Submitted:
30 December 2024
Posted:
31 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Information
2.2. Data Analysis and Statistics
3. Results
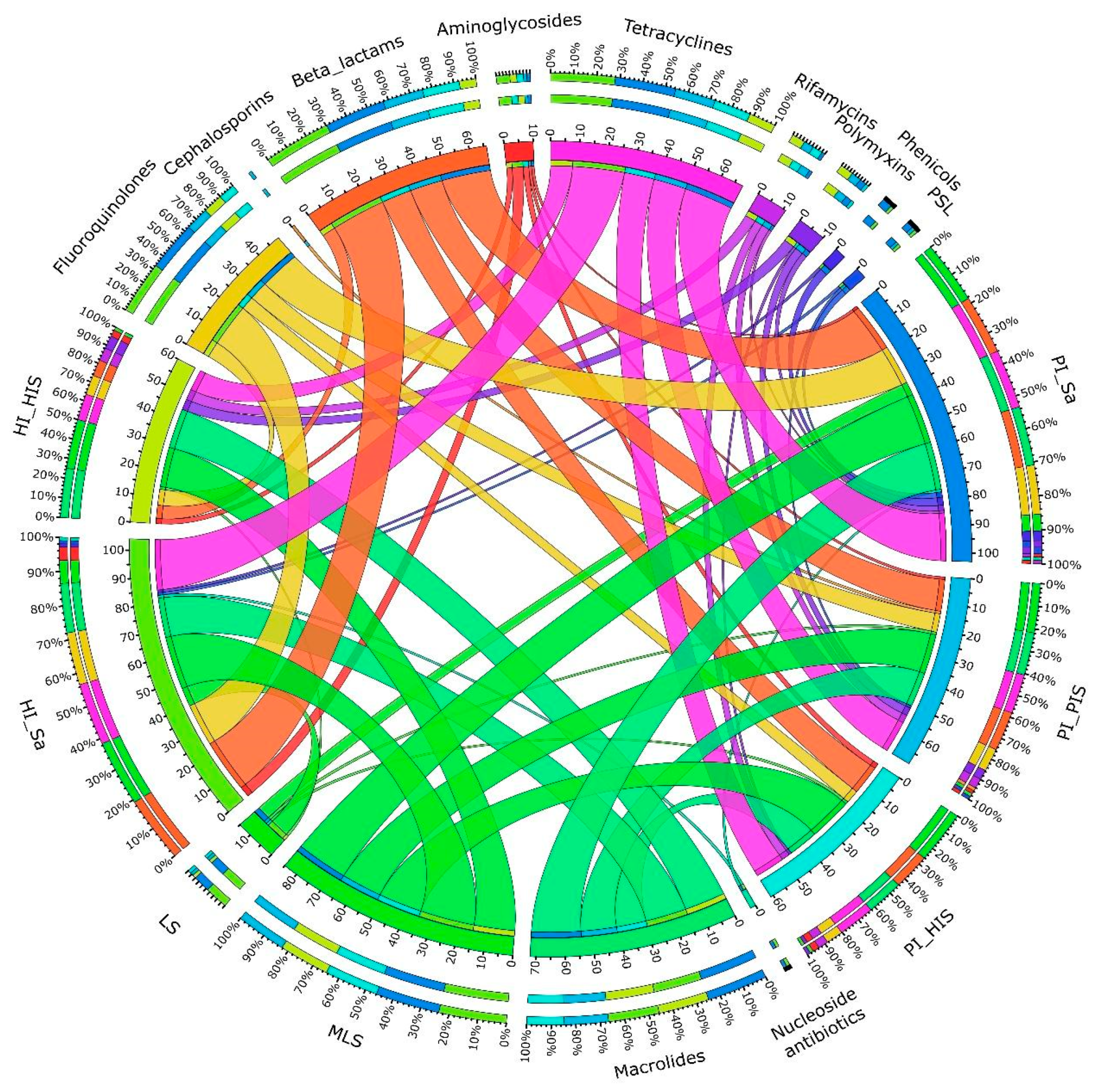
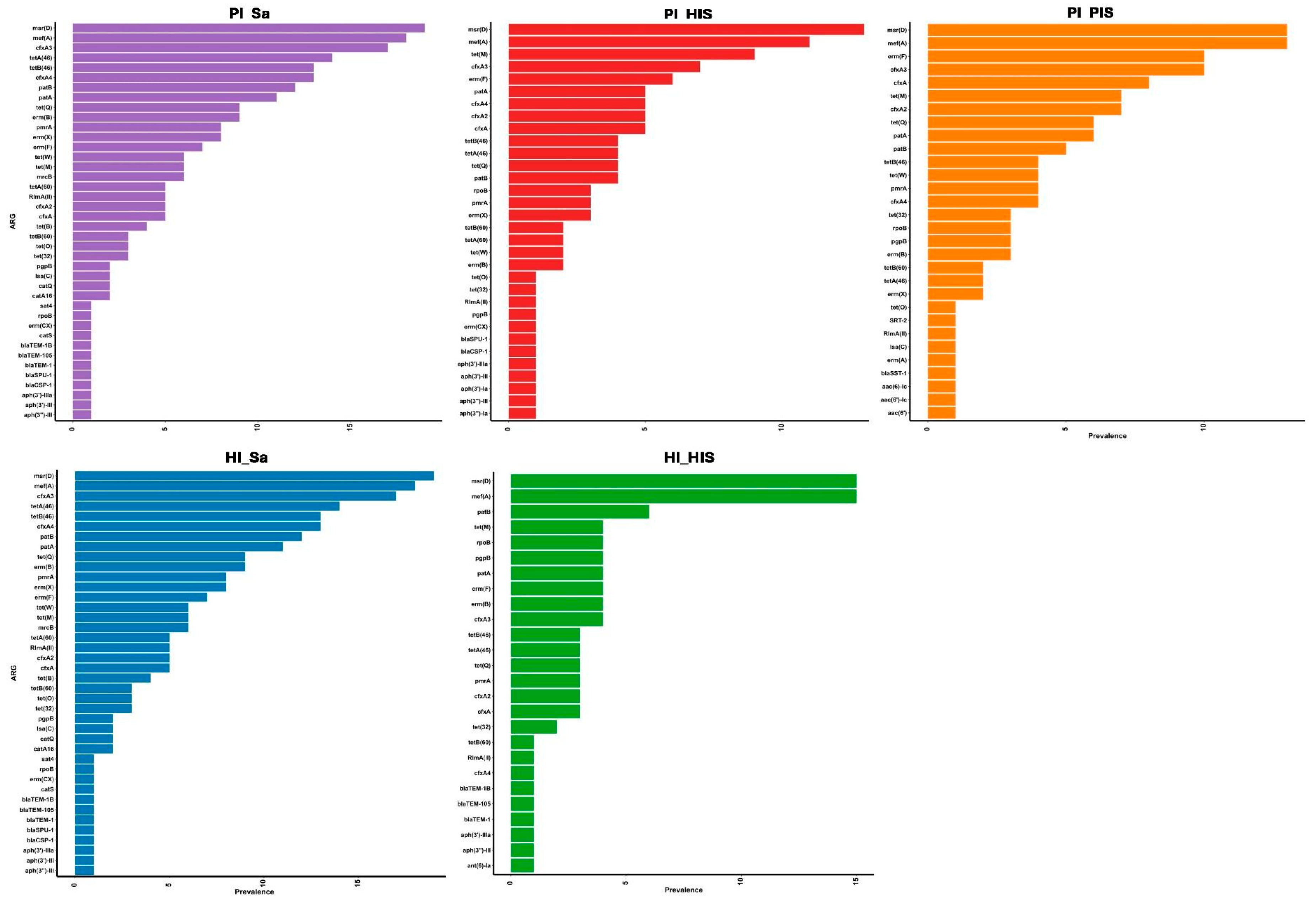
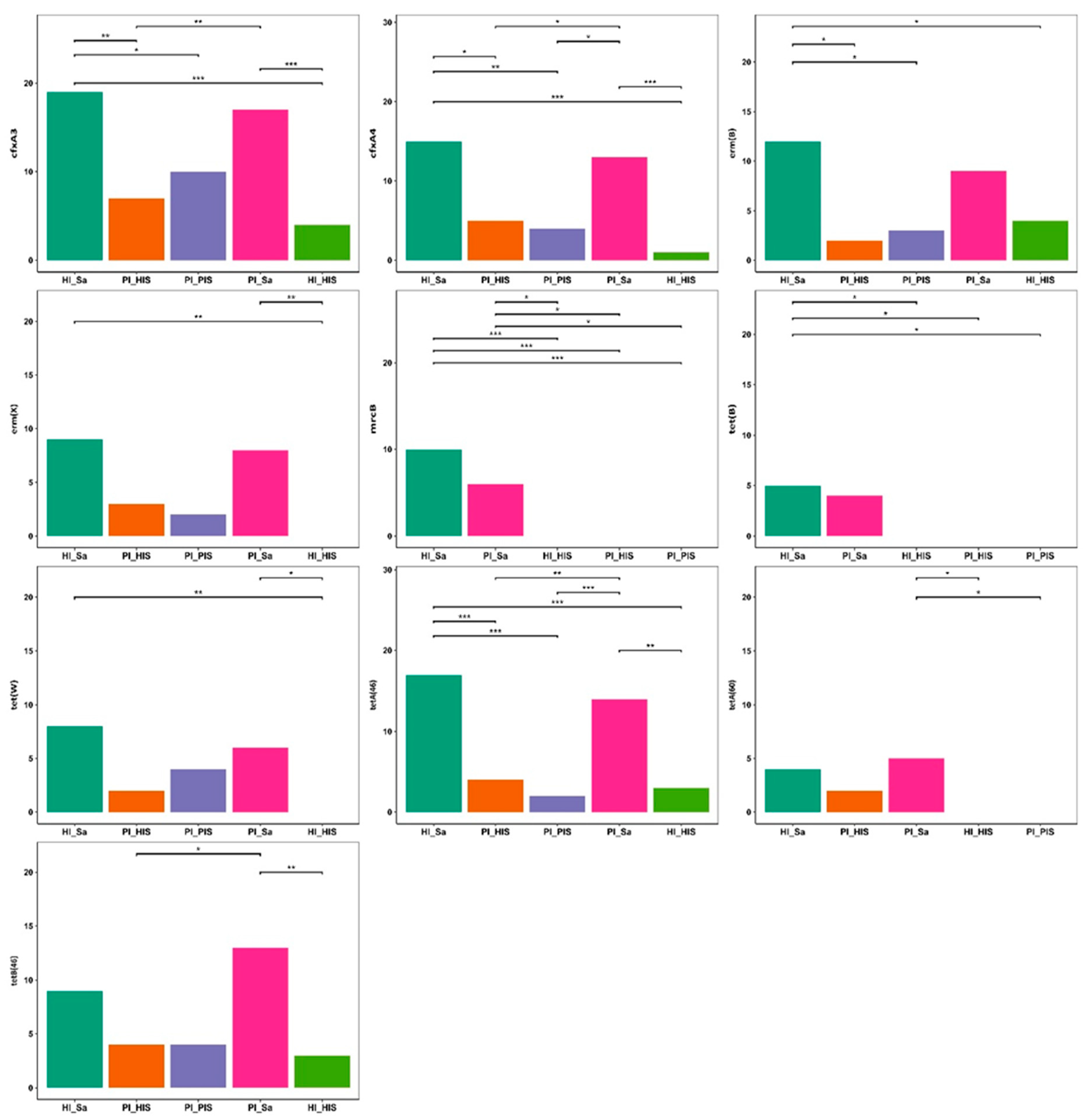
3.1. Detection and Prevalence of ARGs and Plasmids Across Study Groups
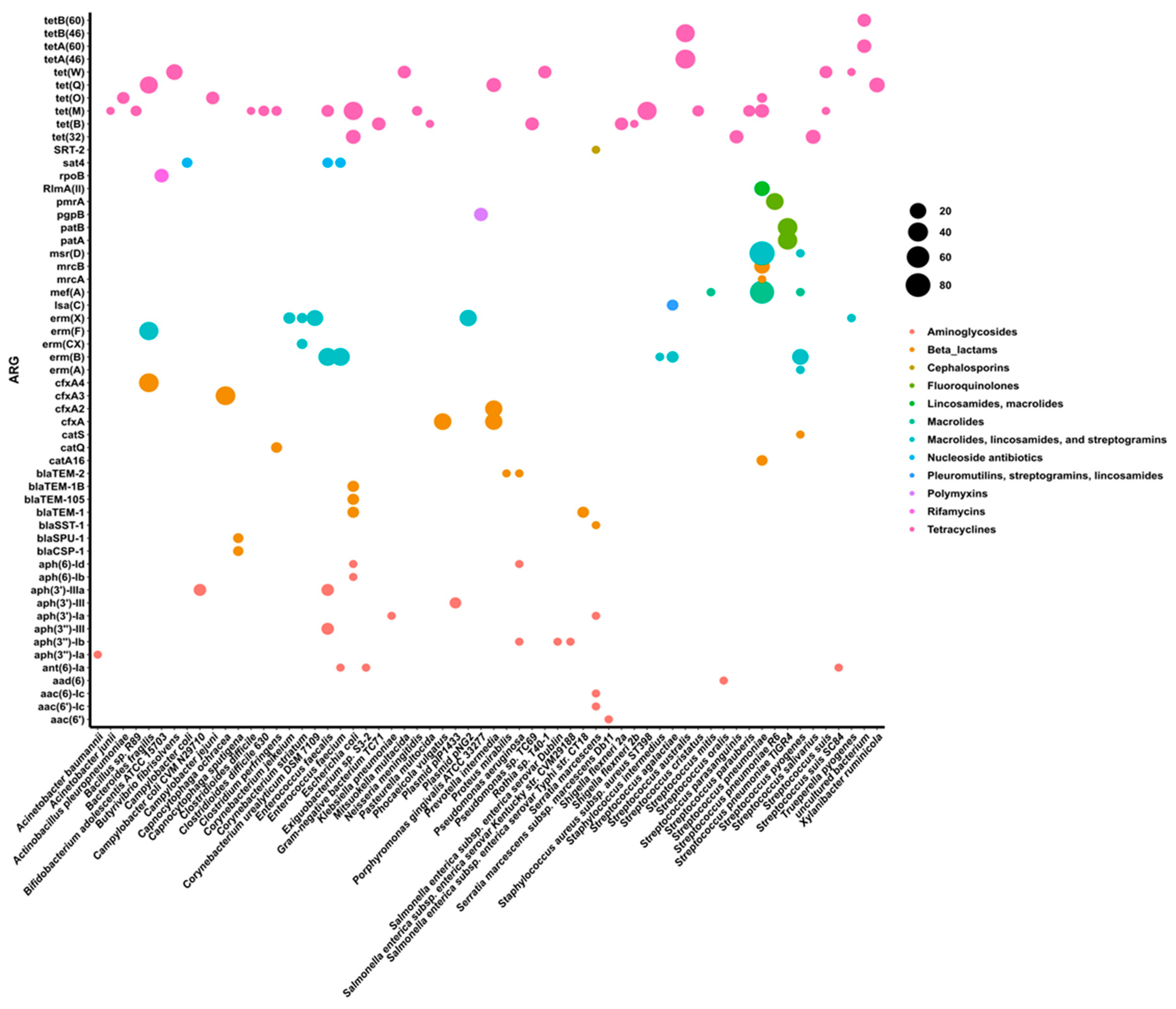
3.2. Taxonomic Assignment of Identified ARGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roberts, A.P.; Mullany, P. Oral Biofilms: A Reservoir of Transferable, Bacterial, Antimicrobial Resistance. Expert Rev. Anti. Infect. Ther. 2010, 8, 1441–1450. [CrossRef]
- Sharma, N.; Bhatia, S.; Sodhi, A.S.; Batra, N. Oral Microbiome and Health. AIMS Microbiol. 2018, 4, 42–66. [CrossRef]
- Olsen, I. Biofilm-Specific Antibiotic Tolerance and Resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12. [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902–903. [CrossRef]
- Bessa, L.J.; Botelho, J.; Machado, V.; Alves, R.; Mendes, J.J. Managing Oral Health in the Context of Antimicrobial Resistance. Int. J. Environ. Res. Public Health 2022, 19, 16448. [CrossRef]
- Kang, Y.; Sun, B.; Chen, Y.; Lou, Y.; Zheng, M.; Li, Z. Dental Plaque Microbial Resistomes of Periodontal Health and Disease and Their Changes after Scaling and Root Planing Therapy. mSphere 2021, 6, 1–20. [CrossRef]
- Sukumar, S.; Roberts, A.P.; Martin, F.E.; Adler, C.J. Metagenomic Insights into Transferable Antibiotic Resistance in Oral Bacteria. J. Dent. Res. 2016, 95, 969–976. [CrossRef]
