Submitted:
13 January 2025
Posted:
14 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Exploring the 3D Protein Landscape: Structural Biology Techniques
2.1. X-ray Crystallography
2.2. Nuclear Magnetic Resonance (NMR)
2.3. Transmission electron microscopy (TEM)
2.4. Cryo-electron microscopy (cryo-EM)
2.5. Small Angle X-ray Scattering (SAXS)
2.6. Cryo-electron tomography (Cryo-ET)
2.7. Emerging Techniques
3. Exploring viral structural proteins
3.1. Envelope glycoproteins
3.2. Viroporins
3.3. Capsid
4. Exploring viral non-structural proteins
4.1. Protease
4.2. Polymerase
4.3. Integrase
4.4. Thymidine kinase (TK)
4.5. Methyltransferase (MTase)
4.6. Helicases
5. Host-targeted antivirals:

6. Rational Drug Design
6.1. De novo designing - a targeted approach with improved features
7. Identifying the Threat of Future – a structural approach:
8. Conclusion and future direction
Author Contributions
Acknowledgement
Conflicts of Interest
References
- Payne, S. Introduction to Animal Viruses. In Viruses; Elsevier, 2017; pp. 1–11.
- Current ICTV Taxonomy Release | ICTV Available online:. Available online: https://ictv.global/taxonomy (accessed on 27 December 2024).
- Wirth, J.; Young, M. The Intriguing World of Archaeal Viruses. PLoS Pathog 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Cvirkaite-Krupovic, V.; Iranzo, J.; Prangishvili, D.; Koonin, E. V. Viruses of Archaea: Structural, Functional, Environmental and Evolutionary Genomics. Virus Res 2018, 244, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Costa, A.R.; van Beljouw, S.P.B.; Fineran, P.C.; Brouns, S.J.J. Approaches for Bacteriophage Genome Engineering. Trends Biotechnol 2023, 41, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin Microbiol Rev 2019, 32. [Google Scholar] [CrossRef]
- Queiroz, V.F.; Tatara, J.M.; Botelho, B.B.; Rodrigues, R.A.L.; Almeida, G.M. de F.; Abrahao, J.S. The Consequences of Viral Infection on Protists. Communications Biology 2024 7:1 2024, 7, 1–15. [Google Scholar] [CrossRef]
- Mimiviruses: Giant Viruses with Novel and Intriguing Features (Review). Available online: https://www.spandidos-publications.com/10.3892/mmr.2022.12723 (accessed on 11 January 2025).
- Coy, S.R.; Gann, E.R.; Pound, H.L.; Short, S.M.; Wilhelm, S.W. Viruses of Eukaryotic Algae: Diversity, Methods for Detection, and Future Directions. Viruses 2018, 10, 487. [Google Scholar] [CrossRef]
- Hough, B.; Steenkamp, E.; Wingfield, B.; Read, D. Fungal Viruses Unveiled: A Comprehensive Review of Mycoviruses. Viruses 2023, 15, 1202. [Google Scholar] [CrossRef]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Janssen, D. Global Plant Virus Disease Pandemics and Epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Bertola, M.; Mutinelli, F. A Systematic Review on Viruses in Mass-reared Edible Insect Species. Viruses 2021, 13, 2280. [Google Scholar] [CrossRef]
- Garrison, A.R.; Alkhovsky, S. V.; Avšič-Županc, T.; Bente, D.A.; Bergeron, É.; Burt, F.; Paola, N. Di; Ergünay, K.; Hewson, R.; Kuhn, J.H.; et al. ICTV Virus Taxonomy Profile: Nairoviridae. Journal of General Virology 2020, 101, 798–799. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. Journal of General Virology 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Weaver, S.C.; Tesh, R.B.; Vasilakis, N. Insect-Specific Virus Discovery: Significance for the Arbovirus Community. Viruses 2015, 7, 4911–4928. [Google Scholar] [CrossRef] [PubMed]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.K.R.; Ebrahimpour, S. A Brief Review of Influenza Virus Infection. J Med Virol 2021, 93, 4638–4646. [Google Scholar] [CrossRef]
- Hutchinson, E.C. Influenza Virus. 2018. [CrossRef]
- Brunker, K.; Mollentze, N. Rabies Virus. Trends Microbiol 2018, 26, 886–887. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic Strategies for COVID-19: Progress and Lessons Learned. Nature Reviews Drug Discovery 2023, 22, 449–475. [Google Scholar] [CrossRef]
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C. Bin; Bernardini, S. The COVID-19 Pandemic. Crit Rev Clin Lab Sci 2020, 365–388. [Google Scholar] [CrossRef]
- Crimi, S.; Fiorillo, L.; Bianchi, A.; D’amico, C.; Amoroso, G.; Gorassini, F.; Mastroieni, R.; Marino, S.; Scoglio, C.; Catalano, F.; et al. Herpes Virus, Oral Clinical Signs and QoL: Systematic Review of Recent Data. Viruses 2019, Vol. 11, Page 463 2019, 11, 463. [Google Scholar] [CrossRef]
- Rechenchoski, D.Z.; Faccin-Galhardi, L.C.; Linhares, R.E.C.; Nozawa, C. Herpesvirus: An Underestimated Virus. Folia Microbiol (Praha) 2017, 62, 151–156. [Google Scholar] [CrossRef]
- Connolly, S.A.; Jardetzky, T.S.; Longnecker, R. The Structural Basis of Herpesvirus Entry. Nature Reviews Microbiology 2020 19:2 2020, 19, 110–121. [Google Scholar] [CrossRef]
- Agut, H.; Bonnafous, P.; Gautheret-Dejean, A. Laboratory and Clinical Aspects of Human Herpesvirus 6 Infections. Clin Microbiol Rev 2015, 28, 313–335. [Google Scholar] [CrossRef] [PubMed]
- de Sanjosé, S.; Brotons, M.; Pavón, M.A. The Natural History of Human Papillomavirus Infection. Best Pract Res Clin Obstet Gynaecol 2018, 47, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M.; Dean, C.L. Human Papillomavirus. Diagnostic Microbiology of the Immunocompromised Host 2016, 177–195. [Google Scholar] [CrossRef]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; De Sanjosé, S.; Fakhry, C.; Monk, B.J.; Stanley, M.A.; Franceschi, S. Carcinogenic Human Papillomavirus Infection. Nature Reviews Disease Primers 2016 2:1 2016, 2, 1–20. [Google Scholar] [CrossRef]
- Yoshimura, K. Current Status of HIV/AIDS in the ART Era. Journal of Infection and Chemotherapy 2017, 23, 12–16. [Google Scholar] [CrossRef]
- Bekker, L.G.; Beyrer, C.; Mgodi, N.; Lewin, S.R.; Delany-Moretlwe, S.; Taiwo, B.; Masters, M.C.; Lazarus, J. V. HIV Infection. Nature Reviews Disease Primers 2023 9:1 2023, 9, 1–21. [Google Scholar] [CrossRef]
- Lévêque, N.; Semler, B.L. A 21st Century Perspective of Poliovirus Replication. PLoS Pathog 2015, 11, e1004825. [Google Scholar] [CrossRef]
- Marzi, A.; Blanco, J.R.; Gibellini, D.; Mbani, C.J.; Pandoua Nekoua, M.; Moukassa, D.; Hober, D. The Fight against Poliovirus Is Not Over. Microorganisms 2023, Vol. 11, Page 1323 2023, 11, 1323. [Google Scholar] [CrossRef]
- Cao, J.; Li, D. Searching for Human Oncoviruses: Histories, Challenges, and Opportunities. J Cell Biochem 2018, 119, 4897–4906. [Google Scholar] [CrossRef]
- Noguera Z., L. P.; Charypkhan, D.; Hartnack, S.; Torgerson, P.R.; Rüegg, S.R. The Dual Burden of Animal and Human Zoonoses: A Systematic Review. PLoS Negl Trop Dis 2022, 16, e0010540. [Google Scholar] [CrossRef]
- Zeller, M.A.; Carnevale de Almeida Moraes, D.; Ciacci Zanella, G.; Souza, C.K.; Anderson, T.K.; Baker, A.L.; Gauger, P.C. Reverse Zoonosis of the 2022–2023 Human Seasonal H3N2 Detected in Swine. npj Viruses 2024 2:1 2024, 2, 1–12. [Google Scholar] [CrossRef]
- Lv, J.X.; Liu, X.; Pei, Y.Y.; Song, Z.G.; Chen, X.; Hu, S.J.; She, J.L.; Liu, Y.; Chen, Y.M.; Zhang, Y.Z. Evolutionary Trajectory of Diverse SARS-CoV-2 Variants at the Beginning of COVID-19 Outbreak. Virus Evol 2024, 10. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Pandemics Throughout History. Front Microbiol 2021, 11, 631736. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. Influenza: The Once and Future Pandemic. http://dx.doi.org/10.1177/00333549101250S305 2010, 125, 15–26. [CrossRef]
- Eisinger, R.W.; Fauci, A.S. Ending the HIV/AIDS Pandemic. Emerg Infect Dis 2018, 24, 413. [Google Scholar] [CrossRef]
- Reflections on 40 Years of AIDS. 2021, 231–245.
- Chan-Yeung, epidemiologyM; Xu, R.; Chan-Yeung, M.; Chan-yeung, M. SARS: Epidemiology. Respirology 2003, 8, S9–S14. [CrossRef]
- Raj, V.S.; Osterhaus, A.D.M.E.; Fouchier, R.A.M.; Haagmans, B.L. MERS: Emergence of a Novel Human Coronavirus. Curr Opin Virol 2014, 5, 58–62. [Google Scholar] [CrossRef]
- Listings of WHO’s Response to COVID-19 Available online:. Available online: https://www.who.int/news/item/29-06-2020-covidtimeline (accessed on 27 December 2024).
- Duggan, A.T.; Perdomo, M.F.; Piombino-Mascali, D.; Marciniak, S.; Poinar, D.; Emery, M. V.; Buchmann, J.P.; Duchêne, S.; Jankauskas, R.; Humphreys, M.; et al. 17th Century Variola Virus Reveals the Recent History of Smallpox. Current Biology 2016, 26, 3407–3412. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.S.; Garon, J.; Seib, K.; Orenstein, W.A. Polio Vaccination: Past, Present and Future. Future Microbiol 2015, 10, 791–808. [Google Scholar] [CrossRef]
- Jacob, S.T.; Crozier, I.; Fischer, W.A.; Hewlett, A.; Kraft, C.S.; Vega, M.A. de La; Soka, M.J.; Wahl, V.; Griffiths, A.; Bollinger, L.; et al. Ebola Virus Disease. Nature Reviews Disease Primers 2020 6:1 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, Z.; Wen, Z.; Liu, Y.; Zeng, C.; Xiao, D.; Ou, M.; Han, Y.; Huang, S.; Liu, D.; et al. Global Epidemiology of Dengue Outbreaks in 1990–2015: A Systematic Review and Meta-Analysis. Front Cell Infect Microbiol 2017, 7, 275966. [Google Scholar] [CrossRef]
- Douam, F.; Ploss, A. Yellow Fever Virus: Knowledge Gaps Impeding the Fight Against an Old Foe. Trends Microbiol 2018, 26, 913–928. [Google Scholar] [CrossRef]
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An Update on Zika Virus Infection. The Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.T.; Halsey, N.A. The Clinical Significance of Measles: A Review. J Infect Dis 2004, 189, S4–S16. [Google Scholar] [CrossRef] [PubMed]
- Weibel Galluzzo, C.; Kaiser, L.; Chappuis, F. Reemergence of Chikungunya Virus. Rev Med Suisse 2015, 11, 1012–1016. [Google Scholar] [CrossRef] [PubMed]
- Turtle, L.; Neurology, T.S.-N.R.; 2018, undefined Japanese Encephalitis—the Prospects for New Treatments. nature.comL Turtle, T SolomonNature Reviews Neurology, 2018•nature.com.
- Campbell, G.L.; Marfin, A.A.; Lanciotti, R.S.; Gubler, D.J. West Nile Virus. Lancet Infectious Diseases 2002, 2, 519–529. [Google Scholar] [CrossRef]
- Baer, G.M. History of Rabies and Global Aspects. 2017, 1–24.
