Submitted:
08 January 2025
Posted:
09 January 2025
You are already at the latest version
Abstract
Pseudomonas aeruginosa is an opportunistic pathogenic bacterium, responsible of several life-threatening infection attributed to its multiple virulence factors and its problematic multi-drug resistance, hence the necessity to find out alternatives such as competitive probiotics. Pediococcus pentosaceus MZF16 is a LAB strain, isolated from traditional dried meat “Ossban”, with high probiotic potential. Our study investigated the capacity of P. pentosaceus MZF16 to counteract P. aeruginosa H103 using several tests on intestinal cells (analysis of cytotoxicity, inflammation, adhesion/invasion) and on the in vivo Caenorhabditis elegans model. The effect of MZF16 on the Quorum sensing of the pathogen was also examined. We found that P. pentosaceus MZF16 was able to reduce H103 cytotoxicity and inflammatory activity, prevented pathogen colonization and translocation across Caco-2/TC7 cells. MZF16 exerted also an anti-virulence effect by attenuating Quorum-sensing (QS) molecules and pyoverdine production and extended C. elegans lifespan. The obtained results highlight the potential of P. pentosaceus MZF16 probiotic strain as an anti-Pseudomonas aeruginosa alternative and establish a basis for elucidating the mechanisms of P. pentosaceus MZF16 involved in countering Pseudomonas aeruginosa virulence.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial strains and culture conditions
2.2. P. aeruginosa H103-gfp and P. pentosaceus MZF16-mCherry construction
