Submitted:
06 January 2025
Posted:
07 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Oral Metagenomic Datasets Acquisition and Information
2.2. Statistical Analysis
2.3. Single Microorganism Classifiers
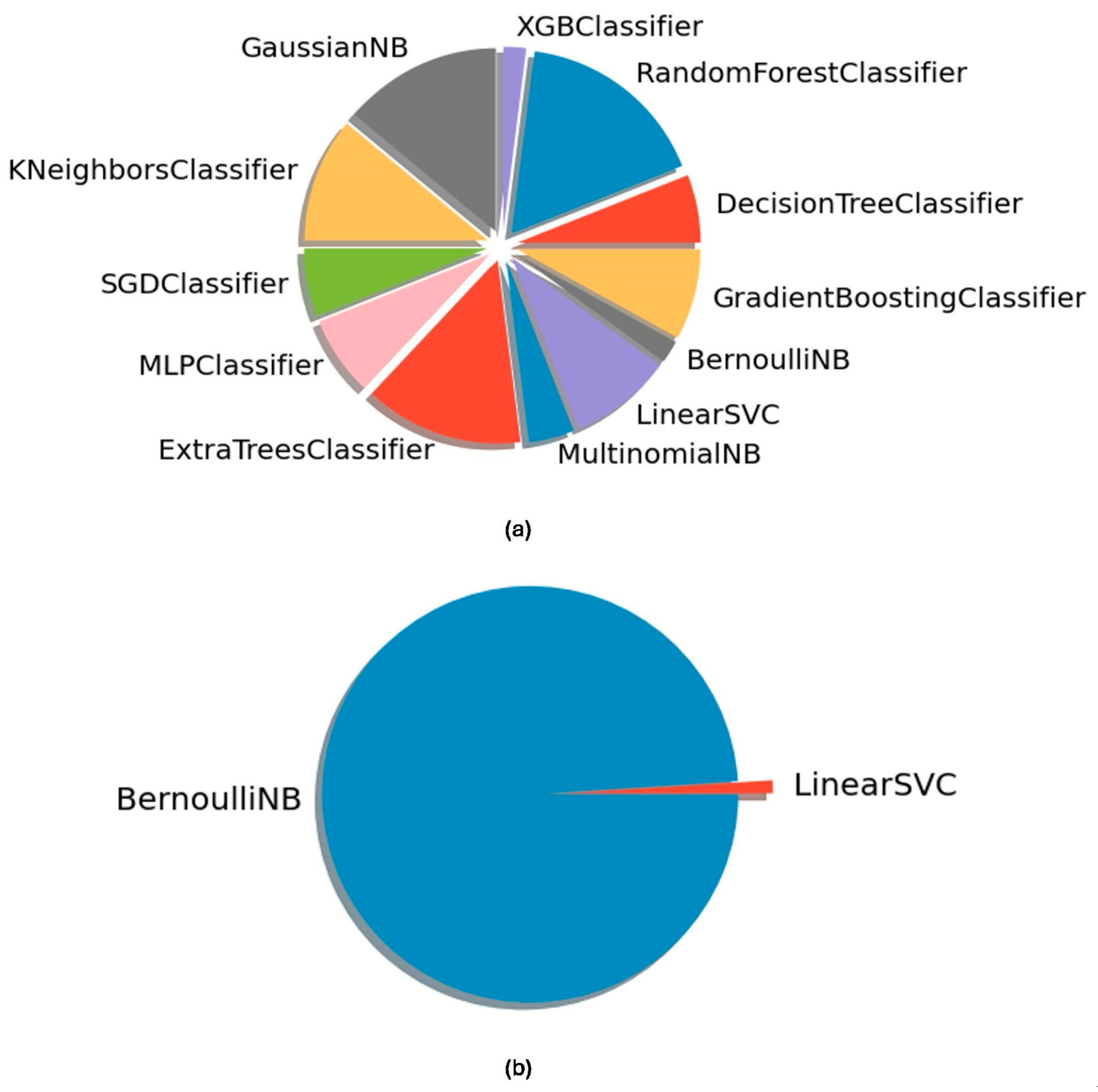
2.4. ML-Based Microbiome Classifiers
2.5. ML-Driven Microorganism Combination Scoring Classifiers
2.6. Classifier Performance Analysis
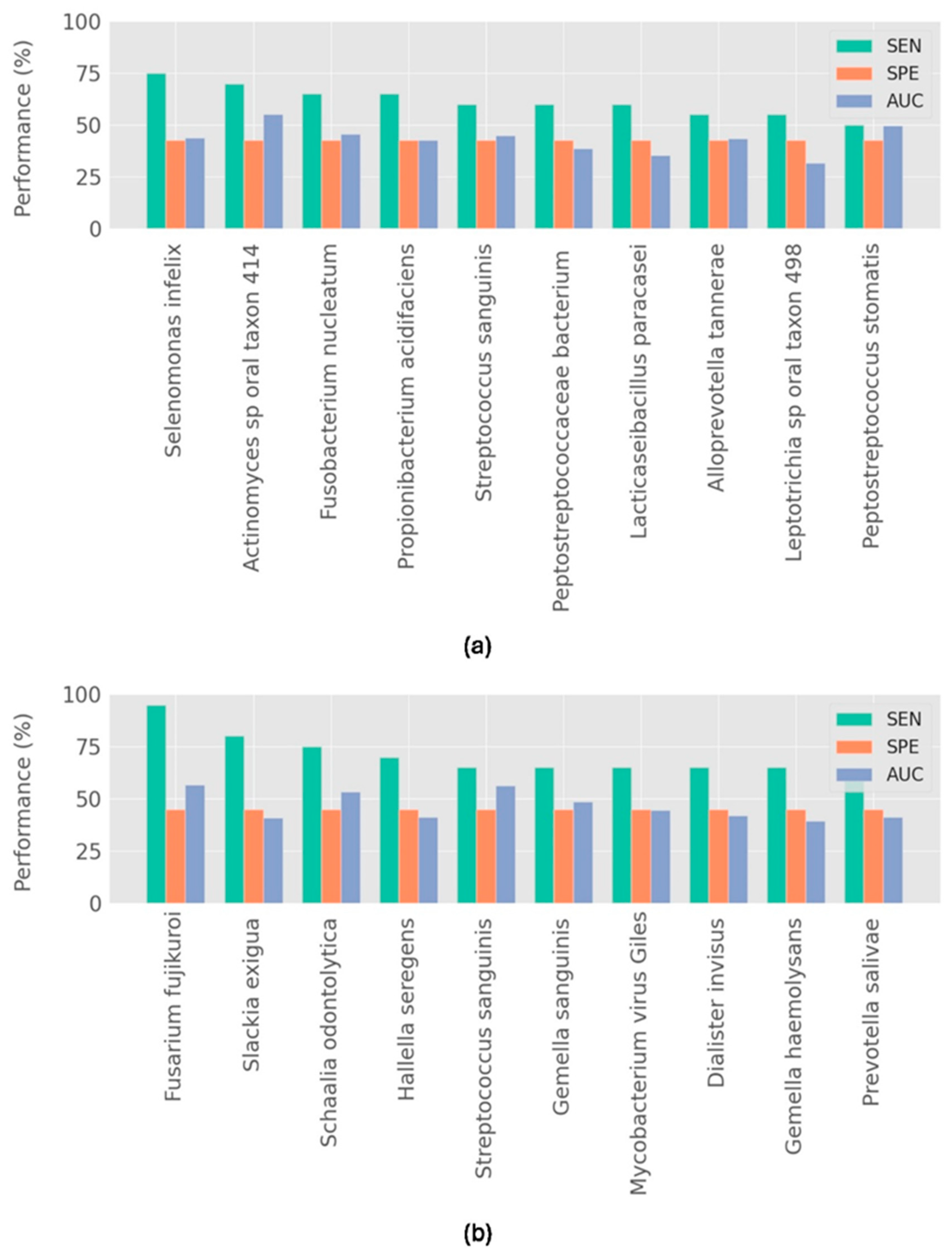
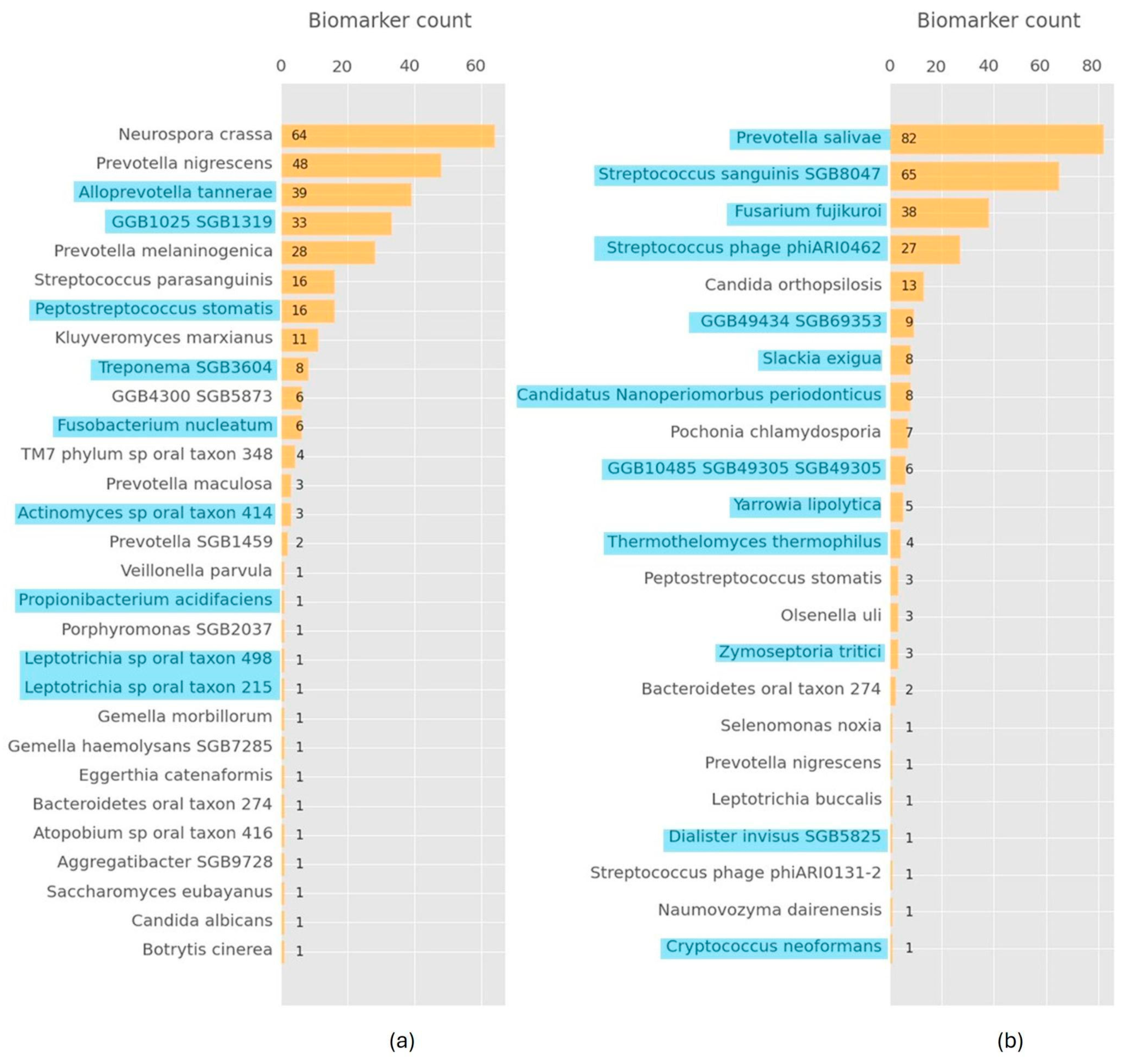
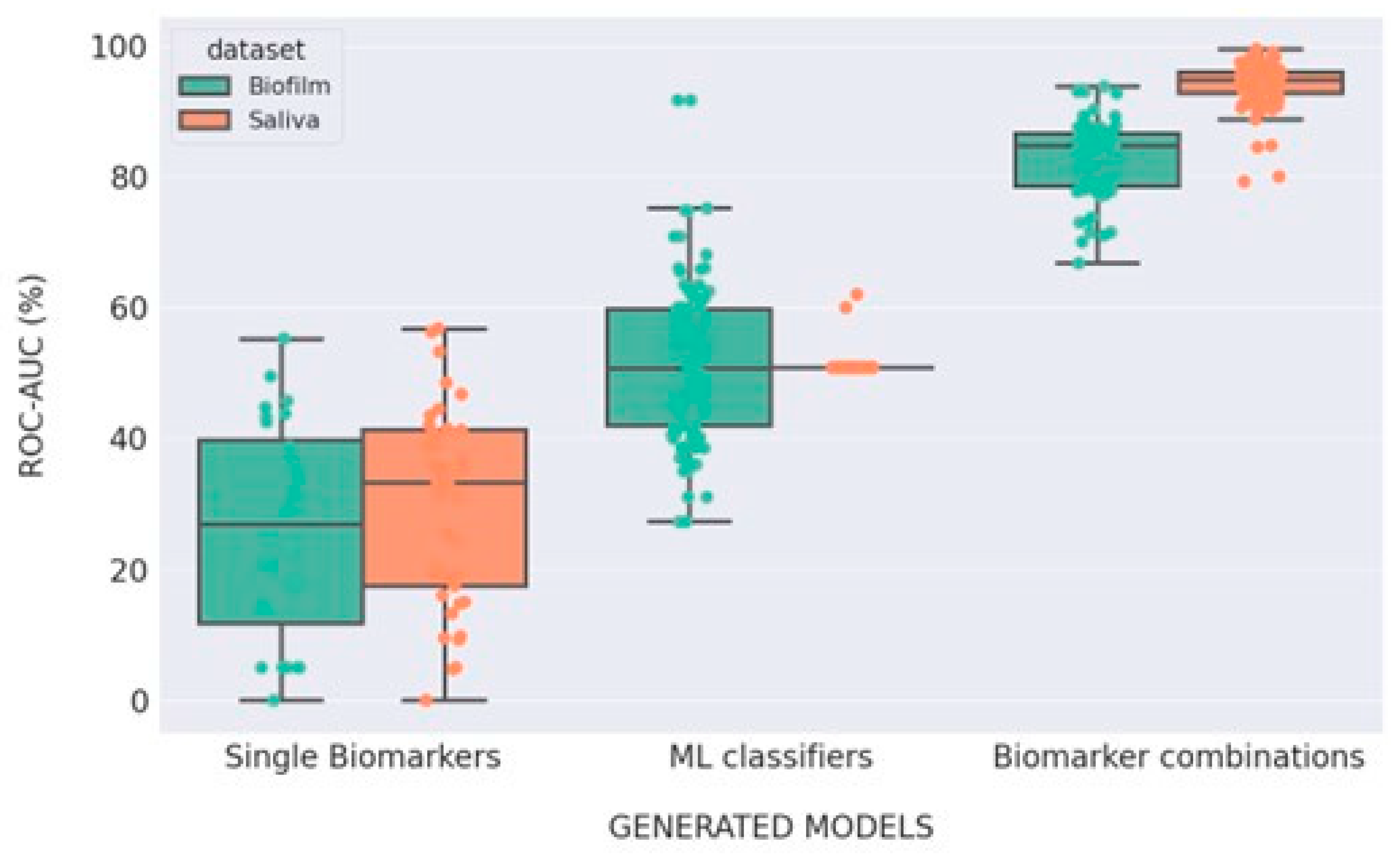
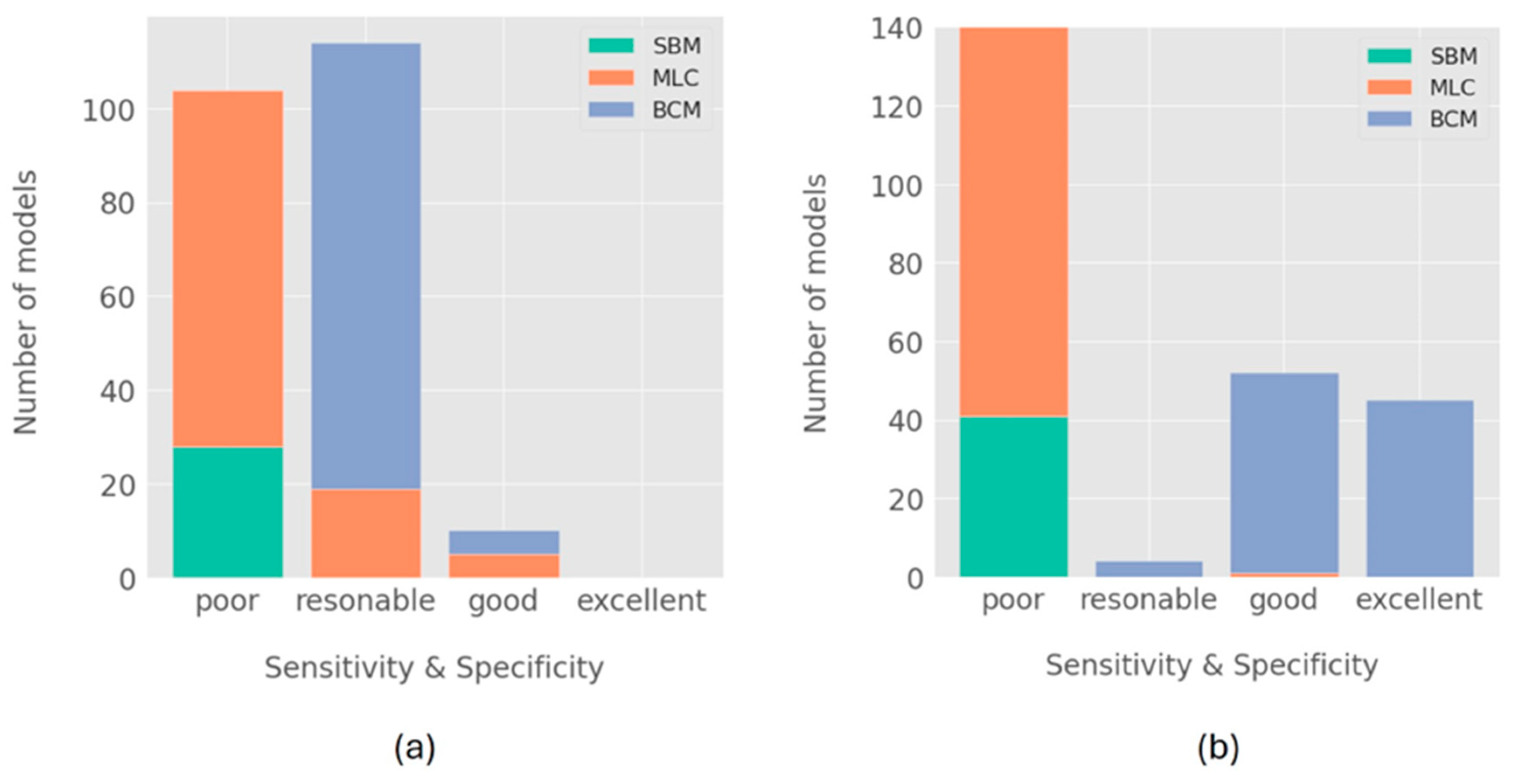
3. Results
Model Performance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kormas, I.; Pedercini, C.; Pedercini, A.; Raptopoulos, M.; Alassy, H.; Wolff, L.F. Peri-Implant Diseases: Diagnosis, Clinical, Histological, Microbiological Characteristics and Treatment Strategies. A Narrative Review. Antibiotics 2020, 9, 1–19. [CrossRef]
- Schwarz, F.; Derks, J.; Monje, A.; Wang, H.L. Peri-Implantitis. J. Clin. Periodontol. 2018, 45, S246–S266. [CrossRef]