- Anderson, A.C.; von Ohle, C.; Frese, C.; Boutin, S.; Bridson, C.; Schoilew, K.; Peikert, S.A.; Hellwig, E.; Pelz, K.; Wittmer, A.; et al. The Oral Microbiota Is a Reservoir for Antimicrobial Resistance: Resistome and Phenotypic Resistance Characteristics of Oral Biofilm in Health, Caries, and Periodontitis. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 1–19. [CrossRef]
- Sukumar, S.; Wang, F.; Simpson, C.A.; Willet, C.E.; Chew, T.; Hughes, T.E.; Bockmann, M.R.; Sadsad, R.; Martin, F.E.; Lydecker, H.W.; et al. Development of the Oral Resistome during the First Decade of Life. Nat. Commun. 2023, 14, 1–14. [CrossRef]
- Sukumar, S.; Rahmanyar, Z.; El Jurf, H.Q.; Akil, W.S.; Hussain, J.; Elizabeth Martin, F.; Ekanayake, K.; Martinez, E. Mapping the Oral Resistome: A Systematic Review. J. Med. Microbiol. 2024, 73, 1–10. [CrossRef]
- Belibasakis, G.N.; Bostanci, N.; Marsh, P.D.; Zaura, E. Applications of the Oral Microbiome in Personalized Dentistry. Arch. Oral Biol. 2019, 104, 7–12. [CrossRef]
- Kim, N.; Ma, J.; Kim, W.; Kim, J.; Belenky, P.; Lee, I. Genome-Resolved Metagenomics : A Game Changer for Microbiome Medicine. Exp. Mol. Med. 2024, 56, pages1501–1512. [CrossRef]
- Thompson, W.; Teoh, L.; Pulcini, C.; Williams, D.; Pitkeathley, C.; Carter, V.; Sanderson, S.; Torres, G.; Walsh, T. Dental Antibiotic Stewardship: Study Protocol for Developing International Consensus on a Core Outcome Set. Trials 2022, 23, 1–6. [CrossRef]
- Teoh, L.; Thompson, W.; Suda, K. Antimicrobial Stewardship in Dental Practice. J. Am. Dent. Assoc. 2020, 151, 589–595. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [CrossRef]