2.3. Caco-2/TC7 cell culture and infection
2.4. Cytotoxicity assay and Interleukin-8 quantification
2.5. In vitro adhesion and invasion assay
2.6. Bacterial translocation and Transepithelial Electrical Resistance (TER) measurement
2.7. Antimicrobial activity test
2.8. Bacterial aggregation assay
2.9. Pyoverdine production measurement
2.10. Extraction and quantification of AHLs and HAQs molecules
2.11. In vivo Caenorhabditis elegans killing assay
2.12. Statistical analysis
3. Results
3.1. Autoaggregation and coaggregation
3.2. Antimicrobial activity test
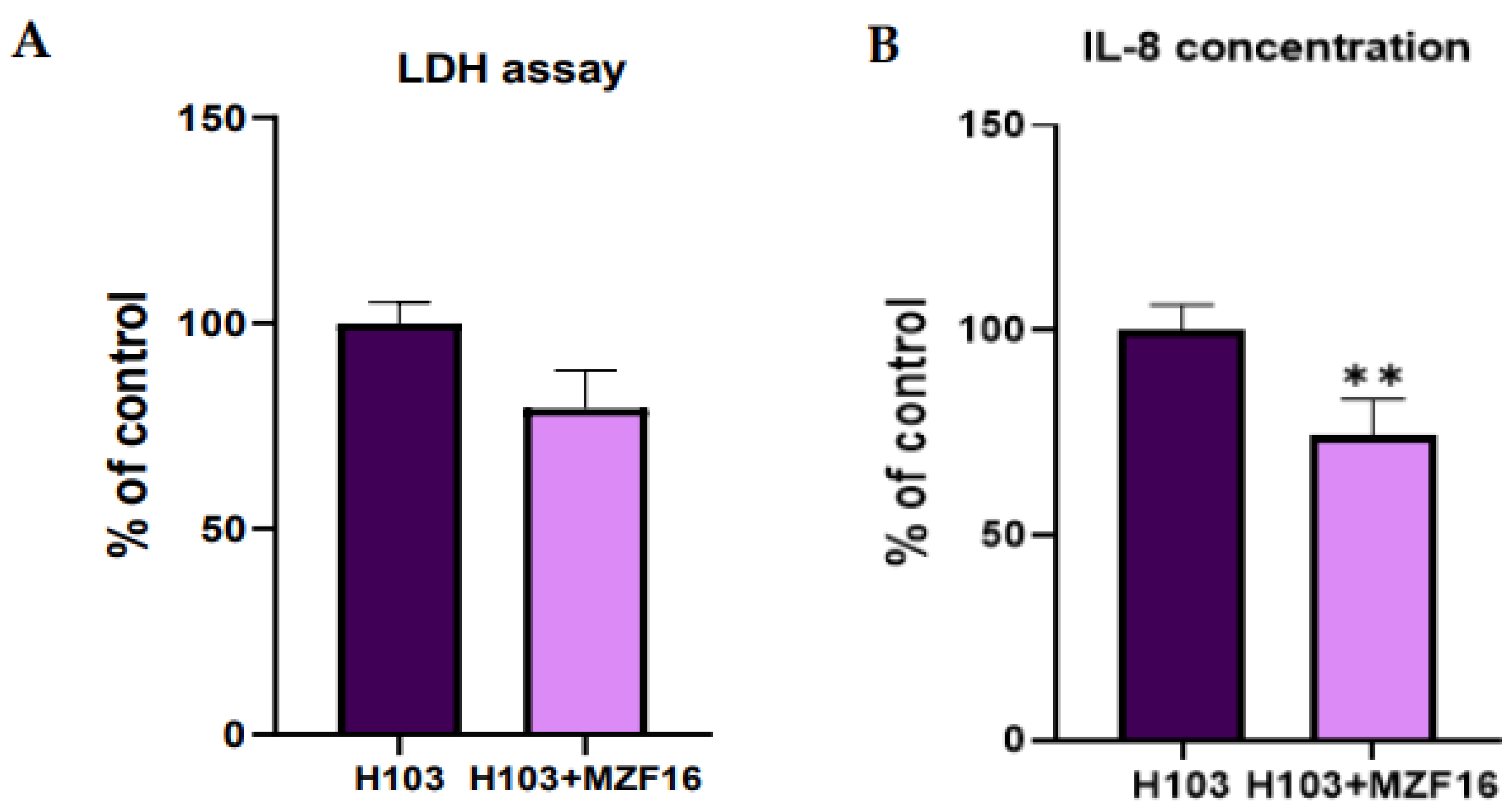
3.3. Cytotoxicity assay and Interleukin-8 quantification
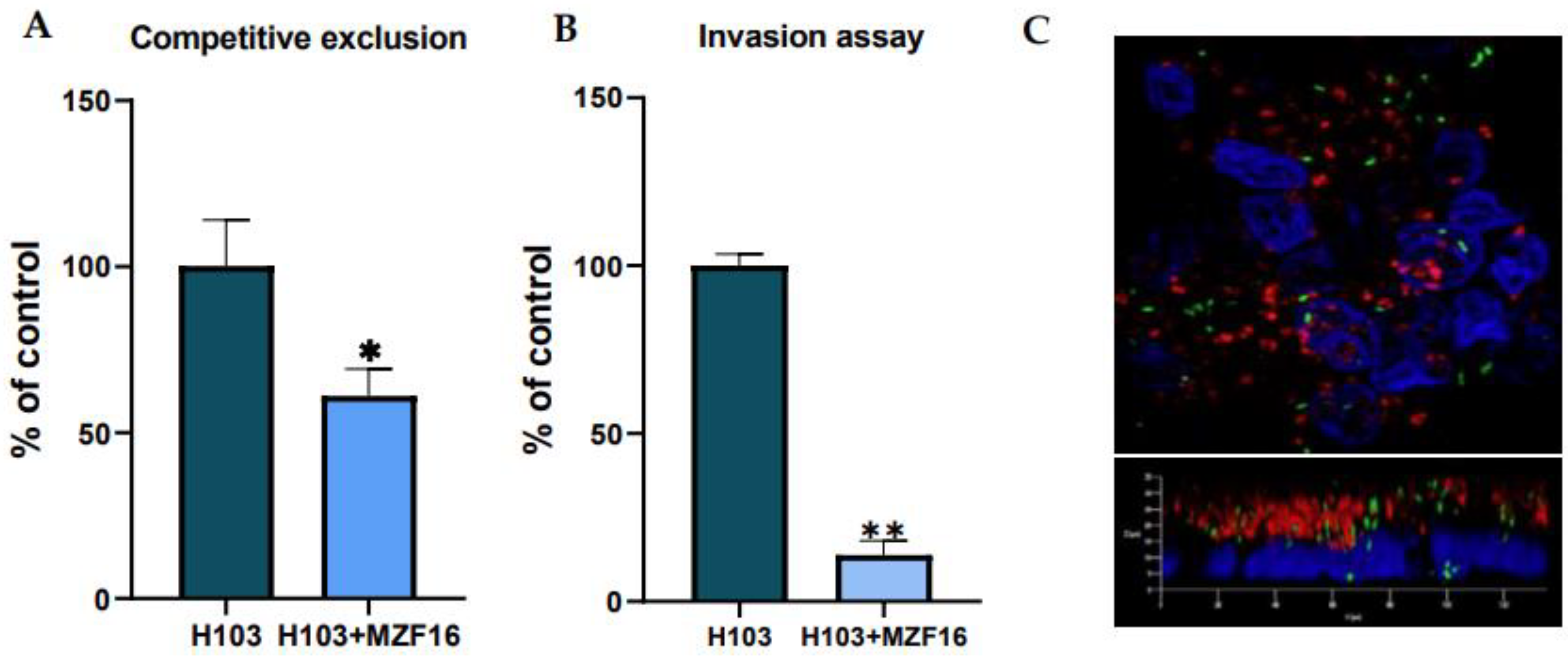
3.4. Adhesion and invasion
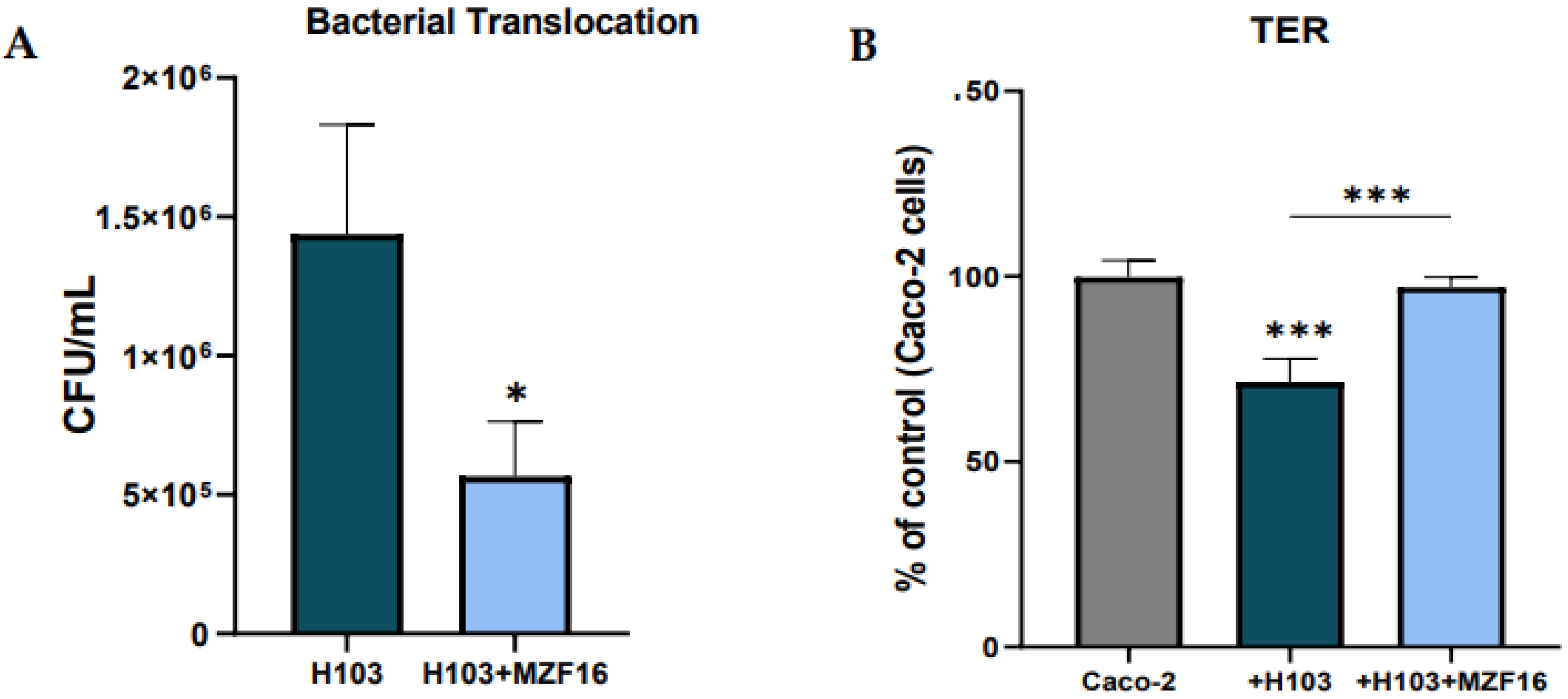
3.5. Transepithelial electric resistance (TER) and translocation
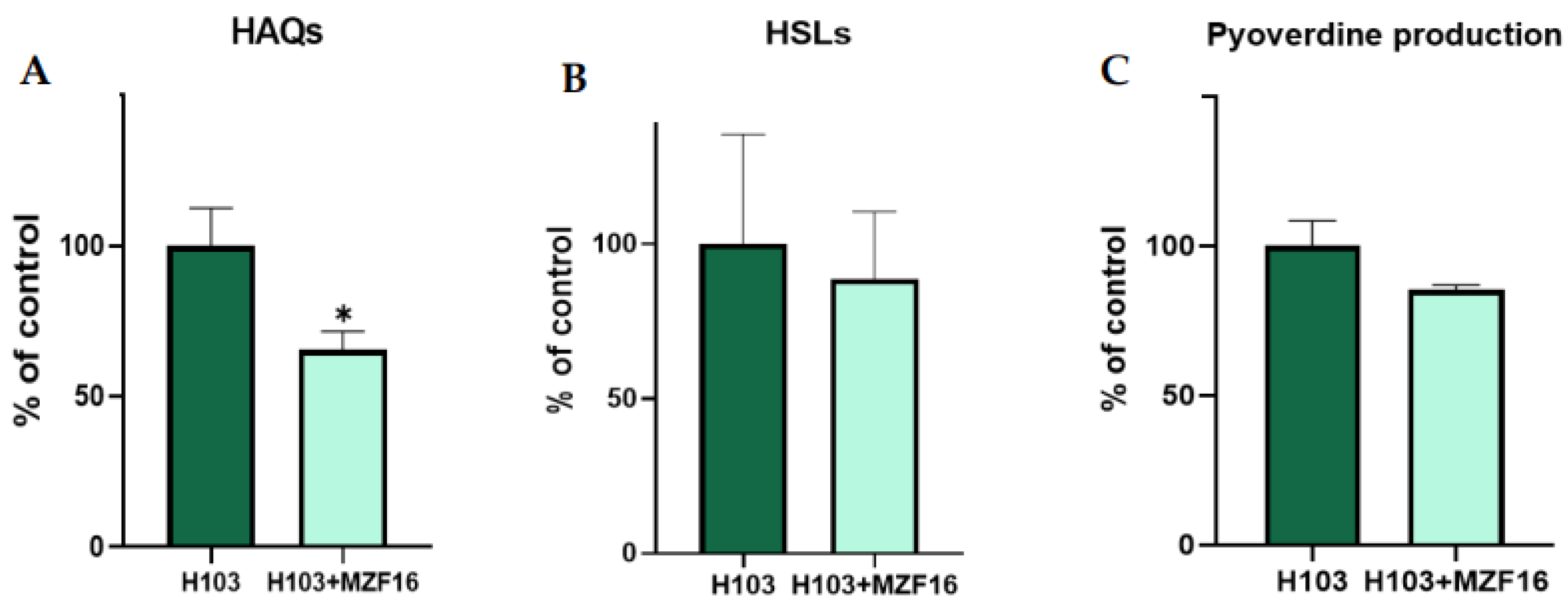
3.6. Quorum-sensing and pyoverdine production
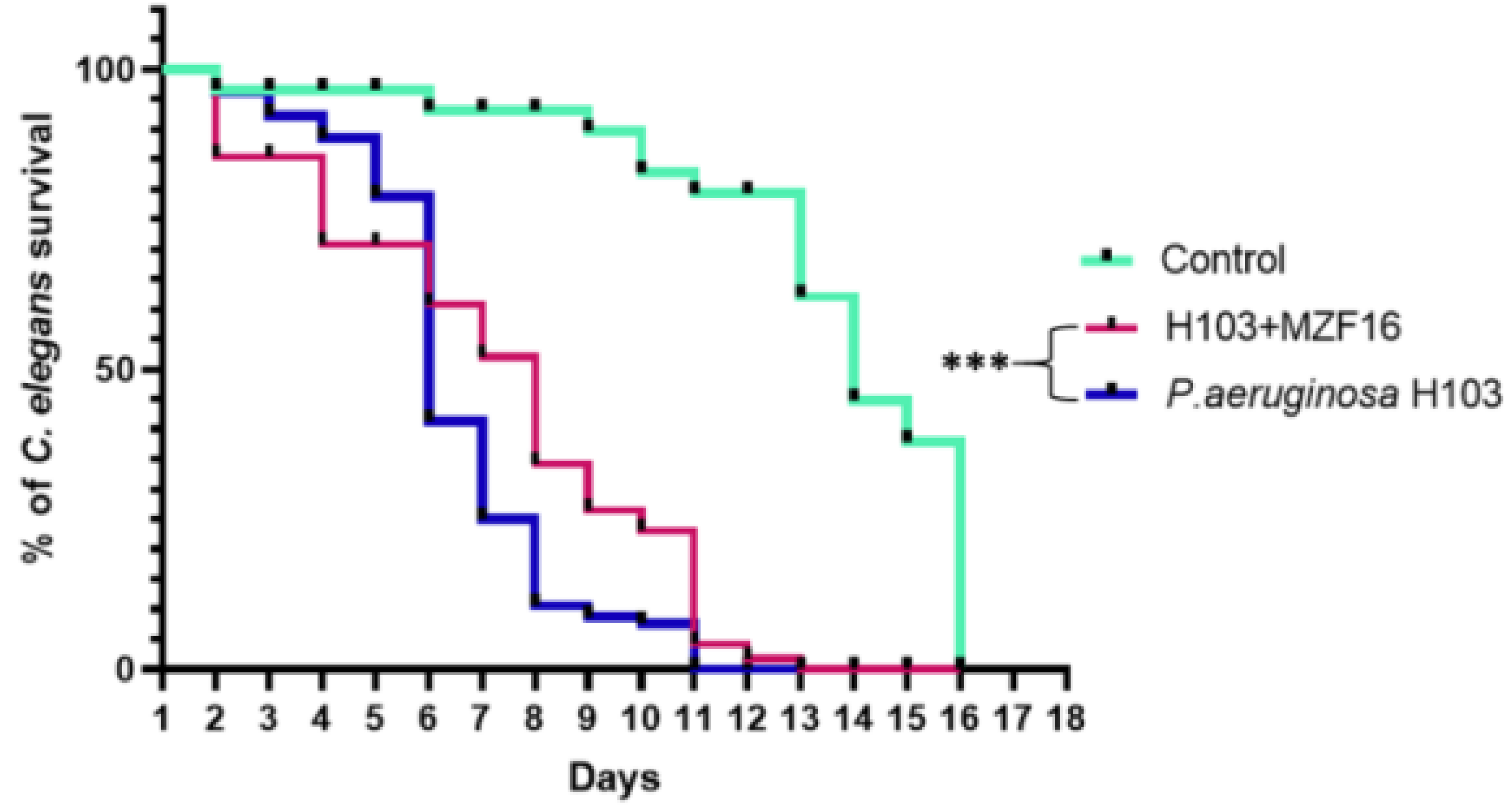
3.7. In vivo virulence test on C. elegans
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health Benefits of Fermented Foods: Microbiota and Beyond. Curr Opin Biotechnol 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Nakamura, R.E.; Nguyen, J.G.; Michels, K.B. Does Consumption of Fermented Foods Modify the Human Gut Microbiota? The Journal of Nutrition 2020, 150, 1680–1692. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.A.; González, S.N.; Alberto, M.R.; Arena, M.E. Human Probiotic Bacteria Attenuate Pseudomonas aeruginosa Biofilm and Virulence by Quorum-Sensing Inhibition. Biofouling 2020, 36, 597–609. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dioso, C.M.; Liong, M.-T.; Nero, L.A.; Khosravi-Darani, K.; Ivanova, I.V. Beneficial Features of Pediococcus: From Starter Cultures and Inhibitory Activities to Probiotic Benefits. World J Microbiol Biotechnol 2022, 39, 4. [Google Scholar] [CrossRef]
- Papagianni, M.; Anastasiadou, S. Pediocins: The Bacteriocins of Pediococci. Sources, Production, Properties and Applications. Microbial cell factories 2009, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Fugaban, J.I.I.; Vazquez Bucheli, J.E.; Park, Y.J.; Suh, D.H.; Jung, E.S.; Franco, B.D.G. de M.; Ivanova, I.V.; Holzapfel, W.H.; Todorov, S.D. Antimicrobial Properties of Pediococcus acidilactici and Pediococcus pentosaceus Isolated from Silage. Journal of Applied Microbiology 2022, 132, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Chelliah, R.; Choi, J.-G.; Hwang, S.; Park, B.-J.; Daliri, E.B.-M.; Kim, S.-H.; Wei, S.; Ramakrishnan, S.R.; Oh, D.-H. In Vitro and in Vivo Defensive Effect of Probiotic LAB against Pseudomonas aeruginosa Using Caenorhabditis Elegans Model. Virulence 2018, 9, 1489–1507. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Kim, B.S.; Kang, S.-S. Bacteriocin of Pediococcus acidilactici HW01 Inhibits Biofilm Formation and Virulence Factor Production by Pseudomonas aeruginosa. Probiotics Antimicrob Proteins 2020, 12, 73–81. [Google Scholar] [CrossRef]
