulated microRNA expression when compared to normal cells. MicroRNA’s direct regulation on target mRNA is affected by complex factors beyond the microRNA. At different stages during carcinogenesis, one microRNA sequence may regulate different targets, which has been termed “microRNA’s differential regulation”. Furthermore, definition of efficient target and related process effects is based on the accordance of changing microRNA regulation and mRNA contribution. The phenomenon of microRNAs regulating different targets at different stages may be universal. Hence, microRNAs are an integral component of genetic regulatory networks due to their role in regulating cellular gene expression at the posttranscriptional level. MicroRNA expression signatures have been shown to be promising biomarkers for the classification or outcome prediction of a wide array of human cancers.
Hu et al, described the first microRNA expression signature reported in cervical cancer [
27]. In this study, a microRNA-based signature was identified for the prediction of cervical cancer survival. The investigators analyzed the expression profiles of 96 cancer-related microRNAs in 102 cervical cancer samples using RT-qPCR-based microRNA assays. Five microRNAs were significantly associated with patient survival (miR-9, miR-21, miR-200a, miR-203, and miR-218). Of the microRNAs analyzed, miR-9 and miR-200a were especially promising; both microRNAs were significantly associated with disease outcome and were good candidates for a prognostic microRNA signature. Functional studies were performed to characterize the effects of these microRNAs in cervical cancer cells. Genome-wide microRNA target prediction, real-time RT-qPCR analysis, and cell migration assays provided evidence that miR-200a affects the metastatic potential of cervical cancer cells by simultaneously suppressing the expression of multiple genes that are important for cell morphology and migration sites. Taken together, these data suggest that miR-9, miR-21, miR-200a, miR-203, and miR-218 may represent a microRNA signature and provide a new biomarker-based strategy for risk stratification of cervical cancer patients.
A study carried out by Pereira et al, evaluated the microRNA expression profiles of a heterogeneous set of cervical tissues from 25 different patients [
28]. They investigated microRNA expression signature in cervical cancer using a microarray platform containing probes for mature microRNAs. They identified eight microRNAs that showed significant downregulation between normal cervical samples and the pre-neoplastic and neoplastic samples; these were miR-26a, miR-29a, miR-99a, miR-143, miR-145, miR-199a, miR-203 and miR-513. Other microRNAs were downregulated from normal to pre-neoplastic cervical samples, but had increased expression in cervical cancer samples, including miR-16, miR-27a, miR-106a, miR-142-5p, miR-197 and miR-205. Among the microRNAs that were upregulated in CIN I, CIN III, and cervical carcinoma samples, were miR-10a, miR-132, miR-148a, miR-196a and miR-302b. These findings highlight the variability of natural microRNA expression profiles and may be explained in part by the biological heterogeneity among cervical samples, which may complicate the use of microRNA profiling in clinical diagnostics.
Cheung et al, reported a distinct microRNA expression signature for CIN and identified individual microRNAs that may be involved in the development of cervical carcinoma [
29]. They performed microdissection of high-grade CIN II/III tissues which were compared with normal cervical epithelium, coupled with RT-qPCR to improve the sensitivity for more accurate of 202 microRNA profiling. Supervised analysis identified 12 highly differentially regulated microRNAs, including miR-9, miR-10a, miR-20b, miR-34b, miR-34c, miR-193b, miR-203, miR-338, miR-345, miR-424, miR-512-5p, and miR-518a, which distinguished the high-grade CIN specimens from normal cervical epithelium. Target prediction analysis revealed that these dysregulated microRNAs mainly control apoptosis signaling pathways and cell cycle regulation. These results support the correlation between this microRNA signature and an independent set of high-grade CIN cases, and this same characteristic signature can also be used to distinguish cervical squamous cell carcinoma from normal controls.
Jia et al, described the first profiling of altered serum microRNAs in cervical cancer patients in order to predict cervical cancer at a relatively early stage [
30]. Serum samples were collected from 213 cervical cancer patients and 158 controls. MicroRNA expression screening was performed by Solexa sequencing and differential expression was validated by RT-qPCR. The analysis revealed 12 markedly upregulated serum microRNAs (miR-21, miR-25, miR-26b, miR-29a, miR-29c, miR-101, miR-140-3p, miR-146b-5p, miR-191, miR-200a, miR-423-3p, and miR-486-5p) in cervical cancer patients compared with controls. Interestingly, a panel of 5 microRNAs (miR-21, miR-25, miR-29a, miR-200a, and miR-486-5p) constituted a more sensitive and specific diagnostic test compared with any single microRNA-based assay. These five serum microRNAs were confirmed as cervical cancer biomarkers. Furthermore, miR-29a and miR-200a exhibited a distinct upregulation in poorly differentiated cases compared with patients with well- or moderately differentiated tumors and might have value in clinical monitoring and prognosis. The results suggest that a genomic signature of five microRNAs identified from genome-wide serum microRNA expression profiling might serve as a fingerprint and non-invasive diagnostic marker for cervical cancer.
A study performed by Zhang et al, also helped establish a serum microRNA signature for diagnosing cervical cancer [
31]. Serum samples were obtained from a cohort of 184 cervical cancer patients, 186 CIN patients and 193 healthy control subjects. RT-qPCR was performed on serum samples to screen a pool of 444 microRNAs at the initial phase, 66 microRNAs at the training phase, and 7 microRNAs at the validation phase. From this analysis, a profile of 4 circulating microRNAs (miR-16-2*, miR-195, miR-497, and miR-2861) was established for diagnosing cervical cancer. The expression of miR-16-2* and miR-497 was upregulated, while expression of miR-195 and miR-2861 was decreased in sera of cervical cancer patients. The 4-microRNA signature showed high accuracy in discriminating individuals with cervical cancer and CIN from healthy controls. These data support the conclusion that this microRNA panel identified in sera of cervical cancer patients can distinguish cervical cancer patients from CIN patients and healthy controls with high sensitivity and specificity. This panel of circulating microRNAs represents novel noninvasive biomarkers for screening and early detection of cervical cancer.
In a study carried out by Zeng et al, a distinct microRNA expression signature was identified for cervical cancer and CIN [
32]. MicroRNA microarrays were performed to compare microRNA expression profiles in cervical cancer, CIN, and normal cervical tissues, and were validated by RT-qPCR. The significance analysis of microarrays of the cervical squamous cell carcinoma tissues relative to the normal cervical tissues showed that miR-15b, miR-16, miR-21, and miR-21*, were the most overexpressed microRNAs, while miR-218 and miR-376 were the most downregulated microRNAs. In CIN III tissues, miR-188-5p, miR-483-5p, miR-663, miR-765, and miR-1300 were the most upregulated microRNAs, while miR-142-3, miR-149, miR-152, miR-218, miR-374b, and miR-376c were the most downregulated microRNAs compared to normal cervical tissues. In the validation analysis, the expression of miR-9, miR-21, and miR-31 were significantly upregulated while the expression of miR-195, miR-199b-5p, miR-218, miR-376a, and miR-497 were significantly downregulated in CIN III and cervical cancer. The most downregulated microRNA was miR-218, which might function as a tumor suppressor in cervical cancer by targeting multiple cancer-related genes, while the most upregulated was miR-21, which likely acts as an oncogene.
MicroRNA expression profiling has also been performed in formalin-fixed paraffin-embedded specimens. How et al, defined a microRNA signature that was prognostic for disease-free survival, which could potentially allow for tailoring of treatment for cervical cancer patients [
33]. However, three different attempts to validate this microRNA signature in an independent cohort of 87 patients with formalin-fixed paraffin-embedded specimens were unsuccessful. Of the 29 significantly differentially expressed microRNAs, a candidate prognostic 9-microRNA signature set was identified in the training set of 79 frozen specimens and only miR-1 and miR-21 were upregulated. The authors proposed three key reasons as to why they could not validate the candidate 9-microRNA prognostic signature for cervical cancer: i) intra-tumor heterogeneity; ii) microRNA expression data from frozen and formalin-fixed paraffin-embedded samples could not be directly compared in this disease; and iii) the current platforms for global microRNA expression profiling are not sufficiently robust. This last point could be resolved by deep-sequencing for microRNA expression profiling. These findings provide an important cautionary tale for application of the 9-microRNA prognostic signature in the clinical oncology setting.