- Belibasakis, G.N.; Charalampakis, G.; Bostanci, N.; Stadlinger, B. Peri-Implant Infections of Oral Biofilm Etiology. In: Donelli, G. (eds) Biofilm-based Healthcare-associated Infections. Advances in Experimental Medicine and Biology, vol 830. Springer, Cham. [CrossRef]
- Renvert, S.; Quirynen, M. Risk Indicators for Peri-Implantitis. A Narrative Review. Clin. Oral Implants Res. 2015, 26, 15–44. [CrossRef]
- Robertson, K.; Shahbazian, T.; MacLeod, S. Treatment of Peri-Implantitis and the Failing Implant. Dent. Clin. North Am. 2015, 59, 329–343. [CrossRef]
- Fu, J.H.; Wang, H.L. Breaking the Wave of Peri-Implantitis. Periodontol. 2000 2020, 84, 145–160. [CrossRef]
- Rekawek, P.; Herbst, E.A.; Suri, A.; Ford, B.P.; Rajapakse, C.S.; Panchal, N. Machine Learning and Artificial Intelligence: A Web-Based Implant Failure and Peri-Implantitis Prediction Model for Clinicians. Int. J. Oral Maxillofac. Implants 2023, 38, 576–582. [CrossRef]
- Mameno, T.; Wada, M.; Nozaki, K.; Takahashi, T.; Tsujioka, Y.; Akema, S.; Hasegawa, D.; Ikebe, K. Predictive Modeling for Peri-Implantitis by Using Machine Learning Techniques. Sci. Rep. 2021, 11, 1–8. [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral Microbiome: Unveiling the Fundamentals. J. oral Maxillofac. Pathol. 2019, 23, 122–128. [CrossRef]