- Sperk, M.; Van Domselaar, R.; Rodriguez, J.E.; Mikaeloff, F.; Sá Vinhas, B.; Saccon, E.; Sönnerborg, A.; Singh, K.; Gupta, S.; Végvári, Á.; et al. Utility of Proteomics in Emerging and Re-Emerging Infectious Diseases Caused by RNA Viruses. J Proteome Res 2020, 19, 4259–4274. [Google Scholar] [CrossRef]
- Çelik, İ.; Saatçi, E.; Eyüboğlu, F.Ö. Emerging and Reemerging Respiratory Viral Infections up to Covid-19. Turk J Med Sci 2020, 50, 557–562. [Google Scholar] [CrossRef]
- Curry, S. Structural Biology: A Century-Long Journey into an Unseen World. Interdisciplinary Science Reviews 2015, 40, 308–328. [Google Scholar] [CrossRef]
- Brooks-Bartlett, J.C.; Garman, E.F. The Nobel Science: One Hundred Years of Crystallography. Interdisciplinary Science Reviews 2015, 40, 244–264. [Google Scholar] [CrossRef]
- Thomas, J.M. Centenary: The Birth of X-Ray Crystallography. Nature 2012, 491, 186–187. [Google Scholar] [CrossRef]
- Shi, Y. A Glimpse of Structural Biology through X-Ray Crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Li, G. Approved Antiviral Drugs over the Past 50 Years. Clin Microbiol Rev 2016, 29, 695–747. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Handing, K.B.; Zimmerman, M.D.; Shabalin, I.G.; Almo, S.C.; Minor, W. X-Ray Crystallography over the Past Decade for Novel Drug Discovery – Where Are We Heading Next? Expert Opin Drug Discov 2015, 10, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Stanley, W.M. ISOLATION OF A CRYSTALLINE PROTEIN POSSESSING THE PROPERTIES OF TOBACCO-MOSAIC VIRUS. Science 1935, 81, 644–645. [Google Scholar] [CrossRef]
- Norrby, E. Nobel Prizes and the Emerging Virus Concept. Arch Virol 2008, 153, 1109–1123. [Google Scholar] [CrossRef]
- Bernal, J.D.; Fankuchen, I. X-RAY AND CRYSTALLOGRAPHIC STUDIES OF PLANT VIRUS PREPARATIONS : I. INTRODUCTION AND PREPARATION OF SPECIMENS II. MODES OF AGGREGATION OF THE VIRUS PARTICLES. J Gen Physiol 1941, 25, 111–146. [Google Scholar] [CrossRef]
- Harrison, S.C.; Olson, A.J.; Schutt, C.E.; Winkler, F.K.; Bricogne, G. Tomato Bushy Stunt Virus at 2.9 A Resolution. Nature 1978, 276, 368–373. [Google Scholar] [CrossRef]
- Bloomer, A.C.; Champness, J.N.; Bricogne, G.; Staden, R.; Klug, A. Protein Disk of Tobacco Mosaic Virus at 2.8 A Resolution Showing the Interactions within and between Subunits. Nature 1978, 276, 362–368. [Google Scholar] [CrossRef]
- Richmond, T.J.; Finch, J.T.; Rushton, B.; Rhodes, D.; Klug, A. Structure of the Nucleosome Core Particle at 7 A Resolution. Nature 1984, 311, 532–537. [Google Scholar] [CrossRef]
- Zheng, H.; Handing, K.B.; Zimmerman, M.D.; Shabalin, I.G.; Almo, S.C.; Minor, W. X-Ray Crystallography over the Past Decade for Novel Drug Discovery -Where Are We Heading Next? Expert Opin Drug Discov 2015, 10, 975–989. [Google Scholar] [CrossRef]
- Schirò, A.; Carlon, A.; Parigi, G.; Murshudov, G.; Calderone, V.; Ravera, E.; Luchinat, C. On the Complementarity of X-Ray and NMR Data. J Struct Biol X 2020, 4. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Handing, K.B.; Zimmerman, M.D.; Shabalin, I.G.; Almo, S.C.; Minor, W. X-Ray Crystallography over the Past Decade for Novel Drug Discovery -Where Are We Heading Next? Expert Opin Drug Discov 2015, 10, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Vincenzi, M.; Leone, M. The Fight against Human Viruses: How NMR Can Help? Curr Med Chem 2021, 28, 4380–4453. [Google Scholar] [CrossRef]
- Yu, H. Extending the Size Limit of Protein Nuclear Magnetic Resonance. Proc Natl Acad Sci U S A 1999, 96, 332–334. [Google Scholar] [CrossRef]
- LaPlante, S.R.; Coric, P.; Bouaziz, S.; França, T.C.C. NMR Spectroscopy Can Help Accelerate Antiviral Drug Discovery Programs. Microbes Infect 2024, 26, 105297. [Google Scholar] [CrossRef]
- Kruger, D.H.; Schneck, P.; Gelderblom, H.R. Helmut Ruska and the Visualisation of Viruses. Lancet 2000, 355, 1713–1717. [Google Scholar] [CrossRef]
- Nagler, F.P.; Rake, G. The Use of the Electron Microscope in Diagnosis of Variola, Vaccinia, and Varicella. J Bacteriol 1948, 55, 45–51. [Google Scholar] [CrossRef]
- Brenner, S.; Horne, R.W. A Negative Staining Method for High Resolution Electron Microscopy of Viruses. Biochim Biophys Acta 1959, 34, 103–110. [Google Scholar] [CrossRef]
- Tyrrell, D.A.J.; Almeida, J.D. Direct Electron-Microscopy of Organ Cultures for the Detection and Characterization of Viruses. Arch Gesamte Virusforsch 1967, 22, 417–425. [Google Scholar] [CrossRef]
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-Electron Microscopy of Viruses. Nature 1984 308:5954 1984, 308, 32–36. [Google Scholar] [CrossRef]
- Schoehn, G.; Chenavier, F.; Crépin, T. Advances in Structural Virology via Cryo-EM in 2022. Viruses 2023, Vol. 15, Page 1315 2023, 15, 1315. [Google Scholar] [CrossRef] [PubMed]
- Dutta, M.; Acharya, P. Cryo-Electron Microscopy in the Study of Virus Entry and Infection. Front Mol Biosci 2024, 11, 1429180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.; Zhang, K. Cryo-EM: A Window into the Dynamic World of RNA Molecules. Curr Opin Struct Biol 2024, 88, 102916. [Google Scholar] [CrossRef] [PubMed]
- Renaud, J.P.; Chari, A.; Ciferri, C.; Liu, W.T.; Rémigy, H.W.; Stark, H.; Wiesmann, C. Cryo-EM in Drug Discovery: Achievements, Limitations and Prospects. Nature Reviews Drug Discovery 2018 17:7 2018, 17, 471–492. [Google Scholar] [CrossRef]
- Boldon, L.; Laliberte, F.; Liu, L. Review of the Fundamental Theories behind Small Angle X-Ray Scattering, Molecular Dynamics Simulations, and Relevant Integrated Application. Nano Rev 2015, 6, 25661. [Google Scholar] [CrossRef]
- Handa, T.; Kundu, D.; Dubey, V.K. Perspectives on Evolutionary and Functional Importance of Intrinsically Disordered Proteins. Int J Biol Macromol 2023, 224, 243–255. [Google Scholar] [CrossRef]
- Barradas-Bautista, D.; Rosell, M.; Pallara, C.; Fernández-Recio, J. Structural Prediction of Protein–Protein Interactions by Docking: Application to Biomedical Problems. Adv Protein Chem Struct Biol 2018, 110, 203–249. [Google Scholar] [CrossRef]
- Zhu, P.; Winkler, H.; Chertova, E.; Taylor, K.A.; Roux, K.H. Cryoelectron Tomography of HIV-1 Envelope Spikes: Further Evidence for Tripod-like Legs. PLoS Pathog 2008, 4. [Google Scholar] [CrossRef]
- Dutta, M.; Acharya, P. Cryo-Electron Microscopy in the Study of Virus Entry and Infection. Front Mol Biosci 2024, 11, 1429180. [Google Scholar] [CrossRef]
- Baumeister, W. Cryo-Electron Tomography: The Power of Seeing the Whole Picture. Biochem Biophys Res Commun 2022, 633, 26–28. [Google Scholar] [CrossRef]
- Meents, A.; Wiedorn, M.O. Virus Structures by X-Ray Free-Electron Lasers. Annu Rev Virol 2019, 6, 161–176. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, K. AlphaFold Gets an Upgrade (and a Nobel). Nature Medicine 2024 30:12 2024, 30, 3393–3393. [Google Scholar] [CrossRef] [PubMed]
- FENNER, F.; BACHMANN, P.A.; GIBBS, E.P.J.; MURPHY, F.A.; STUDDERT, M.J.; WHITE, D.O. Structure and Composition of Viruses. In Veterinary Virology; Elsevier, 1987; pp. 3–19.
- Zheng, B.; Duan, M.; Huang, Y.; Wang, S.; Qiu, J.; Lu, Z.; Liu, L.; Tang, G.; Cheng, L.; Zheng, P. Discovery of a Heparan Sulfate Binding Domain in Monkeypox Virus H3 as an Anti-Poxviral Drug Target Combining AI and MD Simulations. Elife 2024, 13. [Google Scholar] [CrossRef]
- Delogu, I.; Pastorino, B.; Baronti, C.; Nougairède, A.; Bonnet, E.; de Lamballerie, X. In Vitro Antiviral Activity of Arbidol against Chikungunya Virus and Characteristics of a Selected Resistant Mutant. Antiviral Res 2011, 90, 99–107. [Google Scholar] [CrossRef]
- Barrow, E.; Nicola, A. V.; Liu, J. Multiscale Perspectives of Virus Entry via Endocytosis. Virol J 2013, 10, 1–11. [Google Scholar] [CrossRef]
- Kim, A.S.; Diamond, M.S. A Molecular Understanding of Alphavirus Entry and Antibody Protection. Nature Reviews Microbiology 2022 21:6 2022, 21, 396–407. [Google Scholar] [CrossRef]
- Melton, J. V.; Ewart, G.D.; Weir, R.C.; Board, P.G.; Lee, E.; Gage, P.W. Alphavirus 6K Proteins Form Ion Channels. Journal of Biological Chemistry 2002, 277, 46923–46931. [Google Scholar] [CrossRef]
- Button, J.M.; Mukhopadhyay, S. Capsid-E2 Interactions Rescue Core Assembly in Viruses That Cannot Form Cytoplasmic Nucleocapsid Cores. J Virol 2021, 95. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encyclopedia of Virology: Volume 1-5, Fourth Edition 2021, 1–5, 428–440. [Google Scholar] [CrossRef]
- Schlicksup, C.J.; Zlotnick, A. Viral Structural Proteins as Targets for Antivirals. Curr Opin Virol 2020, 45, 43–50. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, B.; Deng, L.; Liang, B.; Ping, J. Virus-Host Interaction Networks as New Antiviral Drug Targets for IAV and SARS-CoV-2. Emerg Microbes Infect 2022, 11, 1371–1389. [Google Scholar] [CrossRef] [PubMed]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Virion Structure and Composition. Fenner and White’s Medical Virology 2017, 27–37. [Google Scholar] [CrossRef]
- Kaur, R.; Neetu; Mudgal, R.; Jose, J.; Kumar, P.; Tomar, S. Glycan-Dependent Chikungunya Viral Infection Divulged by Antiviral Activity of NAG Specific Chi-like Lectin. Virology 2019, 526, 91–98. [CrossRef]
- Kaur, R.; Neetu; Mudgal, R.; Jose, J.; Kumar, P.; Tomar, S. Glycan-Dependent Chikungunya Viral Infection Divulged by Antiviral Activity of NAG Specific Chi-like Lectin. Virology 2019, 526, 91–98. [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encyclopedia of Virology: Volume 1-5, Fourth Edition 2021, 1–5, 428–440. [Google Scholar] [CrossRef]
- Kosik, I.; Yewdell, J.W. Influenza Hemagglutinin and Neuraminidase: Yin–Yang Proteins Coevolving to Thwart Immunity. Viruses 2019, Vol. 11, Page 346 2019, 11, 346. [Google Scholar] [CrossRef]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 Envelope Glycoprotein Biosynthesis, Trafficking, and Incorporation. J Mol Biol 2011, 410, 582–608. [Google Scholar] [CrossRef]
- Wrobel, A.G. Mechanism and Evolution of Human ACE2 Binding by SARS-CoV-2 Spike. Curr Opin Struct Biol 2023, 81, 102619. [Google Scholar] [CrossRef]
- Epand, R.M. Fusion Peptides and the Mechanism of Viral Fusion. Biochimica et Biophysica Acta (BBA) - Biomembranes 2003, 1614, 116–121. [Google Scholar] [CrossRef]
- Zhai, X.; Yuan, Y.; He, W.T.; Wu, Y.; Shi, Y.; Su, S.; Du, Q.; Mao, Y. Evolving Roles of Glycosylation in the Tug-of-War between Virus and Host. Natl Sci Rev 2024, 11. [Google Scholar] [CrossRef]
- Matsuyama, S.; Taguchi, F. Two-Step Conformational Changes in a Coronavirus Envelope Glycoprotein Mediated by Receptor Binding and Proteolysis. J Virol 2009, 83, 11133. [Google Scholar] [CrossRef]
- Marzinek, J.K.; Raghuvamsi Palur, V.; Salem, G.; Chen, F.-C.; Wu, S.-R.; Bond, P.J.; Chao, D.-Y. Uncovering the Conformational Dynamics of Dengue Virus and Its Virus-like Particles as Novel Vaccine Candidates. Biophys J 2023, 122, 508a–509a. [Google Scholar] [CrossRef]
- Katze, M.G.; He, Y.; Gale, M. Viruses and Interferon: A Fight for Supremacy. Nature Reviews Immunology 2002 2:9 2002, 2, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xue, X.; Qiao, S.; Jia, L.; Wen, X.; Wang, Y.; Wang, C.; Li, H.; Cui, J. Umifenovir Epigenetically Targets the IL-10 Pathway in Therapy against Coxsackievirus B4 Infection. Microbiol Spectr 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, K.; Mazmanian, K.; Lim, C. A Strategy for Evaluating Potential Antiviral Resistance to Small Molecule Drugs and Application to SARS-CoV-2. Scientific Reports 2023 13:1 2023, 13, 1–12. [Google Scholar] [CrossRef]
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural Basis of SARS-CoV-2 Omicron Immune Evasion and Receptor Engagement. Science (1979) 2022, 375, 894–898. [Google Scholar] [CrossRef]
- Tang, H.; Ke, Y.; Liao, Y.; Bian, Y.; Yuan, Y.; Wang, Z.; Yang, L.; Ma, H.; Sun, T.; Zhang, B.; et al. Mutational Escape Prevention by Combination of Four Neutralizing Antibodies That Target RBD Conserved Regions and Stem Helix. Virol Sin 2022, 37, 860–873. [Google Scholar] [CrossRef]
- Shih, H.I.; Wang, Y.C.; Wang, Y.P.; Chi, C.Y.; Chien, Y.W. Risk of Severe Dengue during Secondary Infection: A Population-Based Cohort Study in Taiwan. Journal of Microbiology, Immunology and Infection 2024, 57, 730–738. [Google Scholar] [CrossRef]
- Wells, T.J.; Esposito, T.; Henderson, I.R.; Labzin, L.I. Mechanisms of Antibody-Dependent Enhancement of Infectious Disease. Nature Reviews Immunology 2024 25:1 2024, 25, 6–21. [Google Scholar] [CrossRef]
- Sarker, A.; Dhama, N.; Gupta, R.D. Dengue Virus Neutralizing Antibody: A Review of Targets, Cross-Reactivity, and Antibody-Dependent Enhancement. Front Immunol 2023, 14, 1200195. [Google Scholar] [CrossRef]
- Malik, S.; Ahsan, O.; Mumtaz, H.; Tahir Khan, M.; Sah, R.; Waheed, Y. Tracing down the Updates on Dengue Virus—Molecular Biology, Antivirals, and Vaccine Strategies. Vaccines 2023, Vol. 11, Page 1328 2023, 11, 1328. [Google Scholar] [CrossRef]
- Dengvaxia | European Medicines Agency (EMA) Available online:. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/dengvaxia (accessed on 10 January 2025).