A crucial question in building an understanding of microRNA signatures in cervical cancer is whether the HPV E6/E7-dependent maintenance of the growth of HPV-positive cancer cells is linked to specific alterations of the global microRNA network. To this end, Honegger et al, investigated whether the intracellular and exosomal microRNA compositions of HPV-positive cancer cells are dependent on endogenous E6/E7 oncogene expression [
34]. The investigators performed a comprehensive deep-sequencing to describe the influence of the endogenous E6/E7 oncogene expression on the global microRNA composition of HPV-positive cervical cancer cells, both at the intracellular and at the exosomal level. The microRNA candidates were validated by RT-qPCR analyses. E6/E7 silencing significantly affects ten of the 52 most abundant intracellular microRNAs in HeLa cells (HPV18+), downregulating miR-7-5p, miR-17-5p, miR-186-5p, miR-378a-3p, miR-378f, and miR-629-5p, and upregulating miR-23a-3p, miR-23b-3p, miR-27b-3p, and miR-143-3p. The E6/E7-regulated microRNAs are enriched for species involved in the control of cell proliferation, senescence, and apoptosis, suggesting that they contribute to the growth of HPV-positive cancer cells. In exosomes secreted by HeLa cells, a distinct 7-microRNA signature was identified among the most abundant microRNAs, with significant downregulation of let-7d-5p, miR-7-5p, miR-20a-5p, miR-92a-3p, miR-378a-3p, miR-423-3p, and upregulation of miR-21-5p, upon E6/E7 silencing. Several of the E6/E7-dependent exosomal microRNAs have also been linked to the control of cell proliferation and apoptosis. The composition of exosomal and intracellular small RNA fractions differed in that the relative percentage of microRNAs was approximately 50% lower in exosomes. These findings suggest that HPV E6/E7 oncogene expression determines a signature of 7-microRNAs in exosomes secreted from HeLa cells, leading to significant increase in let-7d-5p, miR-7-5p, miR-20a-5p, miR-92a-3p, miR-378a-3p, and miR-423-3p expression, and decrease in miR-21-5p levels.
Xin et al, performed circulating microRNA expression profiling in order to establish a panel of serum microRNA signature for early detection of CIN [
35]. 126 CIN patients and 60 control individuals were recruited in this cohort study. RT-qPCR assays were carried out to detect the expression level of the panel of microRNAs in participants’ serum samples. The expression levels of 4-microRNAs (miR-9, miR-10a, miR-20a, and miR-196a) were significantly upregulated in the serum samples derived from the CIN patients compared with those from the healthy controls. HPV infection status was significantly correlated with the expression levels of microRNAs. This 4-microRNA signature has a high accuracy in discriminating CIN individuals from healthy controls. These findings support the conclusion that these 4-microRNAs function as oncogenes in cervical cancer and their levels in the serum might be closely correlated with the progression of CIN as well as cervical cancer.
Palatnik et al, developed a novel disease-specific transcriptional signature cell-based assay, that use reporter healthy peripheral blood mononuclear cells (PBMC) cultures, to characterize metastatic transcriptional signatures induced by serum from women with cervical cancer [
36]. In this study, serum samples from women with localized or metastatic cervical cancer were used to drive transcription in healthy PBMC reporters, and the responses were read using genome-wide expression profiling. By in silico analysis, 4 candidate microRNAs (miR-23a-3p, miR-23b-3p, miR-193b-5p, and miR-944) were identified that are known to bind and to inhibit the expression of more than one of the differentially expressed genes. Two of these 4-microRNAs (miR-23a-3p and miR-944) were validated by RT-qPCR in a cohort of women with local and metastatic cervical cancer. These findings indicate that a 4-microRNA disease-specific transcriptional signature induced by cervical cancer patient sera in PBMC cultures has the potential to differentiate patients with local versus metastatic disease.
Sun et al, described a new microRNA associated with cervical cancer [
37]. In this study, the authors investigated the functions and targets of only miR-466 in cervical cancer tissues. A total of 157 participants were summarized, including 56 patients with cervical cancer, 60 patients with CIN, and 49 healthy controls. The expression levels of miR-466 in cervical cancer patients and CIN patients were higher than that in healthy controls. MiR-466 expression was higher in patients with lymph node involvement but was not statistically significantly correlated with International Federation of Gynecology and Obstetrics stages, tumor size, or vascular invasion. These data indicate that the aberrant expression of miR-466 is closely associated with the occurrence and development of cervical cancer but may not predict tumor characteristics.
Aftab et al, investigated the expression of selected microRNAs in paired urine, serum, cervical scrape, and tumor tissue specimens from women with CIN and cervical cancer with the goal of determining whether urine microRNAs could be used as reliable non-invasive biomarkers for an early diagnosis and prognosis of cervical cancer [
38]. Expression of three oncomiRs (miR-21, miR-199a, and miR-155-5p) and three tumor suppressor microRNAs (miR-34a, miR-145, and miR-218) selected by a database search, in specimens obtained from cervical pre-cancer and cancer patients, and healthy controls were analyzed using RT-qPCR. The expression of microRNAs was correlated with various clinicopathological parameters. A combination of miR-145-5p, miR-218-5p, and miR-34a-5p in urine yielded 100% sensitivity and 92.8% specificity in distinguishing precancer and cancer patients from healthy controls and correlated well with microRNA profiles of serum and tumor tissue. The expression of miR-34a-5p and miR-218-5p were found to be independent prognostic factors for the overall survival of cervical cancer patients.
Causin et al, reported molecular signatures of microRNAs in cervical precursor lesions from liquid-based cervical cytology and identified molecular pathways potentially associated with cervical cancer progression [
39]. The samples were divided into CIN III or samples with no precursor lesions of cervical cancer. MicroRNA and gene expression profiling were carried out using the nCounter miRNA Expression Assays (NanoString Technology) and Pan Cancer Pathways (NanoString Technology), respectively. A microRNA target prediction was performed by microRNA data integration portal (mirDIP), and molecular pathway interaction was constructed using Cytoscape. Bidirectional in silico analyses and Pearson's correlation were performed to test associations between genes, differentially expressed microRNAs and cervical cancer progression. They identified nine microRNAs with biomarker potential, two were significantly downregulated (miR-381-3p and miR-4531), and seven were significantly upregulated (let-7f-5p, miR-128-2-5p, miR-130a-3p, miR-202-3p, miR-205-5p, miR-323a-5p, and miR-3136-3p) in CIN III. MicroRNA expression patterns were independent of hr-HPV infection. Four microRNAs (miR-130a-3p, miR-205-5p, miR-381-3p, and miR-4531) were identified that could be used as biomarkers for CIN III in liquid-based cytology samples. MicroRNA and gene expression correlations were identified between miR-130a-3p and CCND1 (negative correlation), miR-205-5p and EGFR (positive correlation), and miR-4531 and SMAD2 (negative correlation). This panel of microRNAs may regulate important molecular pathways of carcinogenesis and have potential as biomarkers to distinguish CIN III from healthy cervical tissue in liquid-based cytology samples.
Qi et al, identified microRNAs associated with poor prognosis in patients with cervical cancer, as well as the possible molecular mechanisms these microRNAs regulate [
40]. The microRNA expression profiles and relevant clinical data from patients with cervical cancer were obtained from The Cancer Genome Atlas (TCGA). The selection of prognostic microRNAs was carried out through an integrated bioinformatics approach. The most effective microRNAs with synergistic and additive effects were selected for validation. Three microRNAs (miR-216b-5p, miR-585-5p, and miR-7641) were identified as exhibiting good performance in predicting poor prognosis through additive effects analysis. The functional enrichment analysis suggested that not only pathways traditionally involved in cancer, but also immune system pathways might be important in determining the outcome of the disease.
Liang et al, reported that a three-microRNAs signature predicts survival in cervical cancer [
41]. They identified differential microRNA expression between cervical cancer and normal cervical tissues by analyzing high-throughput microRNA data downloaded from TCGA database. They evaluated the prognostic values of the differentially expressed microRNAs and constructed a three-microRNA signature that could effectively predict patient survival. A total of 78 differentially expressed microRNAs were identified between cervical cancer tissues and matched normal tissues, including 37 up-regulated microRNAs and 41 down-regulated microRNAs. Results revealed a prognostic function of three microRNAs (miRNA-145, miRNA-200c, and miRNA-218-1). Functional enrichment analysis suggested that the target genes of these three microRNAs may be involved in various pathways related to cancer, including MAPK, AMPK, focal adhesion, cGMP-PKG, Wnt, and the mTOR signaling pathway. These findings suggest that this three-microRNA signature could be used as a prognostic marker in cervical cancer.