- Seemann, T. ABRicate: mass screening of contigs for antibiotic resistance genes. 2016. Available online: https://github.com/tseemann/abricate.
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; Doshi, S.; Courtot, M.; Lo, R.; Williams, L.E.; Frye, J.G.; Elsayegh, T.; Sardar, D.; Westman, E.L.; Pawlowski, A.C.; Johnson, T.A.; Brinkman, F.S.; Wright, G.D.; McArthur, A.G. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; Tadesse, D.A.; Morales, C.; Simmons, M.; Tillman, G.; Wasilenko, J.; Folster, J.P.; Klimke, W. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483–19 Erratum in: Antimicrob. Agents Chemother. 2020, 64, e00361–20, doi:10.1128/AAC.00361-20. [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT: A New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [CrossRef]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; Alexander, D.C.; Allen, V.; El Bailey, S.; Bekal, S.; German, G.J.; Haldane, D.; Hoang, L.; Chui, L.; Minion, J.; Zahariadis, G.; Domselaar, V.; Reid-Smith, R.J.; Mulvey, M.R. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O'Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; Wagner, H. vegan: Community Ecology Package; R Package Version 2.6-4, 2020. Available online: https://CRAN.R-project.org/package=vegan.
- Heinze, G.; Ploner, M. logistf: Firth’s Bias-Reduced Logistic Regression. R Package Version 1.23.1, 2020. Available online: https://CRAN.R-project.org/package=logistf.