- Bessa, L.J.; Botelho, J.; Machado, V.; Alves, R.; Mendes, J.J. Managing Oral Health in the Context of Antimicrobial Resistance. Int. J. Environ. Res. Public Health 2022, 19, 16448. [CrossRef]
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The Oral Microbiome - An Update for Oral Healthcare Professionals. Br. Dent. J. 2016, 221, 657–666. [CrossRef]
- Dabdoub, S.M.; Tsigarida, A.A.; Kumar, P.S. Patient-Specific Analysis of Periodontal and Peri-Implant Microbiomes. J. Dent. Res. 2013, 92, 168S-75S. [CrossRef]
- Komatsu, K.; Shiba, T.; Takeuchi, Y.; Watanabe, T.; Koyanagi, T.; Nemoto, T.; Shimogishi, M.; Shibasaki, M.; Katagiri, S.; Kasugai, S.; et al. Discriminating Microbial Community Structure Between Peri-Implantitis and Periodontitis With Integrated Metagenomic, Metatranscriptomic, and Network Analysis. Front. Cell. Infect. Microbiol. 2020, 10, 596490. [CrossRef]
- Kotsakis, G.A.; Olmedo, D.G. Peri-Implantitis Is Not Periodontitis: Scientific Discoveries Shed Light on Microbiome-Biomaterial Interactions That May Determine Disease Phenotype. Periodontol. 2000 2021, 86, 231–240. [CrossRef]