A study carried out by Ferrero et al, investigated the pattern and expression levels of microRNAs in four different biofluids representing potential surrogate tissues for diagnostic and screening programs [
42]. The investigators analyzed data from small RNA-sequencing from 125 plasma-derived exosomes, 48 urine, 31 cervical scrapes, and 39 stool samples collected from healthy subjects. 231 microRNAs were identified that were globally unique among all different specimens, while 11 microRNAs (miR-320a, miR-589-5p, miR-636, miR-1273a, miR-3960, miR-4419a, miR-4497, miR-4709-5p, miR-4792, miR-7641-1, and miR-7641-2) were commonly detected in all types of specimens. Plasma exosome samples were characterized by the highest number of specimen-specific microRNAs (155 microRNAs) followed by stool (55 microRNAs), urine (22 microRNAs), and cervical scrape samples (one microRNA). Considering only the specimen-specific microRNAs, miR-122-5p was the most abundantly expressed in plasma exosome samples, while miR-204-5p, miR-655-5p, and miR-4741 were the most abundantly expressed in stool, urine, and cervical scrapes, respectively. In summary, microRNAs identification by small RNA sequencing provides insight into the constitution of the human miRNome in various biospecimens of healthy individuals with potential for future use in cancer screening programs.
Nair et al, identified a microRNA expression pattern in cervical cancer and proposed a role in carcinogenesis for these microRNAs [
43]. The microRNA expression profile in 24 cervical tissue specimens included 18 cervical cancer and 6 normal cervical tissue samples. Next-generation sequencing and microRNA microarray were used for microRNA profiling in cervical cancer cell lines and tissue samples. A microRNA signature was validated by RT-qPCR and the biological significance was elucidated using various in silico analyses. A total of 15 microRNAs were selected for validation. MiR-21-5p, miR-135b-5p, miR-363-3p, and miR-429 were found to be overexpressed in cervical cancer, whereas miR-136-5p, miR-218-5p, miR-377-3p, miR-497-5p, miR-1184, miR-3196, miR-4687-3p, miR-5587-3p, and miR-5572 were downregulated. MiR-424-5p and miR-4307 were excluded and 13 validated microRNAs were defined as the microRNA signature for cervical cancer. In silico analyses revealed that these microRNAs are mainly involved in PI3K-Akt and mTOR pathways. The results indicate that the hr-HPV associated microRNAs influence similar pathways irrespective of HPV type, thus indicating that mechanism of carcinogenesis may be similar in all hr-HPV infections.
Liu et al, reported a potential role of microRNAs as diagnostic biomarkers in the detection of cervical pre-malignant lesions and cancer [
44]. In this study, 582 patients with cervical diseases and 145 control individuals were recruited. Expression levels of 6 microRNAs (miR-20a, miR-92a, miR-141, miR-183*, miR-210, and miR-944) were found to be significantly upregulated in cervical cancer and premalignant lesions compared to normal cervical samples, suggesting that these are oncogenic microRNAs associated with cervical carcinogenesis. These six microRNAs may be used to distinguish patients with cervical pre-malignant lesions or cancer from healthy individuals, and had a good predictive performance, particularly in cervical pre-malignant lesions. Among the six microRNAs studied, miR-141 was the most sensitive and specific marker for differentiating cancer patients. The aberrant expression of these six microRNAs in CIN suggests that these microRNAs might play an important role during the initial or early stages of cervical cancer development. This microRNA signature offers a superior diagnostic value for the early diagnosis of cervical premalignant lesions and cancer due to its high sensitivity and specificity.
In a hr-HPV triage study, Babion et al, evaluated the clinical value of 8 microRNAs in cervical scrapes from HPV-positive women in cervical cancer screening [
45]. Eight microRNAs were identified with altered expression, 3 with increased expression (miR-9-5p, miR-15b-5p, and miR-28-5p) and 5 with decreased expression (miR-100-5p, miR-125b-5p, miR-149-5p, miR-203a-3p, and miR-375). This was verified by RT-qPCR. The expression patterns detected in cervical tissue samples was reflected in cervical scrapes, with five microRNAs showing significantly different expression between controls, CIN III and cancer. For detection of CIN III in hr-HPV-positive cervical scrapes from the screening population, a microRNA classifier using leave-one-out cross-validation was built. This microRNA classifier consisted of miR-15b-5p and miR-375 and detected a major subset of CIN III, as well as all carcinomas with a specificity of 70%. The CIN III detection rate was improved by combining this two-microRNA classifier with HPV16/18 genotyping. This 2-microRNA classifier had 55% sensitivity at 70% specificity for the detection of CIN III and 100% sensitivity for the detection of cervical squamous cell carcinomas and adenocarcinomas. Thus, these differentially expressed microRNAs may provide an alternative molecular triage strategy in hr-HPV-based cervical screening.
Ma et al, identified a microRNA expression signature capable of predicting survival time for cervical squamous cell carcinoma patients [
46]. The expression of 332 microRNAs in 131 patients (training cohort) and 130 patients (validation cohort) with cervical squamous cell carcinoma were analyzed in the TCGA data portal. The microRNA expression signature was identified and subsequently validated. MicroRNA signature-gene target analysis was performed, followed by the construction of the regulatory network, using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). A total of 345 microRNA signature-target pairs were obtained in the microRNA signature-gene target regulatory network, of which 316 genes were targets of miR-378c and miR-642a. The expression signature of these 2 microRNAs (miR-378c and miR-642a) was found to have significant diagnostic and prognostic value and were associated with survival of cervical carcinoma patients. A network of 316 genes was constructed, which were targets of miR-378c and miR-642a. Functional analysis of target genes showed that the MAPK signaling pathway, VEGF signaling pathway and endocytosis mechanism were significantly enriched signal pathways that covered most genes. These results suggest that this 2-microRNA signature has a potential predictive role in the survival time of cervical squamous cell carcinoma patients.
Bayramoglu et al, analyzed and identified the diagnostic and prognostic use of autophagy-associated microRNAs that are differentially expressed in both cervical squamous cell cancer (SCC) cases and high-grade squamous intraepithelial lesion (HSIL) [
47]. A total of 35 HSIL, 35 cervical SCC and 30 healthy controls were enrolled in the study. Total RNA including microRNAs were isolated from the formalin-fixed paraffin-embedded (FFPE) tissue samples and microRNA expression levels were validated by RT-qPCR. Predicted microRNA targets of autophagy related genes were determined using miRNA-target prediction algorithms. MiR-30c, miR-143, miR-372, and miR-375 were markedly downregulated in HSIL and cervical SCC. MiR-130a was significantly upregulated in the cervical SCC group compared to HSIL and control groups. MiR-30a, miR-520e, miR-548c, and miR-372 were determined to be significant diagnostic markers and were significantly associated with the overall survival of cervical SCC patients. Together, these results indicate that autophagy-associated microRNAs are potentially valuable for the diagnosis and targeted therapy in cervical cancer.
Ma et al, evaluated the utility of a plasma microRNA signature as biomarkers in cervical cancer [
48]. Plasma samples of 97 cervical cancer patients and 87 normal controls were used to identify dysregulation of microRNAs in training, testing, and external validation phases. Sensitivity and specificity of identified individual microRNAs and microRNA panels for the diagnosis of cervical cancer were evaluated. Expression levels of specific microRNAs were also examined in plasma exosomes and tissue samples of cervical cancer patients. Four plasma microRNAs (miR-21-5p, miR-146a-5p, miR-151a-3p, and miR-2110) showed up-regulation in cervical cancer patients. A panel of the four microRNAs were constructed as a potential diagnostic marker for cervical cancer. The expression of miR-21-5p and miR-146a-5p were all up-regulated in cervical cancer tissue specimens, whereas miR-146a-5p, miR-151a-3p, and miR-2110 were up-regulated in plasma exosomes. These findings suggest that this four-microRNA signature identified in peripheral plasma is a promising novel biomarker for the diagnosis of cervical cancer.