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. Available online: https://www.R-project.org/.
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software, PBC: Boston, MA, 2023. Available online: https://posit.co/.
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, 2016; ISBN 978-3-319-24277-4.
- Rofael, S.; Leboreiro Babe, C.; Davrandi, M.; Kondratiuk, A.L.; Cleaver, L.; Ahmed, N.; Atkinson, C.; McHugh, T.; Lowe, D.M. Antibiotic Resistance, Bacterial Transmission and Improved Prediction of Bacterial Infection in Patients with Antibody Deficiency. JAC Antimicrob. Resist. 2023, 5, dlad135. [CrossRef]
- Fox, V.; Santoro, F.; Pozzi, G.; Iannelli, F. Predicted Transmembrane Proteins with Homology to Mef(A) Are Not Responsible for Complementing mef(A) Deletion in the mef(A)–msr(D) Macrolide Efflux System in Streptococcus pneumoniae. BMC Res. Notes 2021, 14, 1–7. [CrossRef]
- Vitali, L.A.; Di Luca, M.C.; Prenna, M.; Petrelli, D. Correlation Between Genetic Features of the mef(A)–msr(D) Locus and Erythromycin Resistance in Streptococcus pyogenes. Diagn. Microbiol. Infect. Dis. 2016, 84, 57–62. [CrossRef]
- Iannelli, F.; Santoro, F.; Santagati, M.; Docquier, J.-D.; Lazzeri, E.; Pastore, G.; Cassone, M.; Oggioni, M.R.; Rossolini, G.M.; Stefani, S.; Pozzi, G. Type M Resistance to Macrolides Is Due to a Two-Gene Efflux Transport System of the ATP-Binding Cassette (ABC) Superfamily. Front. Microbiol. 2018, 9, 1670. [CrossRef]
- Iwahara, K.; Kuriyama, T.; Shimura, S.; Williams, D.W.; Yanagisawa, M.; Nakagawa, K.; Karasawa, T. Detection of cfxA and cfxA2, the β-Lactamase Genes of Prevotella spp., in Clinical Samples from Dentoalveolar Infection by Real-Time PCR. J. Clin. Microbiol. 2006, 44, 172–176. [CrossRef]