- Adnan, M.; Siddiqui, A.J.; Hamadou, W.S.; Ashraf, S.A.; Hassan, M.I.; Snoussi, M.; Badraoui, R.; Jamal, A.; Bardakci, F.; Awadelkareem, A.M.; et al. Functional and Structural Characterization of Pediococcus pentosaceus-Derived Biosurfactant and Its Biomedical Potential against Bacterial Adhesion, Quorum Sensing, and Biofilm Formation. Antibiotics 2021, 10, 1371. [Google Scholar] [CrossRef] [PubMed]
- Barigela, A.; Bhukya, B. Probiotic Pediococcus acidilactici Strain from Tomato Pickle Displays Anti-Cancer Activity and Alleviates Gut Inflammation in-Vitro. 3 Biotech 2021, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Keeratikunakorn, K.; Kaewchomphunuch, T.; Kaeoket, K.; Ngamwongsatit, N. Antimicrobial Activity of Cell Free Supernatants from Probiotics Inhibits against Pathogenic Bacteria Isolated from Fresh Boar Semen. Sci Rep 2023, 13, 5995. [Google Scholar] [CrossRef] [PubMed]
- Yi, E.-J.; Kim, A.-J. Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics (Basel) 2023, 12, 676. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, N.; Huang, T.Y.; Zhong, F.; Peng, G. Pseudomonas Aeruginosa: A Typical Biofilm Forming Pathogen and an Emerging but Underestimated Pathogen in Food Processing. Frontiers in Microbiology 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Allydice-Francis, K.; Brown, P.D. Diversity of Antimicrobial Resistance and Virulence Determinants in Pseudomonas aeruginosa Associated with Fresh Vegetables. Int J Microbiol 2012, 2012, 426241. [Google Scholar] [CrossRef]
- Ohara, T.; Itoh, K. Significance of Pseudomonas aeruginosa Colonization of the Gastrointestinal Tract. Internal Medicine 2003, 42, 1072–1076. [Google Scholar] [CrossRef] [PubMed]
