1. Introduction
Reduced inorganic sulfur compounds (RISCs) are widely distributed in hot springs, sedimentary mud, volcanic craters, and acidic wastewater [1-3]. The dissimilatory sulfur oxidation of RISC is a significant geochemical process, which is carried out exclusively by prokaryotes, and the energy generated during this process is coupled with photosynthesis and respiratory processes [4, 5]. Metallosphaera cuprina, a member of the Sulfolobales order within the archaeal domain, was isolated from the sediment of an acidic hot spring in Tengchong County, Yunnan Province, China. This organism thrives at temperatures ranging from 55 to 75°C and a pH range of 2.5-5.5. M. cuprina is a facultative chemoautotroph that oxidizes S0, pyrite, and tetrathionate to produce sulfuric acid under aerobic conditions while also capable of chemoheterotrophically growing with yeast extract [6-8]. The members of Sulfolobales shows great potential for bioleaching [9, 10].
Archaea with sulfur oxidizing function have been widely observed in the
Sulfolobales order [
11]. Among the
Sulfolobales members,
Metallosphaera [
12],
Acidianus [
13], and
Sulfolobus [
14] are representative sulfur-oxidizing archaea found in high temperature and acidic environments. These species are capable of metabolizing various sulfur compounds, as well as exhibiting heterotrophic and other physiological characteristics that enhance their adaptability to local environments. Previous research on thermophilic archaea has primarily focused on studying enzymes involved in the sulfur oxidation process, resulting in the discovery of a series of enzymes, such as Sulfur Oxygenase Reductase (SOR), Sulfide:Quinone Oxidoreductase (SQR), Sulfite-acceptor Oxidoreductases (SAOR), Thiosulfate:Quinone Oxidoreductase (TQO), and Tetrathionate Hydrolase (TTH) in
Acidianus brierleyi (
Ac. brierleyi) and
Acidianus ambivalens (
Ac. ambivalens) [11, 15].
Meanwhile, there have been numerous studies on the well-established sulfur-oxidizing systems from bacteria [16-18] such as
Acidithiobacillus ferrooxidans (
At.
ferrooxidans),
Acidithiobacillus
caldus (
At.
caldus),
Acidiphilium acidophilum (
Ap. acidophilum). The enzyme-catalyzed reactions in these bacteria are most suitable at pH levels between 2.0–4.0. Studies have reported that TTH of
At. ferrooxidans and
At. caldus are both located in the periplasmic space of cells [19, 20]. The TTH of
At. ferrooxidans hydrolyzes S
4O
62- into S
2O
32-, S
0 and SO
42-, while the TTH of
At. caldus produces S
2O
32-, S
5O
62- and SO
42- [19-22]. In
Thermithiobacillus tepidarius (former
Thiobacillus tepidarius), the TTH was supposed to locate either on or inside the cytoplasmic membrane, oxidize tetrathionate to S
0, SO
32- and other polythionates [
23]. In archaea, the TTH of
Ac. ambivalens is located in the pseudo-periplasmic space of the cell and is connected to the outer membrane of the cell membrane, displaying optimal activity at pH 1.0 and 95°C or above [
15]. TTH of
Acidianus hospitalis (
Ac. hospitalis) is found in extracellular proteins [24, 25]. When exposed to ultraviolet rays and mitogen C stress, the extracellular secretion of
Ac. hospitalis TTH was promoted [
24].
Kanao
et al. utilized Western Blot to examine the regulation of TTH in
Acidithiobacillus thiooxidans in the presence of different substrates. The expression of TTH was significantly higher when cultured with S
4O
62- as substrate compared to that with S
0 and S
2O
32- as substrates.
At. ferrooxidans cultured with S
4O
62- expressed the
tth gene while did not express it when iron was used as the substrate [26, 27]. Through transcriptomic and proteomic analyses, it was observed that
At. caldus expressed the TTH gene when cultured with S
4O
62- as the substrate, but did not when cultured with S
0 alone [
20]. Results from RT-qPCR indicated that the expression level of TTH with S
4O
62- as the substrate in
At. caldus was about 200 times higher than that with S
0 as the substrate [
28]. Protze's Northern Blot experiment demonstrated that when
Ac. ambivalens was cultured with S
4O
62- as the substrate, the transcription level of TTH RNA was significantly higher compared to when cultured with S
0 as the substrate [
15]. Furthermore, TTH activity with cyanolysis could only be detected when S
4O
62- was used as the substrate [
15]. Researches on TTH in archaea primarily focused on the genus
Acidianus. Although high homologous TTH gene sequences have also been identified in the genome of the genus
Metallosphere, they have not been identified in the proteome or extensively studied in the field of biochemistry and molecular biology.