- Yokoyama, S.; Hayashi, M.; Goto, T.; Muto, Y.; Tanaka, K. Identification of cfxA Gene Variants and Susceptibility Patterns in β-Lactamase-Producing Prevotella Strains. Anaerobe 2023, 79, 102688. [CrossRef]
- Serwold-Davis, T.M.; Groman, N.B. Identification of a Methylase Gene for Erythromycin Resistance Within the Sequence of a Spontaneously Deleting Fragment of Corynebacterium diphtheriae Plasmid pNG2. FEMS Microbiol. Lett. 1988, 56, 7–13. [CrossRef]
- Trieu-Cuot, P.; Gerbaud, G.; Lambert, T.; Courvalin, P. In Vivo Transfer of Genetic Information Between Gram-Positive and Gram-Negative Bacteria. EMBO J. 1985, 4, 3583–3587. [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895-903. [CrossRef]
- Karlsson, P.A.; Zhang, T.; Järhult, J.D.; Joffré, E.; Wang, H. Heterogeneity and Metabolic Diversity Among Enterococcus Species During Long-Term Colonization. bioRxiv 2024. [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine: 6th Revision; World Health Organisation: Geneva, 2019.
- Correia, F.; Ribeiro-Vidal, H.; Gouveia, S.; Faria Almeida, R. Prescription of Antibiotic Prophylaxis in Implant Placement Among Portuguese Dentists: A Web Survey. Clin. Oral Implants Res. 2024, 35, 242–250. [CrossRef]
- Becker, K.; Gurzawska-Comis, K.; Klinge, B.; Lund, B.; Brunello, G. Patterns of Antibiotic Prescription in Implant Dentistry and Antibiotic Resistance Awareness Among European Dentists: A Questionnaire-Based Study. Clin. Oral Implants Res. 2024, 35, 771–780. [CrossRef]