- Markou, P.; Apidianakis, Y. Pathogenesis of Intestinal Pseudomonas aeruginosa Infection in Patients with Cancer. Front Cell Infect Microbiol 2014, 3, 115. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Oliver, A.; Blázquez, J. Intrinsic and Environmental Mutagenesis Drive Diversification and Persistence of Pseudomonas Aeruginosa in Chronic Lung Infections. J Infect Dis 2012, 205, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New Insights into Pathogenesis and Host Defenses.
- Chatterjee, M.; Anju, C.P.; Biswas, L.; Anil Kumar, V.; Gopi Mohan, C.; Biswas, R. Antibiotic Resistance in Pseudomonas aeruginosa and Alternative Therapeutic Options. International Journal of Medical Microbiology 2016, 306, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Zommiti, M.; Bouffartigues, E.; Maillot, O.; Barreau, M.; Szunerits, S.; Sebei, K.; Feuilloley, M.; Connil, N.; Ferchichi, M. In vitro Assessment of the Probiotic Properties and Bacteriocinogenic Potential of Pediococcus pentosaceus MZF16 Isolated From Artisanal Tunisian Meat "Dried Ossban". Front Microbiol 2018, 9, 2607–2622. [Google Scholar] [CrossRef] [PubMed]
- Zommiti, M.; Boukerb, A.M.; Feuilloley, M.G.J.; Ferchichi, M.; Connil, N. Draft Genome Sequence of Pediococcus pentosaceus MZF16, a Bacteriocinogenic Probiotic Strain Isolated from Dried Ossban in Tunisia. Microbiol Resour Announc 2019, 8, e00285-19. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Carey, A.M. Outer Membrane of Pseudomonas aeruginosa: Heat- 2-Mercaptoethanol-Modifiable Proteins. Journal of Bacteriology 1979, 140, 902–910. [Google Scholar] [CrossRef]