In this study, we present evidence that M. cuprina Ar-4 cultured with S4O62- displays higher TTH expression level compared with S0 as the sole sulfur source. Additionally, we demonstrate that the TTH protein of M. cuprina Ar-4 mainly present in cytoplasm and periplasm and exhibits an optimum activity at pH of 6.0, distinguishing it from other TTH proteins, which show their activity at pH 1.0–4.0 and exist solely in the periplasm or cytoplasm or extracellular.
2. Results
2.1. Gene Sequence Analysis and Structure Prediction of Tetrathionate Hydrolase of M. cuprina
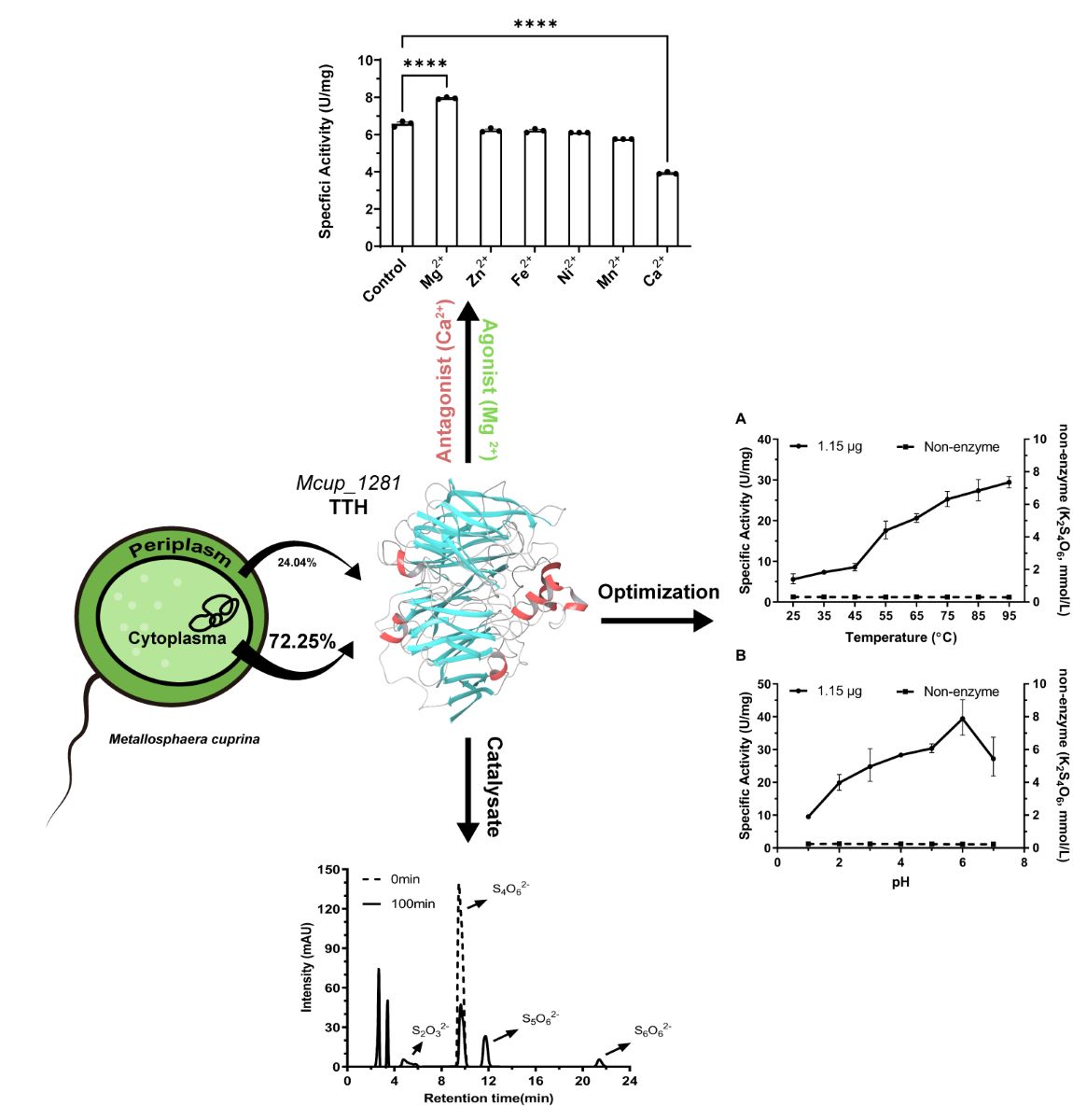
Based on the genome analysis, it is supposed that mcup_1281 of M. cuprina Ar-4 might encode a homologous protein of the TTH1 of Ac. ambivlaens. Further analysis conducted using the ExPASy ProtParam tool and determined that mcup_1281 encoded an open reading frame (ORF) consisting of 537 amino acid residues. The protein did not contain any disulfide bonds and only had one cysteine residue in the sequence. The presumed molecular weight was around 57.4 kDa. Further analysis of the protein’s secondary structure predicted the presence of a transmembrane region. These findings suggest that Mcup_1281 is likely located in the extracellular space and is associated with the cell membrane.