- Ragonnet-Cronin, M.; Nutalai, R.; Huo, J.; Dijokaite-Guraliuc, A.; Das, R.; Tuekprakhon, A.; Supasa, P.; Liu, C.; Selvaraj, M.; Groves, N.; et al. Generation of SARS-CoV-2 Escape Mutations by Monoclonal Antibody Therapy. Nature Communications 2023 14:1 2023, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Malik, Y.S.; Tomar, S. Identification of SARS-CoV-2 Cell Entry Inhibitors by Drug Repurposing Using in Silico Structure-Based Virtual Screening Approach. Front Immunol 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Osterhaus, A.D.M.E.; Treanor, J.J.; Fleming, D.M.; Aoki, F.Y.; Nicholson, K.G.; Bohnen, A.M.; Hirst, H.M.; Keene, O.; Wightman, K. Efficacy and Safety of the Neuraminidase Inhibitor Zanamivir in the Treatment of Influenzavirus Infections. New England Journal of Medicine 1997, 337, 874–880. [Google Scholar] [CrossRef]
- Collins, P.J.; Haire, L.F.; Lin, Y.P.; Liu, J.; Russell, R.J.; Walker, P.A.; Skehel, J.J.; Martin, S.R.; Hay, A.J.; Gamblin, S.J. Crystal Structures of Oseltamivir-Resistant Influenza Virus Neuraminidase Mutants. Nature 2008 453:7199 2008, 453, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Vavricka, C.J.; Li, Q.; Wu, Y.; Qi, J.; Wang, M.; Liu, Y.; Gao, F.; Liu, J.; Feng, E.; He, J.; et al. Structural and Functional Analysis of Laninamivir and Its Octanoate Prodrug Reveals Group Specific Mechanisms for Influenza NA Inhibition. PLoS Pathog 2011, 7, e1002249. [Google Scholar] [CrossRef]
- Pattnaik, G.P.; Chakraborty, H. Entry Inhibitors: Efficient Means to Block Viral Infection. Journal of Membrane Biology 2020, 253, 425–444. [Google Scholar] [CrossRef]
- Beugeling, M.; De Zee, J.; Woerdenbag, H.J.; Frijlink, H.W.; Wilschut, J.C.; Hinrichs, W.L.J. Respiratory Syncytial Virus Subunit Vaccines Based on the Viral Envelope Glycoproteins Intended for Pregnant Women and the Elderly. Expert Rev Vaccines 2019, 18, 935–950. [Google Scholar] [CrossRef]
- Singh, V.A.; Nehul, S.; Kumar, C.S.; Banerjee, M.; Kumar, P.; Sharma, G.; Tomar, S. Chimeric Chikungunya Virus-like Particles with Surface Exposed SARS-CoV-2 RBD Elicits Potent Immunogenic Responses in Mice. bioRxiv 2023, 2023.01.29.526074. [CrossRef]
- Singh, V.A.; Kumar, C.S.; Khare, B.; Kuhn, R.J.; Banerjee, M.; Tomar, S. Surface Decorated Reporter-Tagged Chikungunya Virus-like Particles for Clinical Diagnostics and Identification of Virus Entry Inhibitors. Virology 2023, 578, 92–102. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and Biological Functions. Nature Reviews Microbiology 2012 10:8 2012, 10, 563–574. [Google Scholar] [CrossRef]
- Xia, X.; Cheng, A.; Wang, M.; Ou, X.; Sun, D.; Mao, S.; Huang, J.; Yang, Q.; Wu, Y.; Chen, S.; et al. Functions of Viroporins in the Viral Life Cycle and Their Regulation of Host Cell Responses. Front Immunol 2022, 13, 890549. [Google Scholar] [CrossRef]
- Devantier, K.; Kjær, V.M.S.; Griffin, S.; Kragelund, B.B.; Rosenkilde, M.M. Advancing the Field of Viroporins—Structure, Function and Pharmacology: IUPHAR Review 39. Br J Pharmacol 2024, 181, 4450–4490. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza Virus M2 Protein Has Ion Channel Activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and Biological Functions. Nature Reviews Microbiology 2012 10:8 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and Biological Functions. Nature Reviews Microbiology 2012 10:8 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Surya, W.; Samsó, M.; Torres, J.; Surya, W.; Samsó, M.; Torres, J. Structural and Functional Aspects of Viroporins in Human Respiratory Viruses: Respiratory Syncytial Virus and Coronaviruses. Respiratory Disease and Infection - A New Insight 2013. [CrossRef]
- Das, K. Antivirals Targeting Influenza a Virus. J Med Chem 2012, 55, 6263–6277. [Google Scholar] [CrossRef]
- Thomaston, J.L.; Polizzi, N.F.; Konstantinidi, A.; Wang, J.; Kolocouris, A.; Degrado, W.F. Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters. J Am Chem Soc 2018, 140, 15219–15226. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Castaño-Rodriguez, C.; Aguilella, V.M.; Enjuanes, L. Relevance of Viroporin Ion Channel Activity on Viral Replication and Pathogenesis. Viruses 2015, Vol. 7, Pages 3552-3573 2015, 7, 3552–3573. [Google Scholar] [CrossRef]
- Dey, D.; Siddiqui, S.I.; Mamidi, P.; Ghosh, S.; Kumar, C.S.; Chattopadhyay, S.; Ghosh, S.; Banerjee, M. The Effect of Amantadine on an Ion Channel Protein from Chikungunya Virus. PLoS Negl Trop Dis 2019, 13, e0007548. [Google Scholar] [CrossRef]
- Lamb, R.A. Influenza. Encyclopedia of Virology: Volume 1-5 2008, 1–5, 95–104. [Google Scholar] [CrossRef]
- Scott, C.; Griffin, S. Viroporins: Structure, Function and Potential as Antiviral Targets. Journal of General Virology 2015, 96, 2000–2027. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and Biological Functions. Nature Reviews Microbiology 2012 10:8 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Fatma, B.; Kumar, R.; Singh, V.A.; Nehul, S.; Sharma, R.; Kesari, P.; Kuhn, R.J.; Tomar, S. Alphavirus Capsid Protease Inhibitors as Potential Antiviral Agents for Chikungunya Infection. Antiviral Res 2020, 179, 104808. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, A.; Rao, A.L. The Interplay between Capsid Dynamics and Pathogenesis in Tripartite Bromoviruses. Curr Opin Virol 2021, 47, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Ghaemi, Z.; Gruebele, M.; Tajkhorshid, E. Molecular Mechanism of Capsid Disassembly in Hepatitis B Virus. Proc Natl Acad Sci U S A 2021, 118, e2102530118. [Google Scholar] [CrossRef]
- Mohajerani, F.; Tyukodi, B.; Schlicksup, C.J.; Hadden-Perilla, J.A.; Zlotnick, A.; Hagan, M.F. Multiscale Modeling of Hepatitis B Virus Capsid Assembly and Its Dimorphism. ACS Nano 2022, 16, 13845–13859. [Google Scholar] [CrossRef]
- Koehl, P.; Akopyan, A.; Edelsbrunner, H. Computing the Volume, Surface Area, Mean, and Gaussian Curvatures of Molecules and Their Derivatives. J Chem Inf Model 2023, 63, 973–985. [Google Scholar] [CrossRef]
- Mohajerani, F.; Tyukodi, B.; Schlicksup, C.J.; Hadden-Perilla, J.A.; Zlotnick, A.; Hagan, M.F. Multiscale Modeling of Hepatitis B Virus Capsid Assembly and Its Dimorphism. ACS Nano 2022, 16, 13845–13859. [Google Scholar] [CrossRef]
- Aggarwal, M.; Kaur, R.; Saha, A.; Mudgal, R.; Yadav, R.; Dash, P.K.; Parida, M.; Kumar, P.; Tomar, S. Evaluation of Antiviral Activity of Piperazine against Chikungunya Virus Targeting Hydrophobic Pocket of Alphavirus Capsid Protein. Antiviral Res 2017, 146, 102–111. [Google Scholar] [CrossRef]
- Fatma, B.; Kumar, R.; Singh, V.A.; Nehul, S.; Sharma, R.; Kesari, P.; Kuhn, R.J.; Tomar, S. Alphavirus Capsid Protease Inhibitors as Potential Antiviral Agents for Chikungunya Infection. Antiviral Res 2020, 179. [Google Scholar] [CrossRef]
- HIV-1 Capsid Inhibitors as Antiretroviral Agents.
- Segal-Maurer, S.; DeJesus, E.; Stellbrink, H.-J.; Castagna, A.; Richmond, G.J.; Sinclair, G.I.; Siripassorn, K.; Ruane, P.J.; Berhe, M.; Wang, H.; et al. Capsid Inhibition with Lenacapavir in Multidrug-Resistant HIV-1 Infection. New England Journal of Medicine 2022, 386, 1793–1803. [Google Scholar] [CrossRef]
- Klumpp, K.; Crépin, T. Capsid Proteins of Enveloped Viruses as Antiviral Drug Targets. Curr Opin Virol 2014, 5, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Jia, R.; Wang, M.; Yin, Z.; Cheng, A. Structure and Function of Capsid Protein in Flavivirus Infection and Its Applications in the Development of Vaccines and Therapeutics. Veterinary Research 2021 52:1 2021, 52, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, R.; Zhou, J.; Wang, M.; Yin, Z.; Cheng, A. Capsid-Targeted Viral Inactivation: A Novel Tactic for Inhibiting Replication in Viral Infections. Viruses 2016, Vol. 8, Page 258 2016, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, P.; Mahto, J.K.; Singh, A.; Kumar, P.; Tomar, S. Structural Insights into the RNA Binding Inhibitors of the C-Terminal Domain of the SARS-CoV-2 Nucleocapsid 2024.
- Dhaka, P.; Singh, A.; Choudhary, S.; Peddinti, R.K.; Kumar, P.; Sharma, G.K.; Tomar, S. Mechanistic and Thermodynamic Characterization of Antiviral Inhibitors Targeting Nucleocapsid N-Terminal Domain of SARS-CoV-2. Arch Biochem Biophys 2023, 750, 109820. [Google Scholar] [CrossRef]
- The Isolation and Properties of Crystalline Tobacco Mosaic Virus.