- Yu, X.L.; Chan, Y.; Zhuang, L.; Lai, H.C.; Lang, N.P.; Keung Leung, W.; Watt, R.M. Intra-Oral Single-Site Comparisons of Periodontal and Peri-Implant Microbiota in Health and Disease. Clin. Oral Implants Res. 2019, 30, 760–776. [CrossRef]
- Giok, K.C.; Menon, R.K. The Microbiome of Peri-Implantitis : A Systematic Review of Next-Generation Sequencing Studies. Antibiotics 2023, 12, 1610. [CrossRef]
- Song, L.; Feng, Z.; Zhou, Q.; Wu, X.; Zhang, L.; Sun, Y.; Li, R.; Chen, H.; Yang, F.; Yu, Y. Metagenomic Analysis of Healthy and Diseased Peri-Implant Microbiome under Different Periodontal Conditions: A Cross-Sectional Study. BMC Oral Health 2024, 24, 105. [CrossRef]
- Lumbikananda, S.; Srithanyarat, S.S.; Mattheos, N.; Osathanon, T. Oral Fluid Biomarkers for Peri-Implantitis: A Scoping Review. Int. Dent. J. 2024, 74, 387–402. [CrossRef]
- Roman-Naranjo, P.; Parra-Perez, A.M.; Lopez-Escamez, J.A. A Systematic Review on Machine Learning Approaches in the Diagnosis and Prognosis of Rare Genetic Diseases. J. Biomed. Inform. 2023, 143, 104429. [CrossRef]