AlphaFold2 was used to predict the three-dimensional model. The Mcup_1281 monomer structure appeared to be mainly consisted of beta strands and coil structures and exhibited an eight-bladed β-propeller motif with each blade (I–VIII) consisting of average four antiparallel β-strands. Five α-helices were located outside the β-propeller region. Two of these α-helices (α2-α3) were suggested to participate in dimerization (19). No c-type cytochrome domain was identified in Mcup_1281, unlike the nitrite reductase (Nir PDB 1QKS with RMSD 2.9 Å). The tryptophan residues in the Mcup_1281 were predominantly located within the beta strands region similar to the TTH of
At. ferrooxidans (PDB 6L8A with RMSD 1.5 Å) (
Figure S1).
2.2. The TTH Gene is Expressed BOTH in Tetrathionate and S0 Grown Cells of M. cuprina
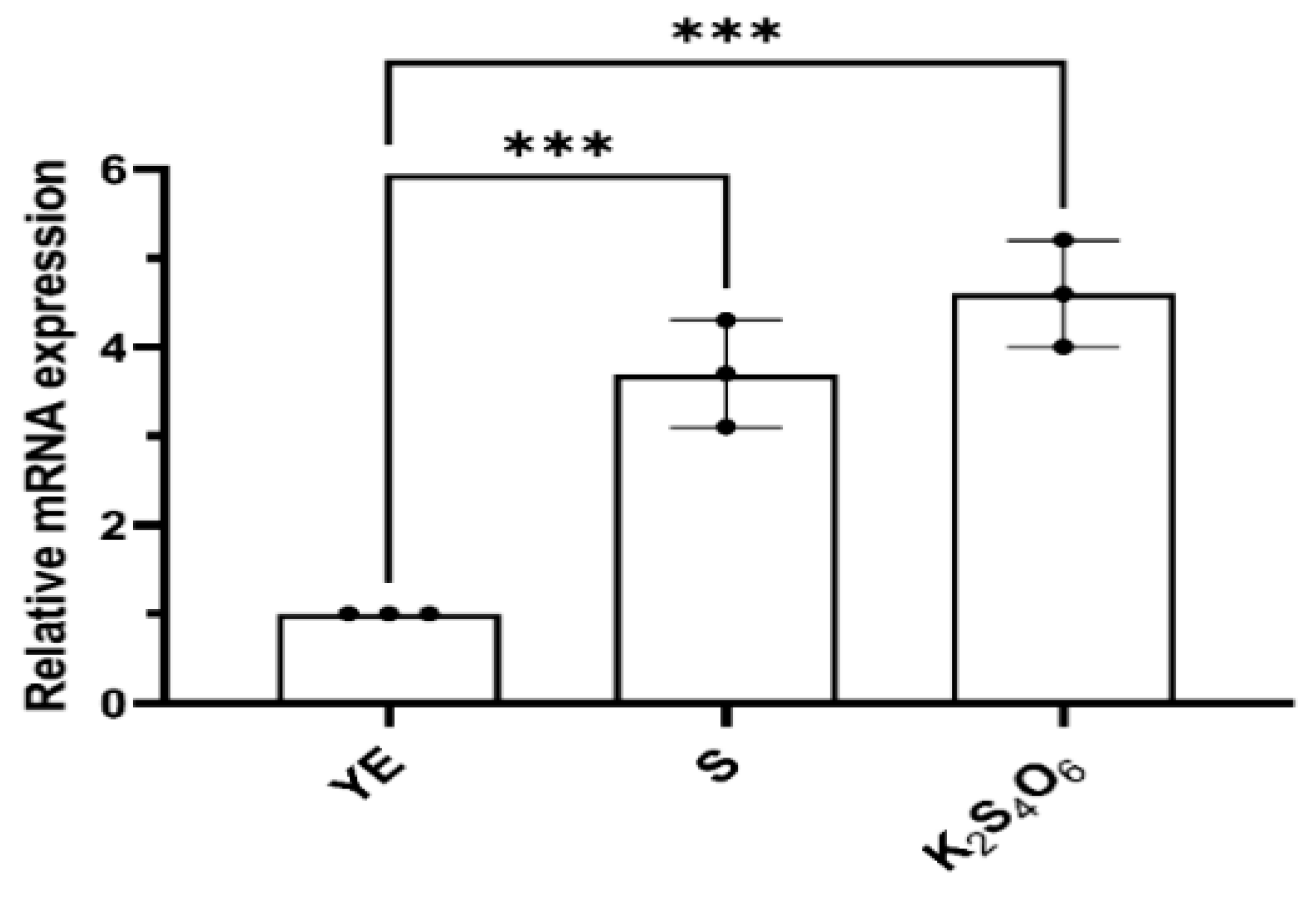
To analyze the transcriptional expression of
mcup_
1281, total RNA from cells cultured in different mediums (5 g S
0, 5 mmol/L K
2S
4O
6, or 2 g/L yeast extract) was extracted respectively. The transcription levels of
mcup_
1281 under different culture conditions were detected via RT-qPCR, and the 16S rRNA gene was used as the reference gene. The results showed that the transcription level of