- Namba, K.; Pattanayek, R.; Stubbs, G. Visualization of Protein-Nucleic Acid Interactions in a Virus: Refined Structure of Intact Tobacco Mosaic Virus at 2.9 Å Resolution by X-Ray Fiber Diffraction. J Mol Biol 1989, 208, 307–325. [Google Scholar] [CrossRef]
- Aggarwal, M.; Tapas, S.; Preeti; Siwach, A.; Kumar, P.; Kuhn, R.J.; Tomar, S. Crystal Structure of Aura Virus Capsid Protease and Its Complex with Dioxane: New Insights into Capsid-Glycoprotein Molecular Contacts. PLoS One 2012, 7. [CrossRef]
- Aggarwal, M.; Dhindwal, S.; Kumar, P.; Kuhn, R.J.; Tomar, S. Trans -Protease Activity and Structural Insights into the Active Form of the Alphavirus Capsid Protease. J Virol 2014, 88, 12242–12253. [Google Scholar] [CrossRef]
- Segal-Maurer, S.; DeJesus, E.; Stellbrink, H.-J.; Castagna, A.; Richmond, G.J.; Sinclair, G.I.; Siripassorn, K.; Ruane, P.J.; Berhe, M.; Wang, H.; et al. Capsid Inhibition with Lenacapavir in Multidrug-Resistant HIV-1 Infection. New England Journal of Medicine 2022, 386, 1793–1803. [Google Scholar] [CrossRef]
- Kanodia, S.; Da Silva, D.M.; Kast, W.M. Recent Advances in Strategies for Immunotherapy of Human Papillomavirus-Induced Lesions. Int J Cancer 2008, 122, 247–259. [Google Scholar] [CrossRef]
- Demmler-Harrison, G.J. ANTIVIRAL AGENTS. Feigin and Cherry’s Textbook of Pediatric Infectious Diseases, Sixth Edition 2009, 3245–3271. [CrossRef]
- Wlodawer, A.; Vondrasek, J. Inhibitors of HIV-1 Protease: A Major Success of Structure-Assisted Drug Design. Annu Rev Biophys Biomol Struct 1998, 27, 249–284. [Google Scholar] [CrossRef]
- FDA-Approved HIV Medicines | NIH Available online:. Available online: https://hivinfo.nih.gov/understanding-hiv/fact-sheets/fda-approved-hiv-medicines (accessed on 27 December 2024).
- Weber, I.T.; Miller, M.; Jaskólski, M.; Leis, J.; Skalka, A.M.; Wlodawer, A. Molecular Modeling of the HIV-1 Protease and Its Substrate Binding Site. Science 1989, 243, 928–931. [Google Scholar] [CrossRef]
- Lapatto, R.; Blundell, T.; Hemmings, A.; Overington, J.; Wilderspin, A.; Wood, S.; Merson, J.R.; Whittle, P.J.; Danley, D.E.; Geoghegan, K.F.; et al. X-Ray Analysis of HIV-1 Proteinase at 2.7 Å Resolution Confirms Structural Homology among Retroviral Enzymes. Nature 1989 342:6247 1989, 342, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Roberts, N.A.; Martin, J.A.; Kinchington, D.; Broadhurst, A. V.; Craig, J.C.; Duncan, I.B.; Galpin, S.A.; Handa, B.K.; Kay, J.; Kröhn, A.; et al. Rational Design of Peptide-Based HIV Proteinase Inhibitors. Science (1979) 1990, 248, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.C.; Duncan, I.B.; Hockley, D.; Grief, C.; Roberts, N.A.; Mills, J.S. Antiviral Properties of Ro 31-8959, an Inhibitor of Human Immunodeficiency Virus (HIV) Proteinase. Antiviral Res 1991, 16, 295–305. [Google Scholar] [CrossRef]
- Kempf, D.J.; Marsh, K.C.; Denissen, J.F.; McDonald, E.; Vasavanonda, S.; Flentge, C.A.; Green, B.E.; Fino, L.; Park, C.H.; Kong, X.P.; et al. ABT-538 Is a Potent Inhibitor of Human Immunodeficiency Virus Protease and Has High Oral Bioavailability in Humans. Proc Natl Acad Sci U S A 1995, 92, 2484–2488. [Google Scholar] [CrossRef]
- Erickson, J.W. Design and Structure of Symmetry-Based Inhibitors of HIV-1 Protease. Perspectives in Drug Discovery and Design 1993, 1, 109–128. [Google Scholar] [CrossRef]
- Dorsey, B.D.; Levin, R.B.; McDaniel, S.L.; Vacca, J.P.; Guare, J.P.; Anderson, P.S.; Huff, J.R.; Darke, P.L.; Zugay, J.A.; Emini, E.A.; et al. L-735,524: The Design of a Potent and Orally Bioavailable HIV Protease Inhibitor. J Med Chem 1994, 37, 3443–3451. [Google Scholar] [CrossRef]
- Vacca, J.P.; Dorsey, B.D.; Schleif, W.A.; Levin, R.B.; Mcdaniel, S.L.; Darke, P.L.; Zugay, J.; Quintero, J.C.; Blahy, O.M.; Roth, E.; et al. L-735,524: An Orally Bioavailable Human Immunodeficiency Virus Type 1 Protease Inhibitor. Proc Natl Acad Sci U S A 1994, 91, 4096–4100. [Google Scholar] [CrossRef]
- Wlodawer, A. Rational Approach to AIDS Drug Design through Structural Biology. Annu Rev Med 2002, 53, 595–614. [Google Scholar] [CrossRef]
- Varney, M.D.; Appelt, K.; Kalish, V.; Reddy, M.R.; Tatlock, J.; Palmer, C.L.; Romines, W.H.; Wu, B.W.; Musick, L. Crystal-Structure-Based Design and Synthesis of Novel C-Terminal Inhibitors of HIV Protease. J Med Chem 1994, 37, 2274–2284. [Google Scholar] [CrossRef]
- Kaldor, S.W.; Kalish, V.J.; Davies, J.F.; Shetty, B. V.; Fritz, J.E.; Appelt, K.; Burgess, J.A.; Campanale, K.M.; Chirgadze, N.Y.; Clawson, D.K.; et al. Viracept (Nelfinavir Mesylate, AG1343): A Potent, Orally Bioavailable Inhibitor of HIV-1 Protease. J Med Chem 1997, 40, 3979–3985. [Google Scholar] [CrossRef]
- Weber, I.T.; Waltman, M.J.; Mustyakimov, M.; Blakeley, M.P.; Keen, D.A.; Ghosh, A.K.; Langan, P.; Kovalevsky, A.Y. Joint X-Ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design. J Med Chem 2013, 56, 5631–5635. [Google Scholar] [CrossRef] [PubMed]
- Chemburkar, S.R.; Bauer, J.; Deming, K.; Spiwek, H.; Patel, K.; Morris, J.; Henry, R.; Spanton, S.; Dziki, W.; Porter, W.; et al. Dealing with the Impact of Ritonavir Polymorphs on the Late Stages of Bulk Drug Process Development. Org Process Res Dev 2000, 4, 413–417. [Google Scholar] [CrossRef]
- McCauley, J.A.; Rudd, M.T. Hepatitis C Virus NS3/4a Protease Inhibitors. Curr Opin Pharmacol 2016, 30, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Love, R.A.; Parge, H.E.; Wickersham, J.A.; Hostomsky, Z.; Habuka, N.; Moomaw, E.W.; Adachi, T.; Hostomska, Z. The Crystal Structure of Hepatitis C Virus NS3 Proteinase Reveals a Trypsin-like Fold and a Structural Zinc Binding Site. Cell 1996, 87, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.L.; Morgenstern, K.A.; Lin, C.; Fox, T.; Dwyer, M.D.; Landro, J.A.; Chambers, S.P.; Markland, W.; Lepre, C.A.; O’Malley, E.T.; et al. Crystal Structure of the Hepatitis C Virus NS3 Protease Domain Complexed with a Synthetic NS4A Cofactor Peptide. Cell 1996, 87, 343–355. [Google Scholar] [CrossRef]
- Barbato, G.; Cicero, D.O.; Nardi, M.C.; Steinkühler, C.; Cortese, R.; De Francesco, R.; Bazzo, R. The Solution Structure of the N-Terminal Proteinase Domain of the Hepatitis C Virus (HCV) NS3 Protein Provides New Insights into Its Activation and Catalytic Mechanism. J Mol Biol 1999, 289, 371–384. [Google Scholar] [CrossRef]
- Llinàs-Brunet, M.; Bailey, M.; Fazal, G.; Goulet, S.; Halmos, T.; Laplante, S.; Maurice, R.; Poirier, M.; Poupart, M.A.; Thibeault, D.; et al. Peptide-Based Inhibitors of the Hepatitis C Virus Serine Protease. Bioorg Med Chem Lett 1998, 8, 1713–1718. [Google Scholar] [CrossRef]
- saalau-bethell, susanne; Woodhead, andrew J.; chessari, gianni; carr, maria; coyle, J.; graham, brent; hiscock, steven; murray, christopher W.; pathuri, puja; rich, sharna J.; et al. Discovery of an Allosteric Mechanism for the Regulation of HcV Ns3 Protein Function. 2012. [CrossRef]
- Choudhary, S.; Nehul, S.; Singh, A.; Panda, P.K.; Kumar, P.; Sharma, G.K.; Tomar, S. Unraveling Antiviral Efficacy of Multifunctional Immunomodulatory Triterpenoids against SARS-COV-2 Targeting Main Protease and Papain-like Protease. IUBMB Life 2024, 76, 228–241. [Google Scholar] [CrossRef]
- Choudhary, S.; Nehul, S.; Nagaraj, S.K.; Narayan, R.; Verma, S.; Sharma, S.; Kumari, A.; Rani, R.; Saha, A.; Sircar, D.; et al. Activity Profiling of Deubiquitinating Inhibitors-Bound to SARS-CoV-2 Papain like Protease with Antiviral Efficacy in Murine Infection Model 2022.
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19. Science (1979) 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved a-Ketoamide Inhibitors. Science (1979) 2020, 368, 409–412. [Google Scholar] [CrossRef]
- FDA Approves First Oral Antiviral for Treatment of COVID-19 in Adults | FDA Available online:. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-oral-antiviral-treatment-covid-19-adults (accessed on 30 December 2024).
- Pagliano, P.; Spera, A.; Sellitto, C.; Scarpati, G.; Folliero, V.; Piazza, O.; Franci, G.; Conti, V.; Ascione, T. Preclinical Discovery and Development of Nirmatrelvir/Ritonavir Combinational Therapy for the Treatment of COVID-19 and the Lessons Learned from SARS-COV-2 Variants. Expert Opin Drug Discov 2023, 18, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Unoh, Y.; Uehara, S.; Nakahara, K.; Nobori, H.; Yamatsu, Y.; Yamamoto, S.; Maruyama, Y.; Taoda, Y.; Kasamatsu, K.; Suto, T.; et al. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J Med Chem 2022, 65, 6499–6512. [Google Scholar] [CrossRef] [PubMed]
- Narwal, M.; Armache, J.P.; Edwards, T.J.; Murakami, K.S. SARS-CoV-2 Polyprotein Substrate Regulates the Stepwise Mpro Cleavage Reaction. Journal of Biological Chemistry 2023, 299. [Google Scholar] [CrossRef]
- Pareek, A.; Kumar, R.; Mudgal, R.; Neetu, N.; Sharma, M.; Kumar, P.; Tomar, S. Alphavirus Antivirals Targeting RNA-Dependent RNA Polymerase Domain of NsP4 Divulged Using Surface Plasmon Resonance. FEBS J 2022, 289, 4901–4924. [Google Scholar] [CrossRef]
- Rani, R.; Long, S.; Pareek, A.; Dhaka, P.; Singh, A.; Kumar, P.; McInerney, G.; Tomar, S. Multi-Target Direct-Acting SARS-CoV-2 Antivirals against the Nucleotide-Binding Pockets of Virus-Specific Proteins. Virology 2022, 577, 1–15. [Google Scholar] [CrossRef]
- Rani, R.; Nehul, S.; Choudhary, S.; Upadhyay, A.; Kumar Sharma, G.; Kumar, P.; Tomar, S.; scientist, S. Revealing and Evaluation of Antivirals Targeting Multiple Druggable Sites of RdRp Complex in SARS-CoV-2. bioRxiv 2023, 2023.07.24.550324. [CrossRef]
- Choi, K.H. Viral Polymerases. Adv Exp Med Biol 2012, 726, 267–304. [Google Scholar] [CrossRef]
- Liu, S.; Knafels, J.D.; Chang, J.S.; Waszak, G.A.; Baldwin, E.T.; Deibel, M.R.; Thomsen, D.R.; Homa, F.L.; Wells, P.A.; Tory, M.C.; et al. Crystal Structure of the Herpes Simplex Virus 1 DNA Polymerase. Journal of Biological Chemistry 2006, 281, 18193–18200. [Google Scholar] [CrossRef]
- Zarrouk, K.; Piret, J.; Boivin, G. Herpesvirus DNA Polymerases: Structures, Functions and Inhibitors. Virus Res 2017, 234, 177–192. [Google Scholar] [CrossRef]
- Gustavsson, E.; Grünewald, K.; Elias, P.; Hällberg, B.M. Dynamics of the Herpes Simplex Virus DNA Polymerase Holoenzyme during DNA Synthesis and Proof-Reading Revealed by Cryo-EM. Nucleic Acids Res 2024, 52, 7292–7304. [Google Scholar] [CrossRef]
- Shankar, S.; Pan, J.; Yang, P.; Bian, Y.; Oroszlán, G.; Yu, Z.; Mukherjee, P.; Filman, D.J.; Hogle, J.M.; Shekhar, M.; et al. Viral DNA Polymerase Structures Reveal Mechanisms of Antiviral Drug Resistance. Cell 2024. [Google Scholar] [CrossRef]
- D’Cruz, O.J.; Uckun, F.M. Dawn of Non-Nucleoside Inhibitor-Based Anti-HIV Microbicides. Journal of Antimicrobial Chemotherapy 2006, 57, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Phenotypic Susceptibility to Nonnucleoside Inhibitors of... : JAIDS Journal of Acquired Immune Deficiency Syndromes.