- Telikani, A.; Gandomi, A.H.; Tahmassebi, A.; Banzhaf, W. Evolutionary Machine Learning: A Survey. ACM Comput. Surv 2021, 54, 1–35. [CrossRef]
- Pais, R.J. Predictive Modelling in Clinical Bioinformatics: Key Concepts for Startups. BioTech 2022, 11, 1–10. [CrossRef]
- Mameno, T.; Wada, M.; Nozaki, K.; Takahashi, T.; Tsujioka, Y.; Akema, S.; Hasegawa, D.; Ikebe, K. Predictive Modeling for Peri-Implantitis by Using Machine Learning Techniques. Sci. Rep. 2021, 11, 1–8. [CrossRef]
- Ghensi, P.; Manghi, P.; Zolfo, M.; Armanini, F.; Pasolli, E.; Bolzan, M.; Bertelle, A.; Dell’Acqua, F.; Dellasega, E.; Waldner, R.; et al. Strong Oral Plaque Microbiome Signatures for Dental Implant Diseases Identified by Strain-Resolution Metagenomics. npj Biofilms Microbiomes 2020, 6. [CrossRef]
- Le, T.T.; Fu, W.; Moore, J.H. Scaling Tree-Based Automated Machine Learning to Biomedical Big Data with a Feature Set Selector. Bioinformatics 2020, 36, 250–256. [CrossRef]