mcup_
1281 increased by 5.4-fold and 4-fold when K
2S
4O
6 and S
0 were used as energy sources compared to the control group (yeast extract as energy source) (
Figure 1). Therefore, we propose that Mcup_1281 plays an important role in the sulfur metabolism of
M. cuprina, and define the Mcup_1281 as TTH
Mc.
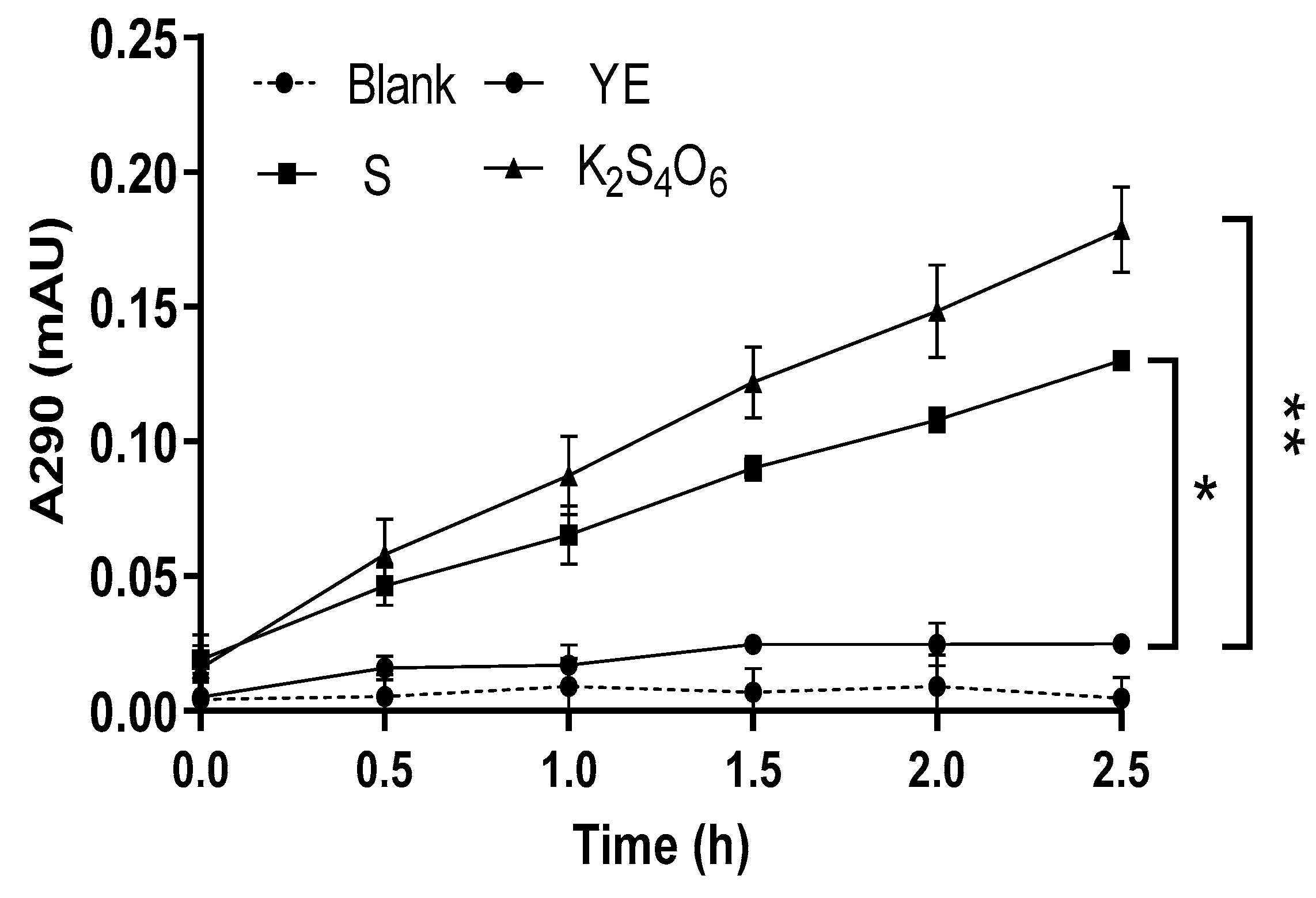
2.3. Preliminary Study on TTH Activity of M. cuprina
The different cell lysates of
Metallosphaera cuprina Ar-4 grown aerobically using three different mediums (Basic Salts Medium added 5 g/L S
0, 5 mmol/L K
2S
4O
6, or 2 g/L yeast extract separately [6, 14], see details at methods section 4.1) were respectively detected by the continuous enzyme activity assay method (detailed as 4.6) [
29]. One unit enzyme activity (U) was defined as the increase of absorbance at 290 nm per minute (∆A
290 min
-1) of cell lysates. When using 1 mmol/L K
2S
4O
6 as the substrate and incubating reaction mixture at 65°C and pH 3.0, the specific enzyme activity (activity of per mg protein) was 0.60 U/mg protein and 0.29 U/mg protein for cells grown on 5 mmol/L K
2S
4O
6 and 5 g/L S
0, respectively. The TTH