- Smerdon, S.J.; Jäger, J.; Wang, J.; Kohlstaedt, L.A.; Chirino, A.J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Structure of the Binding Site for Nonnucleoside Inhibitors of the Reverse Transcriptase of Human Immunodeficiency Virus Type 1. Proc Natl Acad Sci U S A 1994, 91, 3911–3915. [Google Scholar] [CrossRef]
- Merluzzi, V.J.; Hargrave, K.D.; Labadia, M.; Grozinger, K.; Skoog, M.; Wu, J.C.; Shih, C.-K.; Eckner, K.; Hattox, S.; Adams, J.; et al. Inhibition of HIV-1 Replication by a Nonnucleoside Reverse Transcriptase Inhibitor. Science 1990, 250, 1411–1413. [Google Scholar] [CrossRef] [PubMed]
- Omoto, S.; Speranzini, V.; Hashimoto, T.; Noshi, T.; Yamaguchi, H.; Kawai, M.; Kawaguchi, K.; Uehara, T.; Shishido, T.; Naito, A.; et al. Characterization of Influenza Virus Variants Induced by Treatment with the Endonuclease Inhibitor Baloxavir Marboxil. Scientific Reports 2018 8:1 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Omoto, S.; Speranzini, V.; Hashimoto, T.; Noshi, T.; Yamaguchi, H.; Kawai, M.; Kawaguchi, K.; Uehara, T.; Shishido, T.; Naito, A.; et al. Characterization of Influenza Virus Variants Induced by Treatment with the Endonuclease Inhibitor Baloxavir Marboxil. Scientific Reports 2018 8:1 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Chang, S.; Sun, D.; Liang, H.; Wang, J.; Li, J.; Guo, L.; Wang, X.; Guan, C.; Boruah, B.M.; Yuan, L.; et al. Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3Å Resolution. Mol Cell 2015, 57, 925–935. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat Commun 2019, 10. [Google Scholar] [CrossRef]
- FDA Approves First Treatment for COVID-19.
- Yin, W.; Mao, C.; Luan, X.; Shen, D.D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural Basis for Inhibition of the RNA-Dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. Science (1979) 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp): A Molecular Docking Study. Life Sci 2020, 253. [Google Scholar] [CrossRef]
- Sangawa, H.; Komeno, T.; Nishikawa, H.; Yoshida, A.; Takahashi, K.; Nomura, N.; Furuta, Y. Mechanism of Action of T-705 Ribosyl Triphosphate against Influenza Virus RNA Polymerase. Antimicrob Agents Chemother 2013, 57, 5202–5208. [Google Scholar] [CrossRef]
- Painter, G.R.; Natchus, M.G.; Cohen, O.; Holman, W.; Painter, W.P. Developing a Direct Acting, Orally Available Antiviral Agent in a Pandemic: The Evolution of Molnupiravir as a Potential Treatment for COVID-19. Curr Opin Virol 2021, 50, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.J.; Toots, M.; Lee, S.; Lee, M.E.; Ludeke, B.; Luczo, J.M.; Ganti, K.; Cox, R.M.; Sticher, Z.M.; Edpuganti, V.; et al. Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob Agents Chemother 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Peng, R.; Yuan, B.; Wang, M.; Zhao, J.; Fu, L.; Qi, J.; Shi, Y. Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir. Innovation 2021, 2. [Google Scholar] [CrossRef] [PubMed]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Pommier, Y.; Johnson, A.A.; Marchand, C. Integrase Inhibitors to Treat HIV/AIDS. Nat Rev Drug Discov 2005, 4, 236–248. [Google Scholar] [CrossRef]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Marius Clore, G.; Gronenborn, A.M. Solution Structure of the N-Terminal Zinc Binding Domain of HIV-1 Integrase. Nat Struct Biol 1997, 4, 567–577. [Google Scholar] [CrossRef]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and Function of Retroviral Integrase. Nat Rev Microbiol 2022, 20, 20–34. [Google Scholar] [CrossRef]
- Blanco, J.L.; Whitlock, G.; Milinkovic, A.; Moyle, G. HIV Integrase Inhibitors: A New Era in the Treatment of HIV. Expert Opin Pharmacother 2015, 16, 1313–1324. [Google Scholar] [CrossRef]
- Goldgur, Y.; Craigie, R.; Cohen, G.H.; Fujiwara, T.; Yoshinaga, T.; Fujishita, T.; Sugimoto, H.; Endo, T.; Murai, H.; Davies, D.R. Structure of the HIV-1 Integrase Catalytic Domain Complexed with an Inhibitor: A Platform for Antiviral Drug Design. Proc Natl Acad Sci U S A 1999, 96, 13040–13043. [Google Scholar] [CrossRef] [PubMed]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Smith, S.J.; Métifiot, M.; Jaxa-Chamiec, A.; Pommier, Y.; Hughes, S.H.; Cherepanov, P. Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572). Mol Pharmacol 2011, 80, 565–572. [Google Scholar] [CrossRef]
- Ballantyne, A.D.; Perry, C.M. Dolutegravir: First Global Approval. Drugs 2013, 73, 1627–1637. [Google Scholar] [CrossRef]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- New Report Documents Increase in HIV Drug Resistance to Dolutegravir.
- Li, M.; Passos, D.O.; Shan, Z.; Smith, S.J.; Sun, Q.; Biswas, A.; Choudhuri, I.; Strutzenberg, T.S.; Haldane, A.; Deng, N.; et al. Mechanisms of HIV-1 Integrase Resistance to Dolutegravir and Potent Inhibition of Drug-Resistant Variants. Sci Adv 2023, 9. [Google Scholar] [CrossRef]
- Passos, D.O.; Li, M.; Jóźwik, I.K.; Zhao, X.Z.; Santos-Martins, D.; Yang, R.; Smith, S.J.; Jeon, Y.; Forli, S.; Hughes, S.H.; et al. Structural Basis for Strand-Transfer Inhibitor Binding to HIV Intasomes. Science (1979) 2020, 367, 810–814. [Google Scholar] [CrossRef]
- Renzi, G.; Carta, F.; Supuran, C.T. The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Bonnard, D.; Le Rouzic, E.; Eiler, S.; Amadori, C.; Orlov, I.; Bruneau, J.M.; Brias, J.; Barbion, J.; Chevreuil, F.; Spehner, D.; et al. Structure-Function Analyses Unravel Distinct Effects of Allosteric Inhibitors of HIV-1 Integrase on Viral Maturation and Integration. Journal of Biological Chemistry 2018, 293, 6172–6186. [Google Scholar] [CrossRef]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and Function of Retroviral Integrase. Nat Rev Microbiol 2022, 20, 20–34. [Google Scholar] [CrossRef]
- Christ, F.; Voet, A.; Marchand, A.; Nicolet, S.; Desimmie, B.A.; Marchand, D.; Bardiot, D.; Van Der Veken, N.J.; Van Remoortel, B.; Strelkov, S. V.; et al. Rational Design of Small-Molecule Inhibitors of the LEDGF/P75-Integrase Interaction and HIV Replication. Nat Chem Biol 2010, 6, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, L.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Zhu, D.; Zhao, X.; Chen, S.; et al. Alpha-Herpesvirus Thymidine Kinase Genes Mediate Viral Virulence and Are Potential Therapeutic Targets. Front Microbiol 2019, 10, 941. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Visse, R.; Sandhu, G.; Davies, A.; Rizkallah, P.J.; Melitz, C.; Summers, W.C.; Sanderson, M.R. Crystal Structures of the Thymidine Kinase from Herpes Simplex Virus Type-I in Complex with Deoxythymidine and Ganciclovir. Nat Struct Biol 1995, 2, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Wild, K.; Bohner, T.; Aubry, A.; Folkers, G.; Schulz, G.E. The Three-Dimensional Structure of Thymidine Kinase from Herpes Simplex Virus Type 1. FEBS Lett 1995, 368, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Frobert, E.; Ooka, T.; Cortay, J.C.; Lina, B.; Thouvenot, D.; Morfin, F. Herpes Simplex Virus Thymidine Kinase Mutations Associated with Resistance to Acyclovir: A Site-Directed Mutagenesis Study. Antimicrob Agents Chemother 2005, 49, 1055–1059. [Google Scholar] [CrossRef]
- Ramdhan, P.; Li, C. Targeting Viral Methyltransferases: An Approach to Antiviral Treatment for SsRNA Viruses. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Jones, R.; Bragagnolo, G.; Arranz, R.; Reguera, J. Capping Pores of Alphavirus NsP1 Gate Membranous Viral Replication Factories. Nature 2021, 589, 615–619. [Google Scholar] [CrossRef]
- Lampio, A.; Kilpeläinen, I.; Pesonen, S.; Karhi, K.; Auvinen, P.; Somerharju, P.; Kääriäinen, L. Membrane Binding Mechanism of an RNA Virus-Capping Enzyme. Journal of Biological Chemistry 2000, 275, 37853–37859. [Google Scholar] [CrossRef]
- Jones, R.; Hons, M.; Rabah, N.; Zamarreño, N.; Arranz, R.; Reguera, J. Structural Basis and Dynamics of Chikungunya Alphavirus RNA Capping by NsP1 Capping Pores. Proc Natl Acad Sci U S A 2023, 120. [Google Scholar] [CrossRef]
- Bhutkar, M.; Saha, A.; Tomar, S. Viral Methyltransferase Inhibitors: Berbamine, Venetoclax, and Ponatinib as Efficacious Antivirals against Chikungunya Virus. Arch Biochem Biophys 2024, 759. [Google Scholar] [CrossRef]
- Mudgal, R.; Bharadwaj, C.; Dubey, A.; Choudhary, S.; Nagarajan, P.; Aggarwal, M.; Ratra, Y.; Basak, S.; Tomar, S. Selective Estrogen Receptor Modulators Limit Alphavirus Infection by Targeting the Viral Capping Enzyme NsP1. Antimicrob Agents Chemother 2022, 66. [Google Scholar] [CrossRef] [PubMed]
- Mudgal, R.; Mahajan, S.; Tomar, S. Inhibition of Chikungunya Virus by an Adenosine Analog Targeting the SAM-Dependent NsP1 Methyltransferase. FEBS Lett 2020, 594, 678–694. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ray, D.; Zhao, Y.; Dong, H.; Ren, S.; Li, Z.; Guo, Y.; Bernard, K.A.; Shi, P.-Y.; Li, H. Structure and Function of Flavivirus NS5 Methyltransferase. J Virol 2007, 81, 3891–3903. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Sonntag, L.S.; Noble, C.; Nilar, S.H.; Ng, R.H.; Zou, G.; Monaghan, P.; Chung, K.Y.; Dong, H.; Liu, B.; et al. Small Molecule Inhibitors That Selectively Block Dengue Virus Methyltransferase. Journal of Biological Chemistry 2011, 286, 6233–6240. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Cyr, M.; Longenecker, K.; Tripathi, R.; Sun, C.; Kempf, D.J. Crystal Structure of Full-Length Zika Virus NS5 Protein Reveals a Conformation Similar to Japanese Encephalitis Virus NS5. Acta Crystallographica Section:F Structural Biology Communications 2017, 73, 116–122. [Google Scholar] [CrossRef]
- Jia, H.; Zhong, Y.; Peng, C.; Gong, P. Crystal Structures of Flavivirus NS5 Guanylyltransferase Reveal a GMP-Arginine Adduct. J Virol 2022, 96. [Google Scholar] [CrossRef]
- Chen, H.; Lin, S.; Yang, F.; Chen, Z.; Guo, L.; Yang, J.; Lin, X.; Wang, L.; Duan, Y.; Wen, A.; et al. Structural and Functional Basis of Low-Affinity SAM/SAH-Binding in the Conserved MTase of the Multi-Segmented Alongshan Virus Distantly Related to Canonical Unsegmented Flaviviruses. PLoS Pathog 2023, 19. [Google Scholar] [CrossRef]
- Bhutkar, M.; Kumar, A.; Rani, R.; Singh, V.; Pathak, A.; Kothiala, A.; Mahajan, S.; Waghmode, B.; Kumar, R.; Mudgal, R.; et al. SAM-Dependent Viral MTase Inhibitors: Herbacetin and Caffeic Acid Phenethyl Ester, Structural Insights into Dengue MTase. bioRxiv 2024, 2022.05.31.494145. [CrossRef]
- García, L.L.; Padilla, L.; Castaño, J.C. Inhibitors Compounds of the Flavivirus Replication Process. Virol J 2017, 14. [Google Scholar] [CrossRef]