- Gager, Y.; Koppe, J.; Vogl, I.; Gabert, J.; Jentsch, H. Antibiotic Resistance Genes in the Subgingival Microbiome and Implications for Periodontitis Therapy. J. Periodontol. 2023, 94, 1295–1301. [CrossRef]
- Caselli, E.; Fabbri, C.; Accolti, M.D.; Soffritti, I.; Bassi, C.; Mazzacane, S.; Franchi, M. Defining the Oral Microbiome by Whole-Genome Sequencing and Resistome Analysis: The Complexity of the Healthy Picture. BMC Microbiol. 2020, 20, 120. [CrossRef]
- Fu, C.X.; Chen, C.; Xiang, Q.; Wang, Y.F.; Wang, L.; Qi, F.Y.; Zhu, D.; Li, H.Z.; Cui, L.; Hong, W.L.; et al. Antibiotic Resistance at Environmental Multi-Media Interfaces Through Integrated Genotype and Phenotype Analysis. J. Hazard. Mater. 2024, 480, 136160. [CrossRef]
- Scannapieco, F.A. Role of Oral Bacteria in Respiratory Infection. J. Periodontol. 1999, 70, 793–802. [CrossRef]
- Han, Y.W.; Wang, X. Mobile Microbiome: Oral Bacteria in Extra-Oral Infections and Inflammation. J. Dent. Res. 2013, 92, 485–491. [CrossRef]
- Molina, A.; Huck, O.; Herrera, D.; Montero, E. The Association Between Respiratory Diseases and Periodontitis: A Systematic Review and Meta-Analysis. J. Clin. Periodontol. 2023, 50, 842–887. [CrossRef]
- Sánchez-Peña, M.K.; Orozco-Restrepo, L.A.; Suárez-Brochero, Ó.F.; Barrios-Arroyave, F.A. Association Between Oral Health, Pneumonia and Mortality in Patients of Intensive Care. Rev. Med. Inst. Mex. Seguro Soc. 2020, 58, 468–476. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