- Filho, U.L.; Pais, T.A.; Pais, R.J. Facilitating “Omics” for Phenotype Classification Using a User-Friendly AI-Driven Platform: Application in Cancer Prognostics. BioMedInformatics 2023, 3, 1071–1082. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [CrossRef]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and Improving Metagenomic Taxonomic Profiling with Uncharacterized Species Using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ Comput. Sci. 2017, 3, e104. [CrossRef]
- Olson, R.S.; Urbanowicz, R.J.; Andrews, P.C.; Lavender, N.A.; Kidd, L.C.; Moore, J.H. Automating Biomedical Data Science Through Tree-Based Pipeline Optimization. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer, Cham, 2016; Vol. 9597, pp. 123–137 ISBN 9783319312033.
- Pais, R.J.; Lopes, F.; Parreira, I.; Silva, M.; Silva, M.; Moutinho, M.G. Predicting Cancer Prognostics from Tumour Transcriptomics Using an Auto Machine Learning Approach. In Proceedings of the Med. Sci Forum; MDPI: Basel Switzerland, August 8 2023; p. 6.
- Pais, R.J.; Zmuidinaite, R.; Lacey, J.C.; Jardine, C.S.; Iles, R.K. A Rapid and Affordable Screening Tool for Early-Stage Ovarian Cancer Detection Based on MALDI-ToF MS of Blood Serum. Appl. Sci. 2022, 12, 3030. [CrossRef]