- Benarroch, D.; Egloff, M.P.; Mulard, L.; Guerreiro, C.; Romette, J.L.; Canard, B. A Structural Basis for the Inhibition of the NS5 Dengue Virus MRNA 2′-O-Methyltransferase Domain by Ribavirin 5′-Triphosphate. Journal of Biological Chemistry 2004, 279, 35638–35643. [Google Scholar] [CrossRef]
- Milani, M.; Mastrangelo, E.; Bollati, M.; Selisko, B.; Decroly, E.; Bouvet, M.; Canard, B.; Bolognesi, M. Flaviviral Methyltransferase/RNA Interaction: Structural Basis for Enzyme Inhibition. Antiviral Res 2009, 83, 28–34. [Google Scholar] [CrossRef]
- Nencka, R.; Silhan, J.; Klima, M.; Otava, T.; Kocek, H.; Krafcikova, P.; Boura, E. Coronaviral RNA-Methyltransferases: Function, Structure and Inhibition. Nucleic Acids Res 2022, 50, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y. Perspective for Drug Discovery Targeting SARS Coronavirus Methyltransferases: Function, Structure and Inhibition. J Med Chem 2024. [CrossRef] [PubMed]
- Pyle, A.M. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annu Rev Biophys 2008, 37, 317–336. [Google Scholar] [CrossRef]
- Kolykhalov, A.A.; Mihalik, K.; Feinstone, S.M.; Rice, C.M. Hepatitis C Virus-Encoded Enzymatic Activities and Conserved RNA Elements in the 3′ Nontranslated Region Are Essential for Virus Replication In Vivo. J Virol 2000, 74, 2046–2051. [Google Scholar] [CrossRef]
- Jankowsky, E.; Gross, C.H.; Shuman, S.; Pyle, A.M. The DExH Protein NPH-II Is a Processive and Directional Motor for Unwinding RNA. Nature 2000, 403, 447–451. [Google Scholar] [CrossRef]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandão-Neto, J.; Dunnett, L.; Gorrie-stone, T.; Skyner, R.; Fearon, D.; et al. Structure, Mechanism and Crystallographic Fragment Screening of the SARS-CoV-2 NSP13 Helicase. Nat Commun 2021, 12. [Google Scholar] [CrossRef]
- Smelkova, N. V; Borowiec, J.A. Dimerization of Simian Virus 40 T-Antigen Hexamers Activates T-Antigen DNA Helicase Activity. J Virol 1997, 71, 8766–8773. [Google Scholar] [CrossRef]
- Hughes, F.J.; Romanos, M.A. E1 Protein of Human Papillomavirus Is a DNA Helicase/ATPase. Nucleic Acids Res 1993, 21, 5817–5823. [Google Scholar] [CrossRef]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.G.; Lescar, J. Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 Å. J Virol 2005, 79, 10278–10288. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Yang, M.; Yang, J.; Shao, Z.; Gao, Y.; Jiang, X.; Cui, R.; Zhang, Y.; Zhao, X.; et al. Structural and Functional Insights into the Helicase Protein E5 of Mpox Virus. Cell Discov 2024, 10. [Google Scholar] [CrossRef]
- Law, Y.-S.; Wang, S.; Tan, Y.B.; Shih, O.; Utt, A.; Goh, W.Y.; Lian, B.-J.; Chen, M.W.; Jeng, U.-S.; Merits, A.; et al. Interdomain Flexibility of Chikungunya Virus NsP2 Helicase-Protease Differentially Influences Viral RNA Replication and Infectivity. J Virol 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Sun, Y.; Rao, Z. Current Progress in Antiviral Strategies. Trends Pharmacol Sci 2014, 35, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bera, A.K.; Kuhn, R.J.; Smith, J.L. Structure of the Flavivirus Helicase: Implications for Catalytic Activity, Protein Interactions, and Proteolytic Processing. J Virol 2005, 79, 10268–10277. [Google Scholar] [CrossRef] [PubMed]
- Tortorici, M.A.; Duquerroy, S.; Kwok, J.; Vonrhein, C.; Perez, J.; Lamp, B.; Bricogne, G.; Rümenapf, T.; Vachette, P.; Rey, F.A. X-Ray Structure of the Pestivirus NS3 Helicase and Its Conformation in Solution. J Virol 2015, 89, 4356–4371. [Google Scholar] [CrossRef]
- Fang, X.; Lu, G.; Deng, Y.; Yang, S.; Hou, C.; Gong, P. Unusual Substructure Conformations Observed in Crystal Structures of a Dicistrovirus RNA-Dependent RNA Polymerase Suggest Contribution of the N-Terminal Extension in Proper Folding. Virol Sin 2023, 38, 531–540. [Google Scholar] [CrossRef]
- Anindita, P.D.; Halbeisen, M.; Řeha, D.; Tuma, R.; Franta, Z. Mechanistic Insight into the RNA-Stimulated ATPase Activity of Tick-Borne Encephalitis Virus Helicase. Journal of Biological Chemistry 2022, 298. [Google Scholar] [CrossRef]
- Shao, Z.; Su, S.; Yang, J.; Zhang, W.; Gao, Y.; Zhao, X.; Zhang, Y.; Shao, Q.; Cao, C.; Li, H.; et al. Structures and Implications of the C962R Protein of African Swine Fever Virus. Nucleic Acids Res 2023, 51, 9475–9490. [Google Scholar] [CrossRef]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.G.; Lescar, J. Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 Å. J Virol 2005, 79, 10278–10288. [Google Scholar] [CrossRef]
- Gu, M.; Rice, C.M. Three Conformational Snapshots of the Hepatitis C Virus NS3 Helicase Reveal a Ratchet Translocation Mechanism. Proc Natl Acad Sci U S A 2010, 107, 521–528. [Google Scholar] [CrossRef]
- Hutin, S.; Ling, W.L.; Tarbouriech, N.; Schoehn, G.; Grimm, C.; Fischer, U.; Burmeister, W.P. The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y. Sen; Gesell, J.J.; Wyss, D.F. Solution Structure and Backbone Dynamics of an Engineered Arginine-Rich Subdomain 2 of the Hepatitis C Virus NS3 RNA Helicase. J Mol Biol 2001, 314, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Hutin, S.; Ling, W.L.; Tarbouriech, N.; Schoehn, G.; Grimm, C.; Fischer, U.; Burmeister, W.P. The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction. Viruses 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Frick, D.; Lam, A. Understanding Helicases as a Means of Virus Control. Curr Pharm Des 2006, 12, 1315–1338. [Google Scholar] [CrossRef]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J.; et al. Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science (1979) 2013, 341, 1387–1390. [Google Scholar] [CrossRef]
- Wu, B.; Chien, E.Y.T.; Mol, C.D.; Fenalti, G.; Liu, W.; Katritch, V.; Abagyan, R.; Brooun, A.; Wells, P.; Bi, F.C.; et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science (1979) 2010, 330, 1066–1071. [Google Scholar] [CrossRef]
- Haqqani, A.A.; Tilton, J.C. Entry Inhibitors and Their Use in the Treatment of HIV-1 Infection. Antiviral Res 2013, 98, 158–170. [Google Scholar] [CrossRef]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J.; et al. Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science (1979) 2013, 341, 1387–1390. [Google Scholar] [CrossRef]
- Blair, H.A. Ibalizumab: A Review in Multidrug-Resistant HIV-1 Infection. Drugs 2020, 80, 189–196. [Google Scholar] [CrossRef]
- Freeman, M.M.; Seaman, M.S.; Rits-Volloch, S.; Hong, X.; Kao, C.Y.; Ho, D.D.; Chen, B. Crystal Structure of HIV-1 Primary Receptor CD4 in Complex with a Potent Antiviral Antibody. Structure 2010, 18, 1632–1641. [Google Scholar] [CrossRef]
- Bhutkar, M.; Singh, V.; Dhaka, P.; Tomar, S. Virus-Host Protein-Protein Interactions as Molecular Drug Targets for Arboviral Infections. Frontiers in Virology 2022, 2. [Google Scholar] [CrossRef]
- Mahajan, S.; Choudhary, S.; Kumar, P.; Tomar, S. Antiviral Strategies Targeting Host Factors and Mechanisms Obliging +ssRNA Viral Pathogens. Bioorg Med Chem 2021, 46, 116356. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, P.; Singh, A.; Nehul, S.; Choudhary, S.; Panda, P.K.; Sharma, G.K.; Kumar, P.; Tomar, S. Disruption of Molecular Interactions between the G3BP1 Stress Granule Host Protein and the Nucleocapsid (NTD-N) Protein Impedes SARS-CoV-2 Virus Replication. Biochemistry 2024. [Google Scholar] [CrossRef]
- Mahajan, S.; Kumar, R.; Singh, A.; Pareek, A.; Kumar, P.; Tomar, S. Targeting the Host Protein G3BP1 for the Discovery of Novel Antiviral Inhibitors against Chikungunya Virus. bioRxiv 2024, 2022.11.11.516135. [CrossRef]
- De Chassey, B.; Meyniel-Schicklin, L.; Aublin-Gex, A.; André, P.; Lotteau, V. New Horizons for Antiviral Drug Discovery from Virus–Host Protein Interaction Networks. Curr Opin Virol 2012, 2, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Idrees, S.; Chen, H.; Panth, N.; Paudel, K.R.; Hansbro, P.M. Exploring Viral–Host Protein Interactions as Antiviral Therapies: A Computational Perspective. Microorganisms 2024, 12. [Google Scholar] [CrossRef] [PubMed]
- Idrees, S.; Paudel, K.R.; Sadaf, T.; Hansbro, P.M. How Different Viruses Perturb Host Cellular Machinery via Short Linear Motifs. EXCLI J 2023, 22, 1113–1128. [Google Scholar] [CrossRef]
- de Chassey, B.; Meyniel-Schicklin, L.; Vonderscher, J.; André, P.; Lotteau, V. Virus-Host Interactomics: New Insights and Opportunities for Antiviral Drug Discovery. Genome Med 2014, 6. [Google Scholar] [CrossRef]
- Brito, A.F.; Pinney, J.W. Protein-Protein Interactions in Virus-Host Systems. Front Microbiol 2017, 8. [Google Scholar] [CrossRef]
- Kumar, N.; Sharma, S.; Kumar, R.; Tripathi, B.N.; Barua, S.; Ly, H.; Rouse, B.T. Host-Directed Antiviral Therapy. Clin Microbiol Rev 2020, 33. [Google Scholar] [CrossRef]
- Mahajan, S.; Choudhary, S.; Kumar, P.; Tomar, S. Antiviral Strategies Targeting Host Factors and Mechanisms Obliging +ssRNA Viral Pathogens. Bioorg Med Chem 2021, 46. [Google Scholar] [CrossRef]
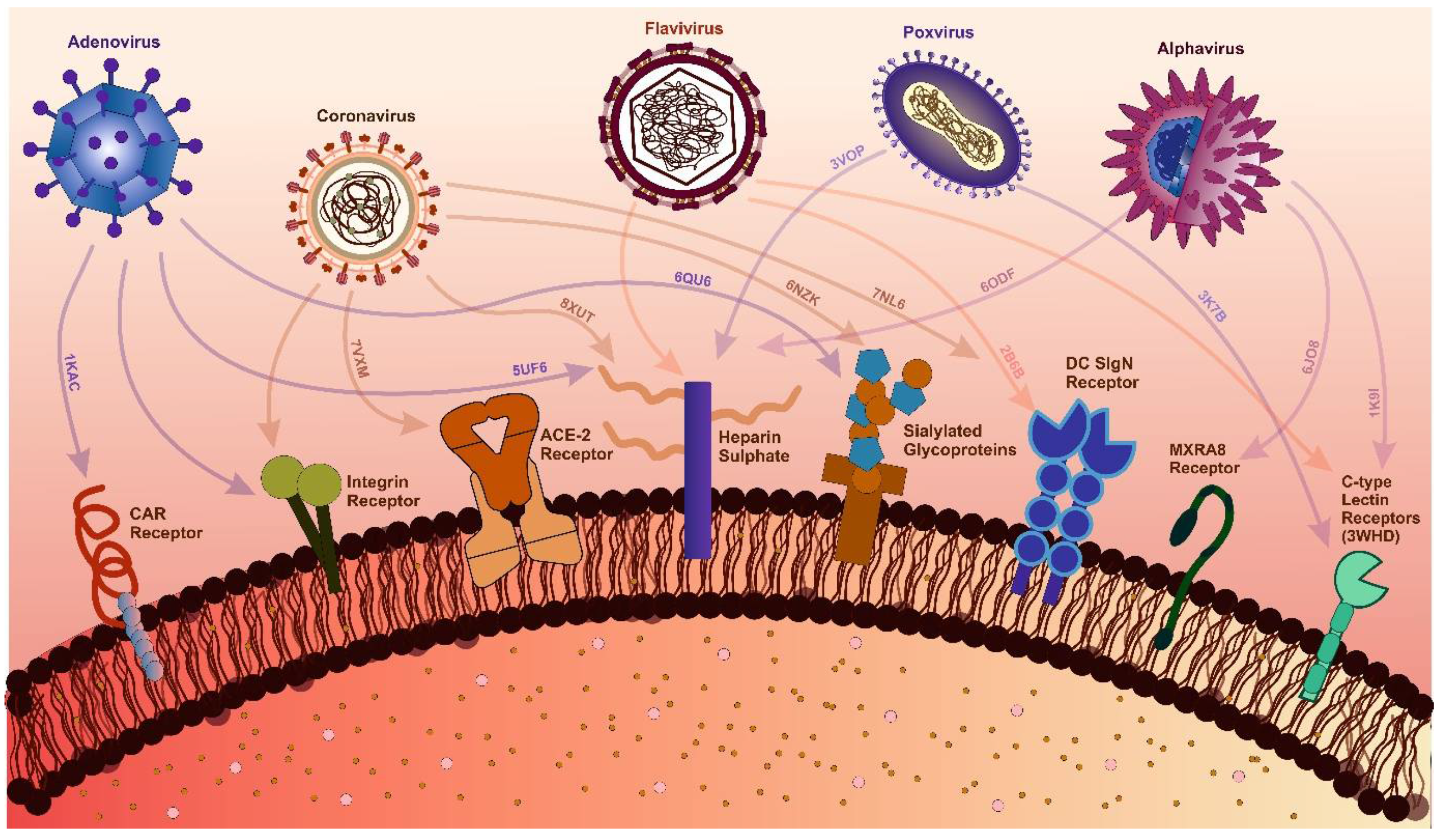
- Eslami, N.; Aghbash, P.S.; Shamekh, A.; Entezari-Maleki, T.; Nahand, J.S.; Sales, A.J.; Baghi, H.B. SARS-CoV-2: Receptor and Co-Receptor Tropism Probability. Curr Microbiol 2022, 79, 1–13. [Google Scholar] [CrossRef]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular Host Factors for SARS-CoV-2 Infection. Nature Microbiology 2021 6:10 2021, 6, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.S.; Diamond, M.S. A Molecular Understanding of Alphavirus Entry and Antibody Protection. Nature Reviews Microbiology 2022 21:6 2022, 21, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.F.; Wu, Y.S.; Poh, C.L. Molecular Mechanisms of Antiviral Agents against Dengue Virus. Viruses 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.N.; Akhtar, R.; Abid, M.; Khan, S.A.; Rehman, Z.U.; Tayyub, M.; Malik, M.I.; Shahzad, M.K.; Mubeen, H.; Qadir, M.S.; et al. The Interactions of Flaviviruses with Cellular Receptors: Implications for Virus Entry. Virology 2022, 568, 77–85. [Google Scholar] [CrossRef]
- Li, H.; Huang, Q.Z.; Zhang, H.; Liu, Z.X.; Chen, X.H.; Ye, L.L.; Luo, Y. The Land-Scape of Immune Response to Monkeypox Virus. EBioMedicine 2023, 87, 104424. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Lee, S.S. A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants With Escape Mutations. Front Immunol 2022, 13, 801522. [Google Scholar] [CrossRef]
- Lemaitre, J.C.; Grantz, K.H.; Kaminsky, J.; Meredith, H.R.; Truelove, S.A.; Lauer, S.A.; Keegan, L.T.; Shah, S.; Wills, J.; Kaminsky, K.; et al. A Scenario Modeling Pipeline for COVID-19 Emergency Planning. Scientific Reports 2021 11:1 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Crucitti, D.; Pérez Míguez, C.; Ángel, J.; Arias, D.; Beltrá, D.; Prada, F.; Orgueira, A.M. De Novo Drug Design through Artificial Intelligence: An Introduction. Frontiers in Hematology 2024, 3, 1305741. [Google Scholar] [CrossRef]