Shi et al, described a microRNA signature consisting of 7-microRNAs for the progression of cervical cancer [
49]. Cervical cancer expression data were obtained from TCGA database. MicroRNAs significantly differentially expressed between early- and advanced-stage samples were identified by expression analysis. An optimal subset of microRNA signature for pathologic stage prediction was delineated using the random forest algorithm and was used for the construction of a cervical cancer-specific support vector machine (SVM) classifier. The roles of microRNA signature in cervical cancer were analyzed by functional annotation. In total, 44 significantly differentially expressed microRNAs were identified. An optimal subset of microRNAs comprising a 7-microRNA signature was identified, including miR-144, miR-147b, miR-218-2, miR-425, miR-451, miR-483, and miR-486. The microRNA signature was used to construct a SVM classifier and exhibited a good performance in predicting pathologic stages of samples. These data suggest that the 7-microRNA signature could potentially serve as a novel diagnostic and prognostic predictor for cervical cancer.
The biological significance of miR-34a, miR-193a, miR-200a, miR-423, and miR-455 as a microRNA expression signature for the prognosis and diagnosis of cervical cancer, and their association with the clinical outcomes of patients was reported by Bozgeyik et al [
50]. Distinct expression profiles of microRNAs in FFPE tissue samples of patients and healthy controls were evaluated using RT-qPCR. The authors identified that miR-34a, miR-200a, and miR-455 were significantly downregulated in cervical squamous cell carcinoma tissues compared to normal cervix tissue from healthy controls. Both miR-34a and miR-455 showed diagnostic potential on receiver operating characteristic analysis, while miR-200a showed no significance. Notably, low expression of miR-34a was markedly associated with the poor overall survival of cervical cancer patients, and miR-34a was found to be an independent prognostic factor. These results underscore the importance of distinct expression of miR-34a, miR-193a, miR-200a, miR-423, and miR-455 as a microRNA signature in cervical squamous cell carcinoma.
Qiu et al. reported a circulating microRNA-based signature for use in diagnosis and prognosis of early-stage cervical cancer [
51]. 112 patients with early-stage cervical cancer, 45 patients with CIN and 90 healthy subjects were recruited. Compared to the normal controls, the expression level of miR-21 was increased, while the levels of miR-125b and miR-370 were decreased in cervical cancer in both Gene Expression Omnibus GSE30656 and TCGA cohort. The expression levels and diagnostic value of microRNAs were validated by RT-qPCR. Their diagnostic and prognostic value for early-stage cervical cancer were further explored. In early-stage cervical cancer patients, compared to patients with CIN and healthy subjects, serum miR-21 was increased, while serum miR-125b and serum miR-370 were decreased. In addition, combining these molecules yielded good performance for differentiating early-stage cervical cancer from CIN or healthy subjects. Moreover, a strong positive association was found between serum miR-21 and lymph node metastasis as well as recurrence of early-stage cervical cancer. Conversely, negative correlations with lymph node metastasis and recurrence were found for serum miR-125b and serum miR-370. These data indicate that combining serum miR-21, miR-125b, and miR-370 as a microRNA-based signature is a promising tool for the detection and prognostication of early-stage cervical cancer.
To synthesize understanding of the role of microRNAs in the progression of cervical cancer, Causin et al, performed a systematic review of cervical cancer microRNA studies [
52]. From 27 published studies, totaling 1721 cases and 1361 noncancerous control tissue samples, 26 differentially expressed microRNAs (DEmiRNAs) were identified in different International Federation of Gynecology and Obstetrics stages of cervical cancer development, all of which were confirmed by RT-qPCR. There were19 downregulated (miR-1, miR-107, miR-132, miR-139-3p, miR-143, miR-195, miR-335-5p, miR-337-3p, miR-361-5p, miR-383-5p, miR-411, miR-424-5p, miR-433, miR-545, miR-573, miR-874, miR-1284, miR-2861, and miR-3941) and seven upregulated (miR-31, miR-92a, miR-93, miR-96-5p, miR-199b-5p, miR-200a, and miR-224) microRNAs in different stages of cervical cancer compared to normal tissue. Some of the dysregulated microRNAs were associated with specific stages of cervical cancer development. The results indicated that DEmiRNAs in different stages of cervical cancer were functionally involved in several key hallmarks of cancer, such as evading growth suppressors, enabling replicative immortality, activating invasion and metastasis, resisting cell death, and sustaining proliferative signaling. Some of the stage-specific microRNAs can also be used as biomarkers for cancer classification and monitoring the progression of cervical cancer.
Cao et al, identified a three-microRNA serum signature for cervical cancer diagnosis [
53]. A total of 29 candidate microRNAs were identified and validated by RT-qPCR. Four microRNAs (miR-20a-5p, miR-21-5p, miR-196a-5p, and miR-218) proposed by previous literature were also analyzed in this study. Interestingly, miR-20a-5p and miR-122-5p showed a significant trend of upregulation in the serum of cervical cancer patients compared with healthy control patients, while miR-133a-3p was downregulated. A subgroup analyses was further performed to evaluate the association between the identified signature and patients’ clinicopathological parameters including the tumor node, metastasis stage and histological type. The three microRNAs were not specific to histological subtype since none of them were differentially expressed between squamous cell carcinoma and adenocarcinoma patients. The expression levels of miR-20a-5p and miR-122-5p were consistently upregulated in cervical cancer patients at early stages (stage I or stage II) and advanced stages (stage III or IV) when compared with control patients. In exosome samples of cervical cancer patients, the expression of exosomal miR-20a-5p and miR-122-5p was significantly increased. These data suggest that these three serum microRNAs (miR-20a-5p, miR-122-5p, and miR-133a-3p) might serve as potential diagnostic biomarkers for cervical cancer patients.
With the purpose of describing the microRNA expression profile in patients receiving treatment, we identified a set of microRNAs that can be used as a molecular signature to predict the pathological response in patients with locally advanced cervical cancer receiving radiation and chemotherapy treatment [
54]. To this end, 41 patients diagnosed with locally advanced cervical cancer were invited to participate. MicroRNA expression profiling was performed by means of a miScript microRNA PCR array. MicroRNA expression profiling identified 101 microRNAs that showed significant differences between pathological non-responders and complete responders. Interestingly, we found that three microRNAs (miR-31-3p, miR-144-3p, and miR-3176) were overexpressed whereas 97 were downregulated in non-responder compared with complete responder patients. Seven differentially expressed microRNAs were selected, and their expression patterns were confirmed in the validation RT-qPCR assays. Thus, miR-31-3p, miR-100-5p, miR-125a-5p, miR-125b-5p, miR-200a-5p, miR-342, and miR-3676 were proposed like as the microRNA signature significantly associated with clinical response. Thus, we report a subset of 101 microRNAs that permit the accurate classification of locally advanced cervical cancer patients regarding clinical response, from which 7-microRNA were validated. This seven-microRNA signature could be used as a molecular biomarker of resistance to radiation and chemotherapy treatment.
The panel of microRNAs that have evidence of roles in the genomic signature in cervical cancer are summarized in
Table 1.
3. Regulatory Genetic Networks Modulated by MicroRNAs and Their Target Genes in Cervical Cancer
In recent years, there has been great effort to elucidate the precise mechanisms involved in carcinogenesis, as well as to identify which genes are involved. Many researchers have attempted to understand how microRNAs act to regulate target genes and what their roles are in the carcinogenesis process. These microRNAs can form feedback or feedforward loops, which play critical roles in various biological processes by regulation of their target genes. Thus, failure of the delicate balance of these regulation circuits between microRNAs and their target genes might result in cancer development.
As the number of reported deregulated microRNAs in cervical cancer has grown, so has the study of regulation of their target genes. The expression profiles of microRNAs and mRNAs and the gene regulation network in cervical tumorigenesis have been analyzed in order to identify candidate molecular markers and key tumorigenic genes in cervical cancer. In a study performed by Ma et al, microRNA and mRNA expression microarrays were used to detect the expression of microRNAs and mRNAs in normal and cancer cervical tissues, and a microRNA-gene network was constructed [
55]. The investigators used the TargetScan 5.0 database to predict the target genes of the microRNAs, analyze their intersection with differentially expressed mRNAs and negatively correlate the intersection with microRNAs. Bioinformatic approaches were used to analyze functions and pathways of the target genes and establish microRNA-gene networks. The analysis showed that 29 microRNAs and 2036 mRNAs were differentially expressed in normal compared with cervical tumor tissues. Among them, 13 microRNAs and 754 mRNAs were up-regulated in cervical tumor tissues and 16 miRNAs, and 1282 mRNAs were down-regulated. Genes targeted by down-regulated microRNAs and genes target by up-regulated microRNAs were involved in 415 and 163 functions, respectively, and in 37 and 17 significant pathways, respectively. A microRNA-gene network was constructed with these data and determined that the microRNAs that regulate the largest number of target genes are miR-15a, miR-20b, and miR-106b. Theses microRNAs are pivotal in the network and have the strongest regulatory effects on differential gene expression in cervical cancer. These finding suggest that the differentially expressed microRNA-gene network plays an important role in many biological functions and signal transduction during the cervical carcinogenesis process.