- Pais, R.J.; Sharara, F.; Zmuidinaite, R.; Butler, S.; Keshavarz, S.; Iles, R. Bioinformatic Identification of Euploid and Aneuploid Embryo Secretome Signatures in IVF Culture Media Based on MALDI-ToF Mass Spectrometry. J. Assist. Reprod. Genet. 2020. [CrossRef]
- Dankers, F.J.W.M.; Traverso, A.; Wee, L.; van Kuijk, S.M.J. Prediction Modeling Methodology. In Fundamentals of Clinical Data Science; Springer International Publishing: Cham, 2019; pp. 101–120.
- Feher, B.; Tussie, C.; Giannobile, W. V Applied Artificial Intelligence in Dentistry: Emerging Data Modalities and Modeling Approaches. 2024. [CrossRef]
- Rekawek, P.; Herbst, E.A.; Suri, A.; Ford, B.P.; Rajapakse, C.S.; Panchal, N. Machine Learning and Artificial Intelligence: A Web-Based Implant Failure and Peri-Implantitis Prediction Model for Clinicians. Int. J. Oral Maxillofac. Implants 2023, 38, 576-582b. [CrossRef]
- Mann, M.; Kumar, C.; Zeng, W.F.; Strauss, M.T. Artificial Intelligence for Proteomics and Biomarker Discovery. Cell Syst. 2021, 12, 759–770.