- Floresta, G.; Zagni, C.; Patamia, V.; Rescifina, A. How Can Artificial Intelligence Be Utilized for de Novo Drug Design against COVID-19 (SARS-CoV-2)? Expert Opin Drug Discov 2023, 18, 1061–1064. [Google Scholar] [CrossRef]
- Patel, C.N.; Mall, R.; Bensmail, H. AI-Driven Drug Repurposing and Binding Pose Meta Dynamics Identifies Novel Targets for Monkeypox Virus. J Infect Public Health 2023, 16, 799–807. [Google Scholar] [CrossRef]
- Slater, A.; Nair, N.; Suétt, R.; Donnchadha, R. Mac; Bamford, C.; Jasim, S.; Livingstone, D.; Hutchinson, E. Visualising Viruses. Journal of General Virology 2022, 103, 001730. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yang, Q.; Ran, T.; Zhang, G.; Li, W.; Zhou, P.; Tang, J.; Dai, M.; Zhong, J.; Chen, H.; et al. Discovery of Orally Bioavailable SARS-CoV-2 Papain-like Protease Inhibitor as a Potential Treatment for COVID-19. Nature Communications 2024 15:1 2024, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Turzynski, V.; Monsees, I.; Moraru, C.; Probst, A.J. Imaging Techniques for Detecting Prokaryotic Viruses in Environmental Samples. Viruses 2021, Vol. 13, Page 2126 2021, 13, 2126. [Google Scholar] [CrossRef] [PubMed]
- Turzynski, V.; Monsees, I.; Moraru, C.; Probst, A.J. Imaging Techniques for Detecting Prokaryotic Viruses in Environmental Samples. Viruses 2021, Vol. 13, Page 2126 2021, 13, 2126. [Google Scholar] [CrossRef]
- Greasley, S.E.; Noell, S.; Plotnikova, O.; Ferre, R.A.; Liu, W.; Bolanos, B.; Fennell, K.; Nicki, J.; Craig, T.; Zhu, Y.; et al. Structural Basis for the in Vitro Efficacy of Nirmatrelvir against SARS-CoV-2 Variants. Journal of Biological Chemistry 2022, 298, 101972. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; Oualid, F. El; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity Profiling and Crystal Structures of Inhibitor-Bound SARS-CoV-2 Papain-like Protease: A Framework for Anti–COVID-19 Drug Design. Sci Adv 2020, 6, 4596–4612. [Google Scholar] [CrossRef]
- Feng, Q.; Cheng, K.; Zhang, L.; Wang, D.; Gao, X.; Liang, J.; Liu, G.; Ma, N.; Xu, C.; Tang, M.; et al. Rationally Designed Multimeric Nanovaccines Using Icosahedral DNA Origami for Display of SARS-CoV-2 Receptor Binding Domain. Nature Communications 2024 15:1 2024, 15, 1–17. [Google Scholar] [CrossRef]
- Dokhale, S.; Garse, S.; Devarajan, S.; Thakur, V.; Kolhapure, S. Rational Design of Antiviral Therapeutics. Computational Methods for Rational Drug Design 2025, 423–443. [Google Scholar] [CrossRef]
- Al-Amran, F.G.; Hezam, A.M.; Rawaf, S.; Yousif, M.G. Genomic Analysis and Artificial Intelligence: Predicting Viral Mutations and Future Pandemics.
- Parvatikar, P.P.; Patil, S.; Khaparkhuntikar, K.; Patil, S.; Singh, P.K.; Sahana, R.; Kulkarni, R. V.; Raghu, A. V. Artificial Intelligence: Machine Learning Approach for Screening Large Database and Drug Discovery. Antiviral Res 2023, 220, 105740. [Google Scholar] [CrossRef]
- KP Jayatunga, M.; Ayers, M.; Bruens, L.; Jayanth, D.; Meier, C. How Successful Are AI-Discovered Drugs in Clinical Trials? A First Analysis and Emerging Lessons. Drug Discov Today 2024, 29, 104009. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in De Novo Drug Design: From Conventional to Machine Learning Methods. International Journal of Molecular Sciences 2021, Vol. 22, Page 1676 2021, 22, 1676. [Google Scholar] [CrossRef] [PubMed]
- Atz, K.; Cotos, L.; Isert, C.; Håkansson, M.; Focht, D.; Hilleke, M.; Nippa, D.F.; Iff, M.; Ledergerber, J.; Schiebroek, C.C.G.; et al. Prospective de Novo Drug Design with Deep Interactome Learning. Nature Communications 2024 15:1 2024, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Bhutkar, M.; Choudhary, S.; Nehul, S.; Kumar, R.; Singla, J.; Kumar, P.; Tomar, S. Structure-Guided Mutations in CDRs for Enhancing the Affinity of Neutralizing SARS-CoV-2 Nanobody. Biochem Biophys Res Commun 2024, 734, 150746. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Choudhary, S.; Bhutkar, M.; Nehul, S.; Ali, S.; Singla, J.; Kumar, P.; Tomar, S. Designing and Bioengineering of CDRs with Higher Affinity against Receptor-Binding Domain (RBD) of SARS-CoV-2 Omicron Variant 2024.
- Sinha, S.; Vegesna, R.; Mukherjee, S.; Kammula, A. V.; Dhruba, S.R.; Wu, W.; Kerr, D.L.; Nair, N.U.; Jones, M.G.; Yosef, N.; et al. PERCEPTION Predicts Patient Response and Resistance to Treatment Using Single-Cell Transcriptomics of Their Tumors. Nature Cancer 2024 5:6 2024, 5, 938–952. [Google Scholar] [CrossRef]
- Mak, K.-K.; Wong, Y.-H.; Pichika, M.R. Artificial Intelligence in Drug Discovery and Development. Drug Discovery and Evaluation: Safety and Pharmacokinetic Assays 2024, 1461–1498. [CrossRef]
- He, H.; He, B.; Guan, L.; Zhao, Y.; Jiang, F.; Chen, G.; Zhu, Q.; Chen, C.Y.C.; Li, T.; Yao, J. De Novo Generation of SARS-CoV-2 Antibody CDRH3 with a Pre-Trained Generative Large Language Model. Nature Communications 2024 15:1 2024, 15, 1–19. [Google Scholar] [CrossRef]
- Dunbar, J.; Krawczyk, K.; Leem, J.; Marks, C.; Nowak, J.; Regep, C.; Georges, G.; Kelm, S.; Popovic, B.; Deane, C.M. SAbPred: A Structure-Based Antibody Prediction Server. Nucleic Acids Res 2016, 44, W474–W478. [Google Scholar] [CrossRef]
- Caradonna, T.M.; Schmidt, A.G. Protein Engineering Strategies for Rational Immunogen Design. npj Vaccines 2021 6:1 2021, 6, 1–11. [Google Scholar] [CrossRef]
- Shanehsazzadeh, A.; McPartlon, M.; Kasun, G.; Steiger, A.K.; Sutton, J.M.; Yassine, E.; McCloskey, C.; Haile, R.; Shuai, R.; Alverio, J.; et al. Unlocking de Novo Antibody Design with Generative Artificial Intelligence. bioRxiv 2024, 2023.01.08.523187. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021 596:7873 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024 630:8016 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De Novo Design of Protein Structure and Function with RFdiffusion. Nature 2023 620:7976 2023, 620, 1089–1100. [Google Scholar] [CrossRef]
- Greasley, S.E.; Noell, S.; Plotnikova, O.; Ferre, R.A.; Liu, W.; Bolanos, B.; Fennell, K.; Nicki, J.; Craig, T.; Zhu, Y.; et al. Structural Basis for the in Vitro Efficacy of Nirmatrelvir against SARS-CoV-2 Variants. Journal of Biological Chemistry 2022, 298, 101972. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Kerry, P.S.; Bance, N.; Niikura, M.; Pinto, B.M. Serendipitous Discovery of a Potent Influenza Virus A Neuraminidase Inhibitor. Angewandte Chemie International Edition 2014, 53, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; Oualid, F. El; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity Profiling and Crystal Structures of Inhibitor-Bound SARS-CoV-2 Papain-like Protease: A Framework for Anti–COVID-19 Drug Design. Sci Adv 2020, 6, 4596–4612. [Google Scholar] [CrossRef]
- Yuan, M.; Zhu, X.; He, W.T.; Zhou, P.; Kaku, C.I.; Capozzola, T.; Zhu, C.Y.; Yu, X.; Liu, H.; Yu, W.; et al. A Broad and Potent Neutralization Epitope in SARS-Related Coronaviruses. Proc Natl Acad Sci U S A 2022, 119, e2205784119. [Google Scholar] [CrossRef]
- Walls, A.C.; Fiala, B.; Schäfer, A.; Wrenn, S.; Pham, M.N.; Murphy, M.; Tse, L. V.; Shehata, L.; O’Connor, M.A.; Chen, C.; et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 2020, 183, 1367–1382.e17. [Google Scholar] [CrossRef]
- Feng, Q.; Cheng, K.; Zhang, L.; Wang, D.; Gao, X.; Liang, J.; Liu, G.; Ma, N.; Xu, C.; Tang, M.; et al. Rationally Designed Multimeric Nanovaccines Using Icosahedral DNA Origami for Display of SARS-CoV-2 Receptor Binding Domain. Nature Communications 2024 15:1 2024, 15, 1–17. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Antanasijevic, A.; Ronk, A.J.; Müller-Kräuter, H.; Watanabe, Y.; Claireaux, M.; Perrett, H.R.; Bijl, T.P.L.; Grobben, M.; Umotoy, J.C.; et al. Lassa Virus Glycoprotein Nanoparticles Elicit Neutralizing Antibody Responses and Protection. Cell Host Microbe 2022, 30, 1759–1772.e12. [Google Scholar] [CrossRef]
- Sesterhenn, F.; Yang, C.; Bonet, J.; Cramer, J.T.; Wen, X.; Wang, Y.; Chiang, C.I.; Abriata, L.A.; Kucharska, I.; Castoro, G.; et al. De Novo Protein Design Enables the Precise Induction of RSV-Neutralizing Antibodies. Science (1979) 2020, 368. [Google Scholar] [CrossRef]
- Marcandalli, J.; Fiala, B.; Ols, S.; Perotti, M.; de van der Schueren, W.; Snijder, J.; Hodge, E.; Benhaim, M.; Ravichandran, R.; Carter, L.; et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 2019, 176, 1420–1431.e17. [Google Scholar] [CrossRef]
- Mousa, J.J.; Sauer, M.F.; Sevy, A.M.; Finn, J.A.; Bates, J.T.; Alvarado, G.; King, H.G.; Loerinc, L.B.; Fong, R.H.; Doranz, B.J.; et al. Structural Basis for Nonneutralizing Antibody Competition at Antigenic Site II of the Respiratory Syncytial Virus Fusion Protein. Proc Natl Acad Sci U S A 2016, 113, E6849–E6858. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen with Individual Antibodies. Science (1979) 2020, 369, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Behre, G.; Hebert, C.; Kumar, P.; Farmer MacPherson, L.; Graham-Clarke, P.L.; De La Torre, I.; Nichols, R.M.; Hufford, M.M.; Patel, D.R.; et al. Bamlanivimab and Etesevimab Improve Symptoms and Associated Outcomes in Ambulatory Patients at Increased Risk for Severe Coronavirus Disease 2019: Results From the Placebo-Controlled Double-Blind Phase 3 BLAZE-1 Trial. Open Forum Infect Dis 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zost, S.J.; Greaney, A.J.; Starr, T.N.; Dingens, A.S.; Chen, E.C.; Chen, R.E.; Case, J.B.; Sutton, R.E.; Gilchuk, P.; et al. Genetic and Structural Basis for SARS-CoV-2 Variant Neutralization by a Two-Antibody Cocktail. Nature Microbiology 2021 6:10 2021, 6, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Casadevall, A.; Franchini, M.; Maggi, F. Sotrovimab: A Review of Its Efficacy against SARS-CoV-2 Variants. Viruses 2024, Vol. 16, Page 217 2024, 16, 217. [Google Scholar] [CrossRef] [PubMed]
- Vishweshwaraiah, Y.L.; Hnath, B.; Rackley, B.; Wang, J.; Gontu, A.; Chandler, M.; Afonin, K.A.; Kuchipudi, S. V; Christensen, N.; Yennawar, N.H.; et al. Adaptation-Proof SARS-CoV-2 Vaccine Design. Adv Funct Mater 2022, 32, 2206055. [Google Scholar] [CrossRef]
- Liu, Q.; Lu, Y.; Cai, C.; Huang, Y.; Zhou, L.; Guan, Y.; Fu, S.; Lin, Y.; Yan, H.; Zhang, Z.; et al. A Broad Neutralizing Nanobody against SARS-CoV-2 Engineered from an Approved Drug. Cell Death & Disease 2024 15:6 2024, 15, 1–9. [Google Scholar] [CrossRef]
- Pathogens Prioritization: A Scientific Framework for Epidemic and Pandemic Research Preparedness.