Sharma et al, completed a systematic analysis of known dysregulated microRNAs involved in cervical cancer and constructed a microRNA-mRNA network in order to identify critical microRNA targets for potential future therapeutic strategies [
56]. The microRNAs reported to be dysregulated in cervical cancer were collected and their targets were predicted using TargetScan, PicTar and miRanda databases. These targets were subsequently compared with previously curated gene datasets involved in cervical cancer to derive the putative target dataset. The authors compared network properties (composed of degree, betweenness centrality, closeness centrality and clustering coefficient) of the putative, validated, and human protein-protein interaction networks. Based on the topological properties, genes were ranked, and it was observed that the gene targets BIRC5 (survivin), HOXA1 and RARB with high Novoseek score in Genecards were enriched in cervical cancer. BIRC5 is an anti-apoptotic protein while HOXA1 and RARB are transcription factors which play critical roles in altering the level of cell cycle and apoptosis-associated proteins. The microRNA-mRNA network was constructed, and it was found that miR-30b and miR-203 may target these genes. This analysis indicates that the genes BIRC5, HOXA1 and RARB are critical targets with an important regulatory role in cervical cancer pathogenesis.
Wang et al, described regulatory genetic networks in cervical cancer which were constructed from databases of differentially expressed microRNA and related genes [
57]. Three regulatory genetic networks were generated; the first was the differentially-expressed network which includes differentially expressed genes, differentially-expressed microRNAs and host genes of differentially expressed microRNAs. The second network was constructed from related genes, related microRNAs and host genes of cervical cancer-related microRNAs. The third network was the global network, which consists of all the elements extracted from the basic source data. In the first network, seven genes and ten microRNAs were experimentally validated as differentially expressed in cervical cancer. A sub-network centered around PTEN was the principal component. TWIST1, miR-214, PTEN and miR-21 function together as an ordered chain of control. MiR-21 targets its regulator PTEN, which formed a self-adapting feedback loop, creating a balance mechanism with STAT3 and miR-21 in the system as components. In another partial network centered on miR-143, TP53 and TGF-B1 jointly have been shown to regulate miR-143. The second, related, network is of a much larger scale and higher complexity than the differentially expressed network. Certain factors are predominant, including microRNAs such as miR-21, miR-23b, miR-34, miR-143, let-7c, and transcription factors of PTEN, TP53, TP63, c-Myc, and k-Ras. Two more self-adapting feedback mechanisms were identified involving let-7c, miR-34, and c-Myc. In addition, c-Myc’s overexpression could alter the suppressive function of let-7c. Meanwhile, another tumor suppressor, miR-34, has a local balance adjustment system with c-Myc as well. In the third network, interactions involving the previously identified transcription factors and their base sequences of 1000-nt were evaluated; these transcription factors were integrated with host genes and differentially expressed microRNAs to include 1000-nt transcription factors in the network system of cervical cancer. Self-adapting feedback was assessed and was found to exist between NF-kB and miR-21. NF-kB regulates miR-21, miR-214, and let-7b. MiR-21 and miR-214 are core biological factors in the cervical cancer network and their appearance here supports the importance of these microRNAs. With the help of miR-21 and mir-214, NF-kB participates in the sub-system centered on PTEN and miR-21. The findings suggest that this method of analysis could be useful to identify more core factors, other parallel networks, and relevant motifs in cervical cancer development and/or other tumorigenesis processes.
Mo et al, developed a SIG (Stochastic process model for Identifying differentially co-expressed Gene pair) ++ algorithm and subsequent analysis strategy to systematically construct microRNA differential regulation mediated networks in the progression of CIN [
58]. They performed a transcriptome analysis of microRNAs and mRNAs for a total of 24 cervical samples in three stages (normal, CIN I, and CIN III) of CIN progression, and they proposed the SIG++ algorithm to detect the microRNA-mRNA pairs with significant regulation change. From this analysis, they proposed the definitions of efficient pair, efficient target, and related effector biological process, as the elemental steps to construct the microRNA differential regulation network. For the course of disease progressing from normal tissue to CIN I stage, and from CIN I stage to CIN III stage, microRNA differential regulatory networks were constructed, founded on the knowledge of human Gene Ontology biological processes to detect efficient targets and related effector biological processes. The resultant data describe pathway enrichment analysis of microRNA regulation. For each stage, the specifically existent microRNA-mRNA regulations are enriched in inflammation; for normal stage, the specific microRNA regulations are particularly enriched in sodium ion transport and glycolysis; from CIN I stage, microRNA regulations have been highly enriched in cell migration and differentiation. For CIN III stage, the specific microRNA regulations are enriched in virus integration. The authors present the concept of microRNA’s differential regulation, describe a novel algorithm for how to identify them during stages of disease development, and construct microRNA differential regulation networks with instructive biological meanings. The relevance of these findings is that regulatory genetic networks enable a better understanding of CIN progression.
To further characterize the role of microRNAs in the progression of cervical cancer, He et al, performed a meta-analysis and gene set enrichment analysis to identify the DEmiRNAs in a microRNA-mRNA interaction network [
59]. In this study, from 85 published reports, which included 3,922 cases of CIN or cancer and 2,099 noncancerous control tissue samples, 63 DEmiRNAs were identified in different stages of cervical cancer development (CIN I-III and cervical cancer). To illustrate the impact of microRNAs on the pathogenesis of cervical cancer, a microRNA-mRNA interaction network of selected pathways was built by integrating viral oncoproteins, dysregulated microRNAs and their predicted and validated targets. The results indicated that the deregulated microRNAs at the different stages of cervical carcinogenesis were functionally involved in several key cancer related pathways, such as cell cycle, p53 and Wnt signaling pathways. These data indicate that some of the stage-specific microRNAs can be used as biomarkers for cancer classification and monitoring the progression of cancer.
Kori et al, performed a meta-analysis of cervical cancer-associated transcriptome data and identified reporter biomolecules at the RNA (microRNA- mRNA), protein (receptor, transcription factor), and metabolite levels through the integration of gene expression profiles with genome-scale biomolecular networks [
60]. Considering the intertwined structure of signaling, regulatory and metabolic processes within a cell, they employed three genome-scale biomolecular networks (protein-protein interaction (PPI), metabolic, and post-transcriptional regulatory networks) to analyze cervical cancer. Accordingly, a meta-analysis of the cervical cancer associated transcriptomic datasets was performed by taking into consideration five independent studies and a total of 236 samples, and the core information about differentially expressed genes (DEGs) was obtained by statistical analyses. Gene set over-representation analyses were performed on core DEGs to identify significantly enriched pathways and Gene Ontology terms. The differential expression profiles of all reporter biomolecules were cross-validated in independent RNA-Seq and microRNA-Seq datasets, and the prognostic power of several reporter biomolecules was also demonstrated. In summary, this study reports candidate biomolecules that can be considered as diagnostic/prognostic biomarkers or potential therapeutic targets for further experimental and clinical trials for cervical cancer.
Functional genomics data provides opportunities to study aberrant microRNA-mRNA interactions. Paul et al, developed an algorithm, termed relevant and functionally consistent microRNA-mRNA modules (RFCM3), to elucidate these interactions in cervical cancer [
61]. RFCM3 integrates microRNA and mRNA expression data in cervical cancer for identification of potential microRNA-mRNA regulatory modules. It selects a set of microRNA-mRNA modules by maximizing relation of mRNAs with microRNA and functional similarity between selected mRNAs. Later, using the knowledge of the microRNA-microRNA synergistic network, different modules are fused and finally a set of modules are generated containing several microRNAs as well as mRNAs. The effectiveness of the proposed approach over other existing methods was demonstrated on microRNA and mRNA expression data of cervical cancer with respect to enrichment analyses and other standard matrices. The RFCM3 module was found to generate more robust, integrated, and functionally enriched microRNA-mRNA modules in cervical cancer. These data suggest that the RFCM3 module is a powerful tool to study biological pathways containing multiple microRNAs and mRNAs in cervical cancer.