- Lof, M.; Janus, M.M.; Krom, B.P. Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms. J. Fungi 2017, 3, 40. [CrossRef]
- Patil, R.; Ajagunde, J.; Khan, S.; Kannuri, S.; Gandham, N.; Mukhida, S. Rhino-Orbital Cerebral Mycosis: A Case Series of Non-Mucorales in COVID Patients. Access Microbiol. 2023, 5, 1–19. [CrossRef]
- Könönen, E.; Fteita, D.; Gursoy, U.K.; Gursoy, M. Prevotella Species as Oral Residents and Infectious Agents with Potential Impact on Systemic Conditions. J. Oral Microbiol. 2022, 14, 2079814. [CrossRef]
- Takahiko, N.; Takahiko, S.; Keiji, K.; Shunsuke, M.; Tatsuro, K.; Takashi, N.; Ryota, K.; Yasuo, T.; Takanori, I. Characterizing Bacterial Communities among Healthy, Peri-Implant Mucositis, and Peri-Implantitis Statuses by 16S RRNA Gene Amplicon Sequencing. J. Stomatol. Soc. 2024, 91, 8–18. [CrossRef]
- Verdugo, F.; Castillo, A.; Castillo, F.; Uribarri, A. Epstein–Barr Virus Associated Peri-Implantitis: A Split-Mouth Study. Clin. Oral Investig. 2015, 19, 535–543. [CrossRef]
- Koyanagi, T.; Sakamoto, M.; Takeuchi, Y.; Ohkuma, M.; Izumi, Y. Analysis of Microbiota Associated with Peri-Implantitis Using 16S RRNA Gene Clone Library. J. Oral Microbiol. 2010, 2, 5104. [CrossRef]
| Characteristics | Biofilm Dataset | Saliva Dataset |
|---|---|---|
| Sample number | 60 | 40 |
| Number of patients | 401 | 401 |
| Total healthy implants (HI) | 40 | 20 |
| Implants with PI | 20 | 20 |
| Healthy implants from patients with PI | 20 | 0 |
| Bacteria | 5961 | 5961 |
| Fungus | 521 | 521 |
| Virus | 5861 | 5861 |
| Classifier rank |
Algorithm type |
Model input data (abundances) |
Sensitivity (%) |
Specificity (%) |
AUC (%) |
|---|---|---|---|---|---|
| 1 | Biomarker Scoring |
Alloprevotella tannerae Neurospora crassa Prevotella nigrescens |
95 | 80 | 89 |
| 2 | Biomarker Scoring |
Prevotella nigrescens Prevotella melaninogenica TM7 phylum sp oral taxon 348 |
90 | 80 | 88 |
| 3 | Biomarker Scoring |
Alloprevotella tannerae TM7 phylum sp oral taxon 348 Botrytis cinerea Prevotella nigrescens |
90 | 83 | 83 |
| 4 | Biomarker Scoring |
Alloprevotella tannerae Neurospora crassa Streptococcus parasanguinis |
90 | 80 | 85 |
| 5 | Biomarker Scoring |
Alloprevotella tannerae Neurospora crassa Streptococcus parasanguinis |
90 | 80 | 84 |
| 6 | MLPClassifier | Entire microbiome | 90 | 85 | 75 |
| 7 | GaussianNB | Entire microbiome | 80 | 98 | 65 |
| 8 | SGDClassifier | Entire microbiome | 85 | 80 | 66 |
| 9 | GaussianNB | Entire microbiome | 80 | 85 | 62 |
| 10 | GaussianNB | Entire microbiome | 80 | 85 | 61 |
| Classifier rank |
Algorithm type |
Model input data (abundances) |
Sensitivity (%) |
Specificity (%) |
AUC (%) |
|---|---|---|---|---|---|
| 1 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis GGB10485 SGB49305 GGB49434 SGB69353 |
100 | 95 | 99 |
| 2 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis Pochonia chlamydosporia GGB10485 SGB49305 |
100 | 95 | 98 |
| 3 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis GGB10485 SGB49305 |
100 | 95 | 97 |
| 4 | Biomarker Scoring |
Streptococcus phage phiARI0462 Fusarium fujikuroi |
95 | 95 | 97 |
| 5 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis Yarrowia lipolytica |
95 | 95 | 96 |
| 6 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis Pochonia chlamydosporia |
95 | 95 | 96 |
| 7 | Biomarker Scoring |
Prevotella salivae Streptococcus phage phiARI0462 Fusarium fujikuroi |
95 | 95 | 96 |
| 8 | Biomarker Scoring |
Streptococcus phage phiARI0462 Fusarium fujikuroi |
95 | 95 | 96 |
| 9 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis Pochonia chlamydosporia |
95 | 95 | 95 |
| 10 | Biomarker Scoring |
Prevotella salivae Streptococcus sanguinis Pochonia chlamydosporia |
95 | 95 | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