- Bardhan, M.; Ray, I.; Roy, S.; Roy, P.; Thanneeru, P.; Twayana, A.R.; Prasad, S.; Bardhan, M.; Anand, A. Disease X and COVID-19: Turning Lessons from India and the World into Policy Recommendations. Ann Med Surg (Lond) 2024, 86, 5914–5921. [Google Scholar] [CrossRef]
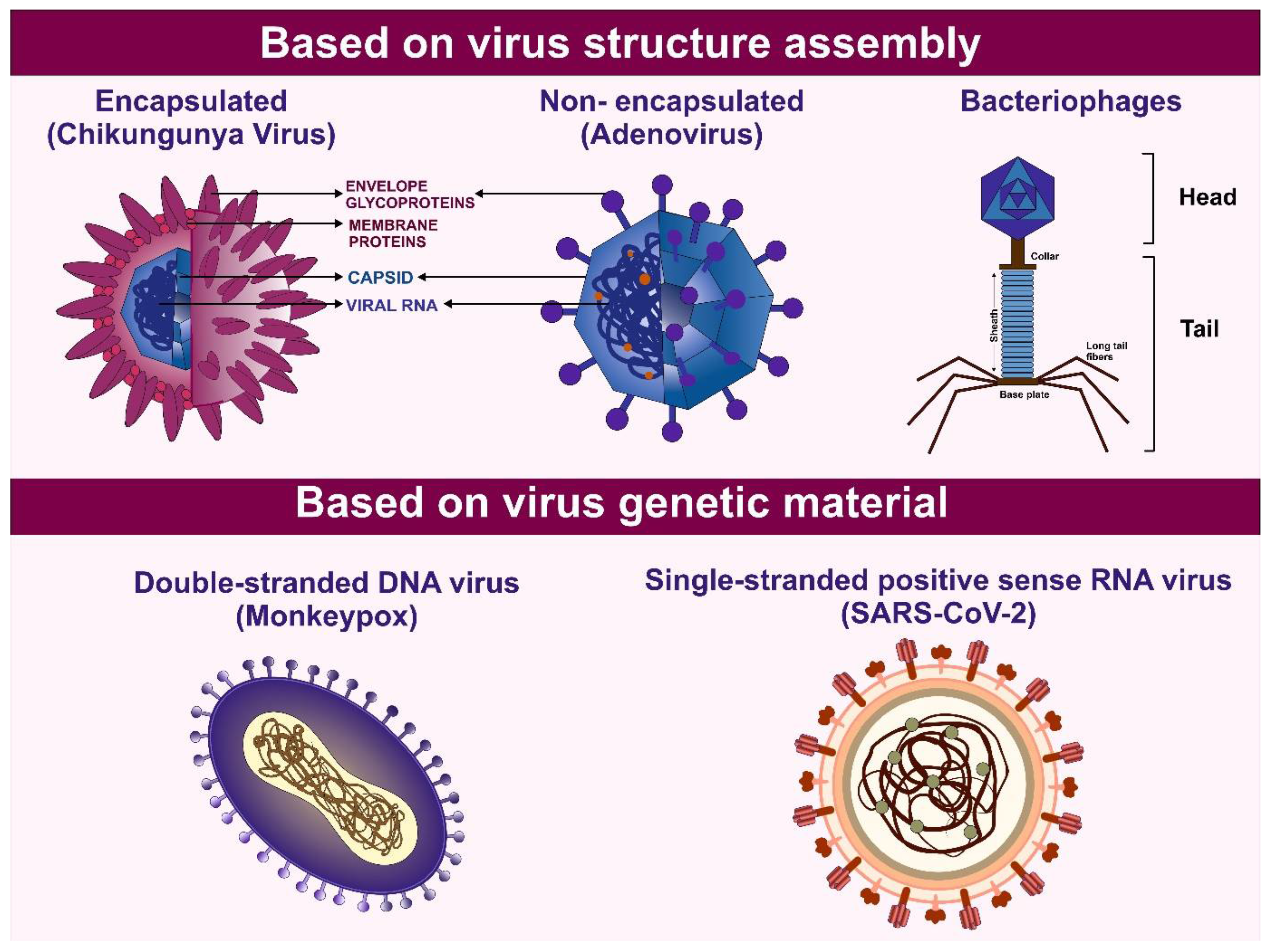
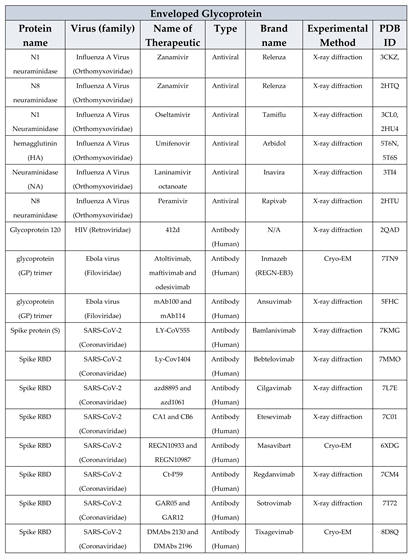
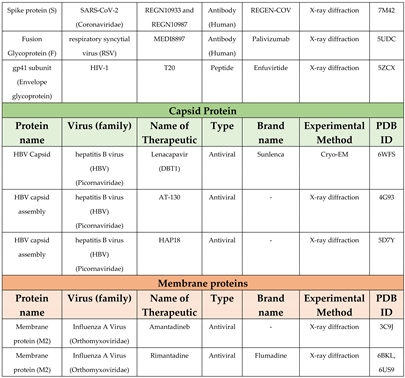
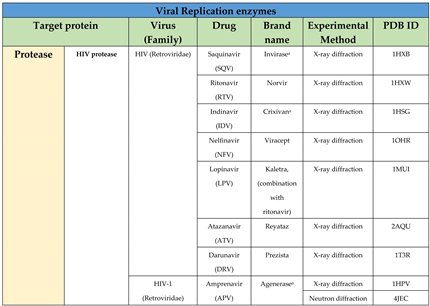
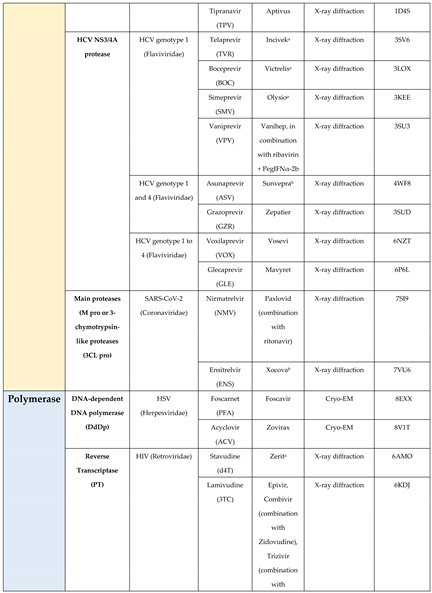
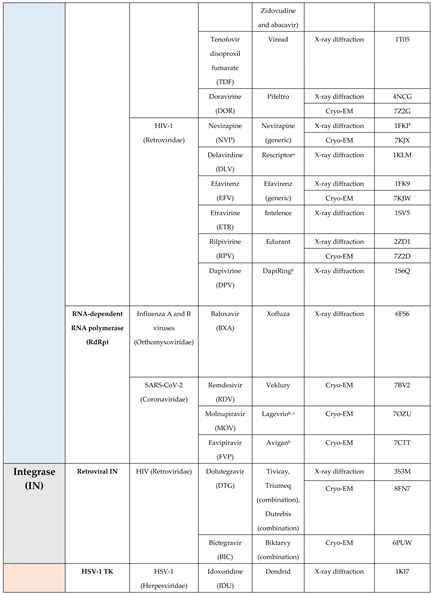
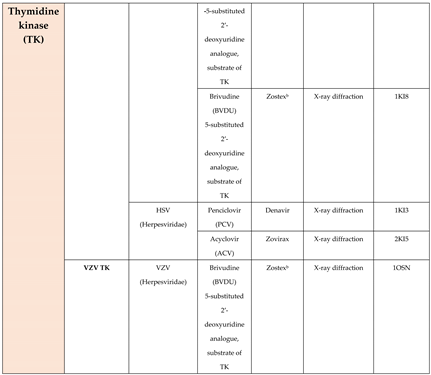
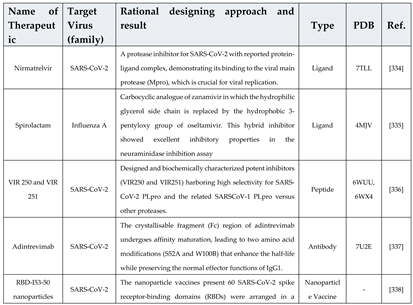
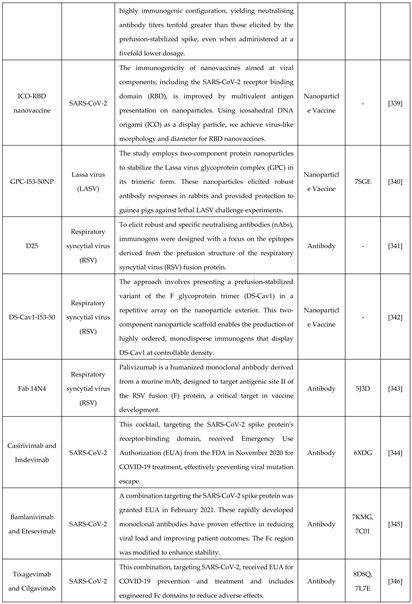
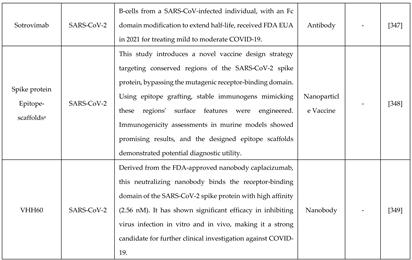
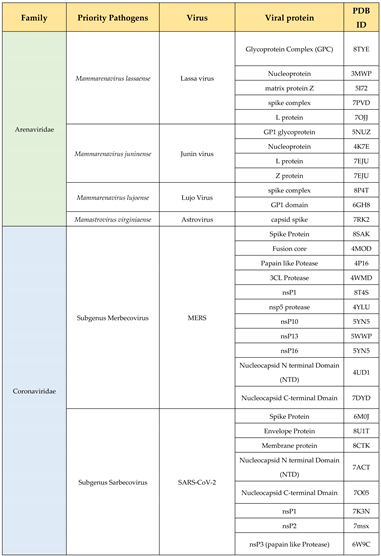
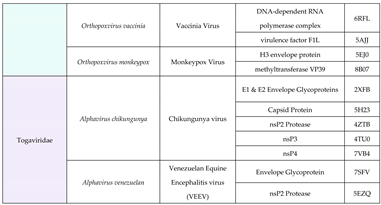
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