Liu et al, reported a microRNA-mRNA regulatory network with prognostic value in cervical cancer [
62]. MicroRNA and mRNA expression profiles were downloaded from the TCGA database. DEmiRNAs and DEmRNAs were obtained from "Empirical Analysis of Digital Gene Expression Data in R (EdgeR)" package. With Cytoscape software, a PPI network was established to identify hub genes which were used to build a microRNA-hub gene network. A prognostic signature based on hub genes was constructed by Cox regression analysis, and its prognostic value was assessed by a nomogram. The relationship between immune cell infiltration and the three genes in the prognostic model was investigated by using the CIBERSORT algorithm. 5096 DEmRNAs and 114 DEmiRNAs were screened out between healthy cervix and cervical cancer tissues. Then, 102 target DEmRNAs of upregulated DEmiRNAs and 150 target DEmRNAs of downregulated DEmiRNAs were identified. The network analysis identified 10 top upregulated and 10 top downregulated hub genes. The upregulated DEmiRNAs were mostly enriched in pathways in cancer, proteoglycans in cancer, and focal adhesion, while the downregulated DEmiRNAs were focused on pathways in cancer, cell cycle, and viral carcinogenesis. The microRNA-hub gene network showed that most hub genes could be potentially modulated by miR-23b-3p, miR-106b-5p, and miR-200c-3p. To validate these candidate DEmiRNAs, their prognostic roles in cervical cancer were assessed by using the ‘‘survival’’ package. Four upregulated DEmiRNAs (miR-106b-5p, miR-200c-3p, miR-210-5p, and miR-425-5p) and 13 downregulated DEmiRNAs (miR-23b-3p, miR-29c-5p, miR-99a-5p, miR-101-3p, miR-143-5p, miR-145-3p, miR-145-5p, miR-181c-5p, miR-502-5p, miR-504-5p, miR-505-5p, miR-532-5p, and miR-6507-5p) were associated with cervical cancer prognosis. Furthermore, a prognostic gene signature was established based on enhancer of zeste homolog 2 (EZH2), fms-related tyrosine kinase 1 (FLT1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). These three genes were also found to be related to the infiltration of six types of immune cells, including dendritic cells, macrophages M0 and M1, mast cells, and monocytes. In summary, this bioinformatics analysis improves our understanding of the microRNA-mRNA network regulating cervical cancer development and proposes a prognostic gene signature for cervical cancer patients.
Comprehensive bioinformatics analyses have been used to integrate microRNA profiling and gene expression to characterize the detailed molecular mechanisms of cervical cancer. Differential expression analysis, enrichment analysis, PPI, network analysis, lncRNAs (long non-coding RNAs), and ceRNA (competing endogenous RNAs) network analysis have been studied to unravel the microRNAs, genes and pathways related to the cervical carcinogenesis process. The lncRNAs and ceRNAs are a class of RNA molecules with transcripts over 200 nucleotides in length, which do not encode proteins. Instead, they regulate the expressions of genes at various levels including epigenetic and transcriptional. In these types of studies, the differentially expressed microRNAs and differentially expressed genes regulatory network, PPI network and transcription factor, target regulatory network has been constructed. Furthermore, lncRNAs associated with DEmiRNAs were analyzed to establish the lncRNA-microRNA-target-ceRNA network. To interpret the biologic interactome of these regulatory networks, Zong et al [
63] used the Gene Ontology biological process and KEGG pathway for enrichment analysis. 18 DEmiRNAs and 620 DEGs were identified. DEmiRNAs were enriched in 35 KEGG pathways, including PI3K-Akt signaling pathway (involving miR-451a). DEGs were enriched in various functions, such as DNA replication (involving E2F7) and angiogenesis (involving EREG). There were 120 nodes and 216 interaction pairs in the DEmiRNA-DEGs regulatory network, and miR-106b-5p had the greatest degree. EREG and E2F7 were regulated by miR-148a-3p and miR-451a, respectively. Interestingly, E2F7 was identified in the transcription factor-target regulatory network regulating CDC6. There were 15 lncRNAs, 11 microRNAs and 90 DEGs in the ceRNA network. Specially, miR-148a-3p interacted with lncRNA HOTAIR in the ceRNA network. These data identify E2F7, EREG, miR-106b-5p, and miR-451a as genetic factors related to cervical cancer development.
Mei et al, reported reliable and effective biomarkers for diagnosis and prognosis of cervical cancer [
64]. Microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database to search potential microRNA-mRNA networks in cervical cancer. GO term enrichment and KEGG pathway analyses were conducted to reveal the underlying functions and pathways of DEGs. A total of 209 DEGs, including 115 upregulated genes and 94 downregulated genes, were identified using the Robust Rank Aggreg R package in the differential expression analysis. Small molecule drugs were screened in the Connectivity Map database based on DEGs, and three, thioguanosine, apigenin, and trichostatin A, were identified as potential new drugs to treat cervical cancer. 9 upregulated microRNAs (let-7c-5p, miR-10b-5p, miR-101-3p, miR101-5p, miR-195-5p, mir-204-5p, miR-377-5p, miR-497-5p, and mir-6507-5p) and 12 downregulated microRNAs (miR-7-5p, miR-15b-5p, miR-16-2-3p, miR-18a-3p, miR-18a-5p, miR-31-5p, miR-130b-3p, miR-130b-5p, miR-183-5p, miR-203a-3p, miR-3934-5p, and miR-4638-3p) were identified as candidate DEmiRNAs. Based on the negative regulation between DEmiRNAs and target genes, a regulatory network between the DEmiRNAs and their corresponding genes was constructed. Two microRNAs (mir-101-3p and mir-6507-5p) were identified as hub DEmiRNAs associated with the prognosis of cervical cancer patients. A five-gene signature (APOBEC3B, DSG2, CXCL8, ABCA8, and PLAGL1) was selected by prognosis signature analysis and might be used as an independent prognostic index to stratify risk subgroups for patients with cervical cancer. The risk score of the prognostic model was also found to be associated with immune cell infiltration, including mast cell activation, resting natural killer cells, resting dendritic cells, T regulatory cells (Tregs), and T follicular helper cells. These findings indicate that the microRNA-mRNA regulatory network and this prognostic model are of great clinical significance in the prognosis and treatment of cervical cancer.
4. MicroRNA-mRNA Regulatory Modules in Cervical Cancer
The regulation of genes by microRNAs has drawn particular attention. The identification of microRNA-mRNA regulatory modules will aid in deciphering the aberrant transcriptional regulatory networks in cervical cancer, and though it represents a bioinformatics challenge, it will advance understanding of the role of known biomarkers like microRNAs and mRNAs in various pathways in cervical cancer.
There are several techniques reported in the literature for integration analysis of microRNA-mRNA networks [
65]. Correlation-based techniques have several disadvantages. These techniques assume that one microRNA affects only one mRNA, an assumption that is no longer accurate [
66]. Linear modeling-based techniques have been developed to address this point. In this approach, mRNA expressions are modeled as linear combinations of microRNAs, and the Bayesian algorithm is applied to discover hidden microRNA targets. The linear modeling techniques also use different distribution techniques, integrating sequence and structure information from previous study [
67]. Another approach used for the integration of microRNA and mRNA interactions is the Bayesian network technique [
68]. This method performs an integrated analysis using differentially expressed microRNAs and mRNAs through Bayesian network technique. Due to the large amount of biological data available, it is necessary to develop a scalable solution, and the Bayesian network-based machine learning model could be a valid strategy [
69].
All events that take place in a living system happen within a specific biological organization. This concept has motivated the development of statistical approaches to microRNA and mRNA integration. Investigators have evaluated different microRNA-mRNA expression data using statistical approaches, without any other prior knowledge, and developed a method to distinguish different tissues. Using a similar approach, Nersisyan et al, developed a new tool to generate microRNA-gene-transcription factor networks [
70]. Another method that generates microRNA-mRNA networks is the probability learning-based technique. In this approach, the interaction probabilities of known microRNA-mRNA pair are estimated [
71]. However, for this operation to be performed robustly and affectively, more than one source of information is needed. The non-negative matrix factorization technique is another important machine learning method. In this technique, microRNA and gene expression profiles are jointly analyzed from different information sources and additional network data are simultaneously integrated to generate significant microRNA-mRNA groups [
72].