- Cheng, H.P.; Walker, G.C. Succinoglycan Is Required for Initiation and Elongation of Infection Threads during Nodulation of Alfalfa by Rhizobium Meliloti. J Bacteriol 1998, 180, 5183–5191. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Mohedano, M.L.; Pardo, M.Á.; López, P. β-Glucan-Producing Pediococcus parvulus 2.6: Test of Probiotic and Immunomodulatory Properties in Zebrafish Models. Front Microbiol 2018, 9, 1684. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Adhesion and Aggregation Properties of Probiotic and Pathogen Strains. Eur Food Res Technol 2008, 226, 1065–1073. [Google Scholar] [CrossRef]
- Fletcher, M.P.; Diggle, S.P.; Cámara, M.; Williams, P. Biosensor-Based Assays for PQS, HHQ and Related 2-Alkyl-4-Quinolone Quorum Sensing Signal Molecules. Nat Protoc 2007, 2, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.W.; Mahajan-Miklos, S.; Ausubel, F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa Used to Model Mammalian Bacterial Pathogenesis. Proc Natl Acad Sci U S A 1999, 96, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Blier, A.-S.; Veron, W.; Bazire, A.; Gerault, E.; Taupin, L.; Vieillard, J.; Rehel, K.; Dufour, A.; Le Derf, F.; Orange, N.; et al. C-Type Natriuretic Peptide Modulates Quorum Sensing Molecule and Toxin Production in Pseudomonas aeruginosa. Microbiology (Reading) 2011, 157, 1929–1944. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of Antibiotic Resistance Pseudomonas aeruginosa in Intensive Care Unit; a Critical Review. Genes Dis 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Choi, M.K.; Choi, S. In Vitro and in Vivo Anti-Clostridial Activity of Newly Isolated Pediococcus acidilactici SPM138 against Clostridium Difficile. Anaerobe 2020, 61, 102146. [Google Scholar] [CrossRef] [PubMed]
- Vasiee, A.; Falah, F.; Behbahani, B.A.; Tabatabaee-yazdi, F. Probiotic Characterization of Pediococcus Strains Isolated from Iranian Cereal-Dairy Fermented Product: Interaction with Pathogenic Bacteria and the Enteric Cell Line Caco-2. Journal of Bioscience and Bioengineering 2020, 130, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Ye, P.; Lei, Q.; Cheng, Y.; Yu, H.; Du, J.; Pan, H.; Cao, Z. In Vitro Probiotic Properties of Pediococcus pentosaceus L1 and Its Effects on Enterotoxigenic Escherichia Coli-Induced Inflammatory Responses in Porcine Intestinal Epithelial Cells. Microb Pathog 2020, 144, 104163. [Google Scholar] [CrossRef] [PubMed]
- Barnett, A.M.; Roy, N.C.; Cookson, A.L.; McNabb, W.C. Metabolism of Caprine Milk Carbohydrates by Probiotic Bacteria and Caco-2:HT29−MTX Epithelial Co-Cultures and Their Impact on Intestinal Barrier Integrity. Nutrients 2018, 10, 949. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol Adv 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial Quorum Sensing. Cell 2018, 174, 1328–1328.e1. [Google Scholar] [CrossRef]