Additional approaches use rule induction-based techniques based on information theory. Generally, data obtained from more than one data source need to be integrated with a rule induction-based technique to find microRNA-mRNA group modules [
73]. Gamberger et al, used the CN2-SD system as the rule generation system to identify microRNA-mRNA groups [
74]. The main characteristic of this tool is the microRNA-mRNA grouping methodology. The maTE platform adopts a biological grouping methodology, while miRcorrNet platform uses correlation information to generate the groups. Additionally, there are other techniques that differ from previous approaches. For instance, MiR Module Net generates microRNA-mRNA group integration by using statistical information [
75].
A useful tool is miRNet (
https://www.mirnet.ca/), an easy-to-use, web-based platform designed to help elucidate microRNA functions by integrating users’ data with existing knowledge via network-based visual analytics [
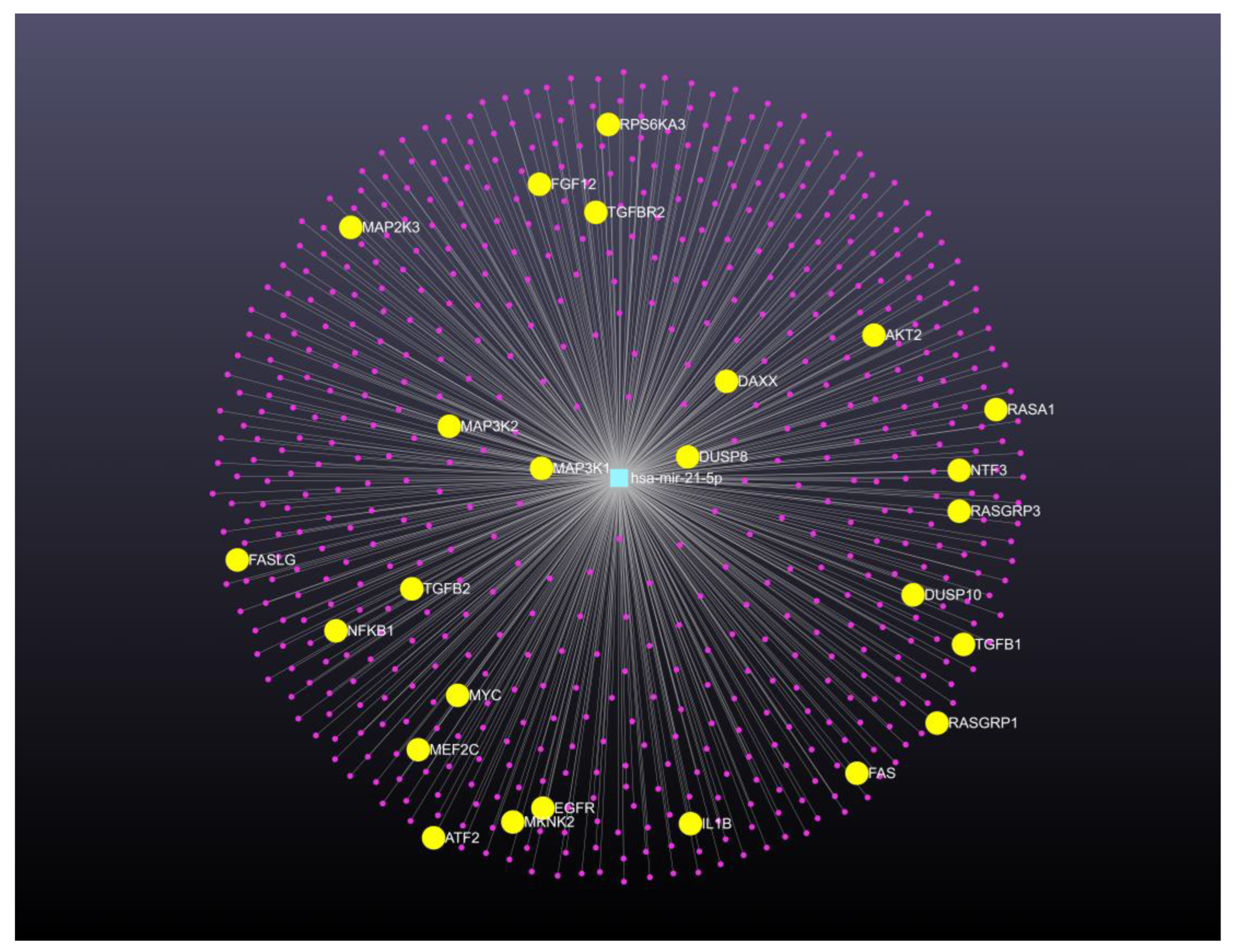
76]. To address emerging bioinformatics needs and challenges, miRNet version 2.0 allows users to easily create complex microRNA-centric networks for systems level interpretation of microRNA functions and gene regulation. With the goal of analyzing the pathways of regulation by microRNA-gene networks in cervical cancer, we used the features in the miRNet 2.0 website platform. We used data from cervical cancer from Table I to identify the role of the relevant oncomiR miR-21-5p in the pathogenesis of cervical cancer. In this analysis, the microRNA target analysis is performed by searching relevant microRNA-gene networks, even if the platform has other advanced functions like a transcription factor-gene database. The network was manually built by using Cytoscape. We reconstructed and visualized the network in miRNet 2.0 by using the microRNA module, for oncomiR for miR-21-5p, which is the most overexpressed microRNA in cervical cancer.
The resulting network is comprised of 1 node (genes: 612; microRNAs: 1) and 612 edges. For better visual exploration, a degree cutoff of 1.0 was applied. As shown in
Figure 1, the microRNA-gene regulatory network is displayed at the center of the network viewer page in “default layout”. It illustrates several interactions between microRNAs (center zone) and target genes (middle layer). The nodes are sorted by degree centrality measures in the node explorer table. In this case, miRNet 2.0 confirmed the detection of important targets according to their degree measures. We also performed functional enrichment and module analysis on the whole network. The analysis shows other networks with different pathways, for instance from MAPK signaling pathway, after the edge thickness is manually increased. The results of the functional enrichment analysis using the KEGG database are displayed in the function explorer table. The MAPK signaling pathway analysis has 25 hits (AKT2, ATF2, DAXX, DUSP8, DUSP10, EGFR, FAS, FASLG, FGF12, IL1B, MEF2C, MAP3K1, MAP2K3, MAP3K2, MKNK2, MYC, NF-KB1, NTF3, RASA1, RASGRP1, RASGRP3, RPS6KA3, TGF-B1, TGF-B2, and TGFBR2), which may be potential targets regulated by miR-21-5p and have been supported by several publications. Interestingly, a deep analysis allowed us to identify MAP3K1, which is involved in the MAPK signaling pathway, as a bioinformatic target of miR-21-5p. A search in NCBI showed that human MAP3K1 regulation by miR-21-5p has not yet been currently reported experimentally. The protein encoded by the MAP3K1 gene is a serine/threonine kinase and is part of signal transduction cascades, including the ERK and JNK kinase pathways, as well as the NF-kappa-B pathway, which are relevant in carcinogenesis processes. MAP3K1 protein is activated by autophosphorylation and requires magnesium as a cofactor in phosphorylating other proteins. In addition, MAP3K1 has E3 ligase activity in its N-terminus and phospho-kinase activity in its C-terminus [
77]. Compared to the original network, this module is much more digestible while keeping the important nodes and connections. This case example highlights that with only a few mouse clicks, we can easily create comprehensive regulatory networks to gain a more holistic view of microRNA mediated regulation as well as to identify important modules for more in-depth analysis. In summary, this platform is easy to use to identify as yet unreported target genes that may be functionally regulated by microRNAs.
5. Gene Therapy Clinical Trials with MicroRNAs for Uterine Cancers
If lesions are not identified and treated at the pre-cancerous stages, treatment options are limited for women with metastatic or recurrent cervical cancer. Unfortunately, only up to one third of patients with metastatic and recurrent disease will respond to radiation or chemotherapy, and these responses are short-lived, on the order of months. Gene therapy strategies targeted to HPV products represent new treatment options for cervical cancer and also other tumors in which HPV participates as a cancer promoter. Currently, it is well known that HPV E6 and E7 interact with a plethora of cellular proteins, both nuclear and cytoplasmic, and participate in molecular pathways involved in the activation and establishment of the tumor phenotype. Hence, it is feasible to design experimental strategies aimed to block the expression of viral oncoproteins. In this regard, several efforts are focusing on the development of HPV therapeutic vaccines based in viral vectors, such as Herpes simplex virus, adenoviral and adeno-associated viral vectors, lentiviral vectors, and measles virus; as well as in non-viral systems, such as shRNA, siRNA, microRNA, naked plasmid DNA-based vaccines, lipid-based and polymeric nanoparticles, CRISPR/Cas9, HPV peptide and proteins, specific tumor, and dendritic cells. Some of these approaches have been designed to deliver target gene via lentiviral vectors [
78,
79].
The standard treatment for advanced or metastatic cervical cancer is concomitant cisplatin-based and radiotherapy regimens. These approaches have been found to deliver better clinical outcomes as compared to either chemotherapy alone or combined with hysterectomy [
80,
81]. However, primary or acquired chemo- and/or radio-resistance is a serious treatment failure in which patients experience systemic toxicity and side-effects that contribute greatly to disease recurrence, progression, and mortality. Cisplatin-based concurrent chemotherapy mechanisms of action include DNA damage induction and activation of various genes involved in DNA damage repair. Radiotherapy is a therapeutic modality that uses ionizing radiation to induce damage in unwanted cells. The main goal of these approaches consists in delivering a precise dose of drug or radiation in a target volume, such as tumor, promoting the tumor cells eradication with as minimal damage as possible to surrounding normal tissues. Besides the chemo- and/or radiotherapy dose are standardized among patients, local recurrences are common and can occur even when modern techniques are used [
82,
83].
The concept of personalized medicine has become increasingly popular, and it is even considered essential in multiple clinical conditions. In the oncology setting, there is increasing evidence that microRNAs can influence the way that cells respond to drugs or ionizing radiation, making them more chemo- or radio- sensitive or resistant through several specific pathways. These include modifying DNA repair pathways which interfere with cell cycle checkpoints activation, tumor microenvironment and apoptosis. Some microRNAs are involved in controlling cell cycle progression, tumor microenvironment, apoptosis, and chemo- radio-related signals pathway. Thus, microRNAs may be viewed as promising biomarkers capable of predicting chemo or radiation response and used to develop a customized treatment plan for each patient, ultimately opening a new therapeutic window for personalized intervention in cancer patients. The identification of new competitive and accurate biomarkers and genomic signatures is needed to improve the accuracy of diagnosis and prognosis and to identify the optimal treatment for cervical cancer. The recent development of microRNA targeted therapy has provided new research opportunities to identify novel strategies that can modify a patient's tumor biology and therapeutic response, which will facilitate the development of new targeted strategies for cervical cancer treatment.
Table 2 summarizes in systematic format information about uterine cancer microRNA gene therapy clinical trials worldwide. The data were compiled from the official clinical trials database from the National Library Medicine from National Institute of Health US [
84]. In many clinical trials, safety and tolerability were evaluated in combination with gene therapy strategies. Several studies are active, and/or recruiting participants, while others are completed.
The ClinicalTrials.gov identifier: NCT04087785 identifies a subset of microRNAs with prognostic value associated with early-stage cervical cancer patients treated with radical hysterectomy and bilateral pelvic lymphadenectomy, with positive lymph node metastasis. The identification of global microRNA profiles was performed using GeneChip miRNA 3.0 array from Affymetrix. FFPE tissue samples of patients with a diagnosis of early-stage cervical cancer treated by radical hysterectomy with lymphadenectomy were collected. To obtain the microRNA profile, the processed samples were divided into 2 groups, samples with and without lymph node metastasis. Differentially expressed microRNAs were identified using a cutoff value of p <0.01 and a fold change of 1.5. The microRNAs that met these criteria were classified as over-expressed or under-expressed. The results have not been posted for this study.
The ClinicalTrials.gov identifier: NCT03824613 is a prospective feasibility study with the addition of a retrospective cohort study on the expression of microRNA in urine from endometrial cancer patients. The data will allow determine how well microRNA correlates with final histology, for example, if microRNA profile may determine the subtype of endometrial cancer. The results have not been posted for this study.
The ClinicalTrials.gov identifier: NCT04845425 is an observational study with a retrospective cohort. The authors propose evaluating the microRNA expression profile to identify novel potential biomarkers to better stratify endometrial cancer patients, considering the molecular alterations of endometrial cancer from TCGA. In addition, the authors will integrate molecular results with clinical-pathological data. Although the TCGA has identified four distinct prognostic groups of endometrial carcinomas based on molecular alterations, carcinomas with no specific molecular profile present a wide variability in molecular alterations and biological aggressiveness. Given that the study aims to evaluate the miRNA expression profile to identify novel potential biomarkers to better stratify the endometrial carcinoma patients, considering the molecular status [
85].
The ClinicalTrials.gov identifier: NCT03776630 is a non-randomized, open label and multicenter project to study ovarian and endometrial cancer. For endometrial cancer, the aim is to validate the 5-microRNA index assessed in plasma samples as a diagnostic marker to assess the risk of lymph node metastases. For ovarian cancer, the data aim is to validate the previous finding on the prognostic value of the pre-/post-treatment variation of miR-200b plasma concentrations with regards to progression-free survival. The study will investigate the links of the 5-microRNA index with classical predictors of lymph node involvement in the context of endometrial cancer. A second aim is to validate multiplexed homogenous microRNA detection based on RCA-FRET compared to conventional RT-qPCR in plasma samples. The results have not been posted for this study.
In the ClinicalTrials.gov identifier: NCT01119573, microRNAs are studied as biomarkers in tissue samples from patients with stage I or stage III endometrial cancer. The objective is to identify microRNA expression patterns associated with lymph node metastasis in samples from patients with endometrial cancer. The banked tumor tissue specimens have been analyzed for microRNA expression profiling by microarray analysis and RT-PCR assays. The results have not been posted for this study.
The ClinicalTrials.gov identifier: NCT04010487 explores the driving genes and the molecular mechanism of malignant transformation of adenomyosis. The eutopic endometrium, and normal adenomyosis tissues samples were acquired from FFPE tissue from patients with pathologically conformed endometrial carcinoma arising in adenomyosis (EC-AIA). The normal eutopic endometrium and normal adenomyosis tissue were obtained by laser microdissection. The driving genes and potential molecular mechanism of EC-AIA will be identified by the technology of whole exome sequencing and transcriptomics (RNA-sequencing). Furthermore, the alteration of RNA expression, including mRNA, microRNA, and lncRNA will be compared between eutopic and ectopic endometrium, and cancer tissues by transcriptome sequencing. The results have not been posted for this study.
The ClinicalTrials.gov identifier: NCT02983279 aims to determine whether short term calorie restriction will affect tumor biology in biopsy proven breast, endometrial or prostate cancers, which will positively impact biomarkers including serum miR-21, an oncomiR known to impact cancer outcomes. The changes in miR-21 expression assessed in serum will be evaluated by a two-sided paired t-test. Initial cancer biopsy specimen for genomic analysis will be comped to definitive surgical specimens. Moreover, the trial will investigate measurable changes induced by caloric restriction on both patient and tumor characteristics. Patient’s nutritional status will be assessed, their caloric needs will be calculated, and local recurrence, progression free survival, distant metastases and overall survival will be assessed. The results have not been posted for this study.
The ClinicalTrials.gov identifier: NCT05292573 is a study that will prospectively enroll a total of 1000 patients with simple hyperplasia/complex hyperplasia without atypia and evaluate progression to endometrial cancer with or without metformin intervention by linking with national health databases from Health and Welfare Data Science Center in Taiwan. The analysis of whole genome sequencing data of those patients who progressed and did not progress will carry out. The microRNA panel in tissues and sera will be analyzed from CMRPG3G1511-3 data of microRNA panel on endometrial cancer progression rate.
The ClinicalTrials.gov identifier: NCT03742869 is a multi-omics study of uterine cervical adenocarcinoma patients with and without HPV infections. Peripheral venous blood, and samples of cancer tissue and tissue adjacent to cancer will be collected from eligible patients. The alteration of patterns of RNA expression, including mRNA, microRNA, and lncRNA, will be compared between patients with and without HPV integration. The multi-omics profiles include genome wide association study (GWAS), whole exome sequencing, analysis of transcriptomics and metabolomics. The HPV integration status will be interpreted by GWAS. The results have not been posted for this study.