- Wade, D.S.; Calfee, M.W.; Rocha, E.R.; Ling, E.A.; Engstrom, E.; Coleman, J.P.; Pesci, E.C. Regulation of Pseudomonas Quinolone Signal Synthesis in Pseudomonas aeruginosa. J Bacteriol 2005, 187, 4372–4380. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pérez, S.P.; Solis, C.S.; López-Bucio, J.S.; Valdez Alarcón, J.J.; Villegas, J.; Reyes-De la Cruz, H.; Campos-Garcia, J. Pathogenesis in Pseudomonas aeruginosa PAO1 Biofilm-Associated Is Dependent on the Pyoverdine and Pyocyanin Siderophores by Quorum Sensing Modulation. Microb Ecol 2023, 86, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Ghanei-Motlagh, R.; Mohammadian, T.; Gharibi, D.; Menanteau-Ledouble, S.; Mahmoudi, E.; Khosravi, M.; Zarea, M.; El-Matbouli, M. Quorum Quenching Properties and Probiotic Potentials of Intestinal Associated Bacteria in Asian Sea Bass Lates Calcarifer. Marine Drugs 2020, 18, 23. [Google Scholar] [CrossRef]
- Devi, S.; Chhibber, S.; Harjai, K. Optimization of Cultural Conditions for Enhancement of Anti-Quorum Sensing Potential in the Probiotic Strain Lactobacillus rhamnosus GG against Pseudomonas aeruginosa. 3 Biotech 2022, 12, 133. [Google Scholar] [CrossRef]
- Kiymaci, M.E.; Altanlar, N.; Gumustas, M.; Ozkan, S.A.; Akin, A. Quorum Sensing Signals and Related Virulence Inhibition of Pseudomonas aeruginosa by a Potential Probiotic Strain’s Organic Acid. Microbial Pathogenesis 2018, 121, 190–197. [Google Scholar] [CrossRef]
| Pediococcus pentosaceus MZF16 | |
|---|---|
| Autoaggregation | 44.74% ± 2.58 |
| Coaggregation with P. aeruginosa H103 | 45.77% ± 3.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