Mc activity with K
2S
4O
6 as the energy source was almost 2 times higher than that with S
0. However, TTH
Mc activity was not detected in the cell lysate cultured in yeast extract (
Figure 2). The results were in line with the transcription results of
Figure 1.
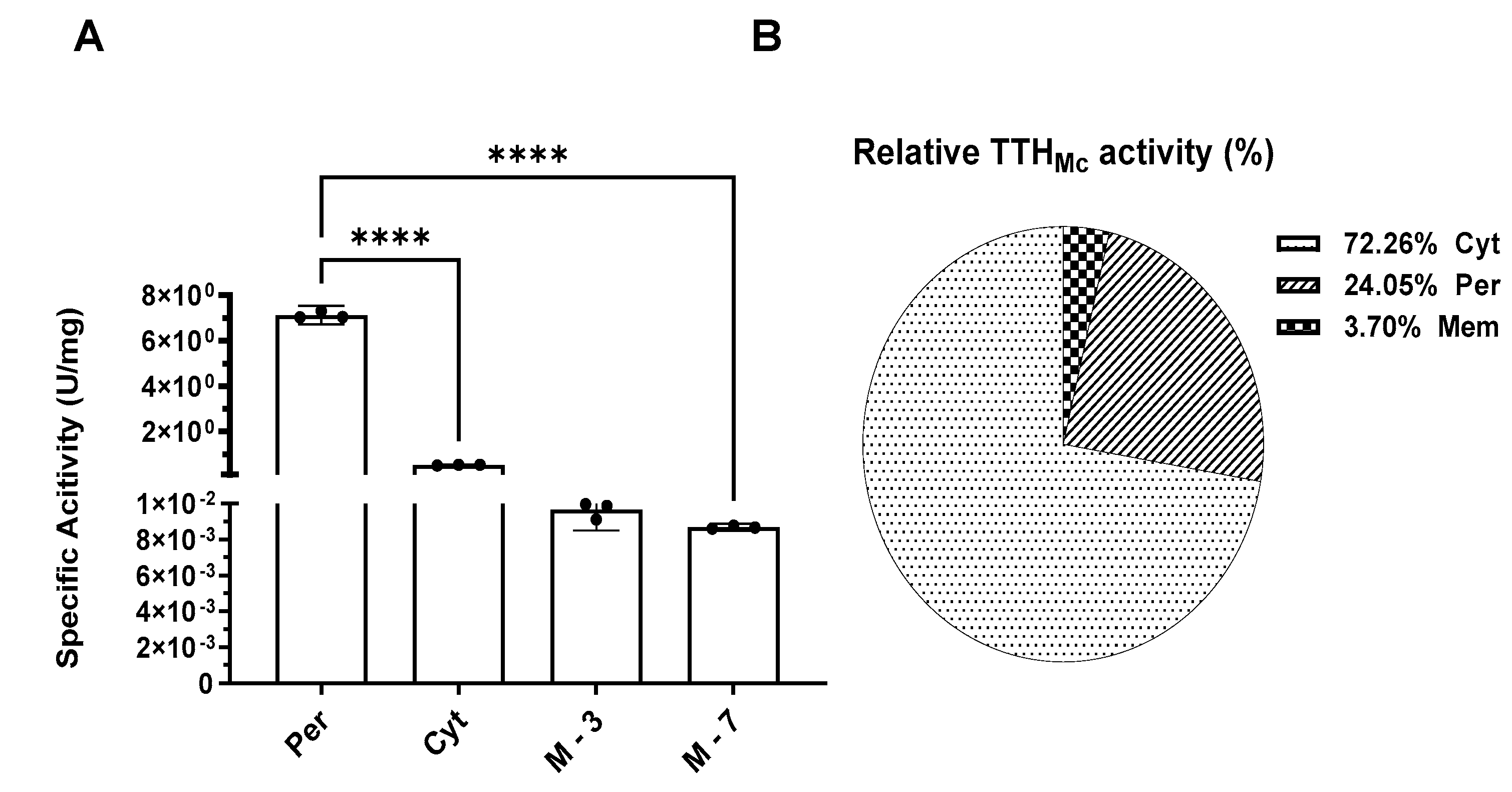
2.4. Tetrathionate Hydrolase Primarily Located in Cytoplasm and Periplasm
Discontinuous method (see 4.6.2) [
30] was used to detect the tetrathionate hydrolase (TTH) activity of Mcup_1281 in various cell fractions, including cell-free culture supernatant, cytoplasm, periplasm, and membrane fraction. Pyrophosphatase, as the cytoplasm marker, was used for identified different cell fractions. TTH activity was found in the cell-free culture supernatant, while its activity did not increase when cells were stressed with 5% mitomycin C, different from TTH of
Acidianus hospitalis [
24]. The cytoplasm, periplasm, and membrane proteins presented 1.172 U, 0.391 U, and 0.059 U of TTH activity respectively, which accounted for 72.26%, 24.05%, and 3.70% of the total TTH activity. The specific activity of the periplasmic fraction was 7.727 U/mg, of the cytoplasmic fraction was 0.526 U/mg, and 0.01 U/mg for the membrane at pH 3.0 and 0.009 U/mg for the membrane at pH 7.0 (
Figure 3). The higher specific activity of the periplasmic indicated that less amount TTH in periplasm than cytoplasm, while the least TTH activity and the specific activity supported that TTH did not locate on membrane. The results indicated that TTH
Mc was primarily located in the cytoplasm with less amount presence in the periplasm and barely any in the membrane (
Table S1).
2.5. Purification of the Tetrathionate Hydrolase from Whole Cell
Silver-stained SDS gels of different purification steps confirmed the presence of TTH band at about 54 kDa in
M. cuprina Ar-4 (
Figure S2). The specific activity of TTH in the total cell extract of
M. cuprina Ar-4 grown on 5 mmol/L K
2S
4O
6 was found to be 0.724 U/mg. After four chromatographic purification steps including DEAE anion exchange column chromatography, hydrophobic chromatography, size exclusion chromatography, and Q-sepharose chromatography, the specific activity increased to 11.67 U/mg protein, representing a 26.70-fold purification with 1.24% recovery (
Table 1 &
Figure S3).
2.6. Tetrathionate Hydrolase of M. cuprina is an Acid Resisting Thermophilic Enzyme
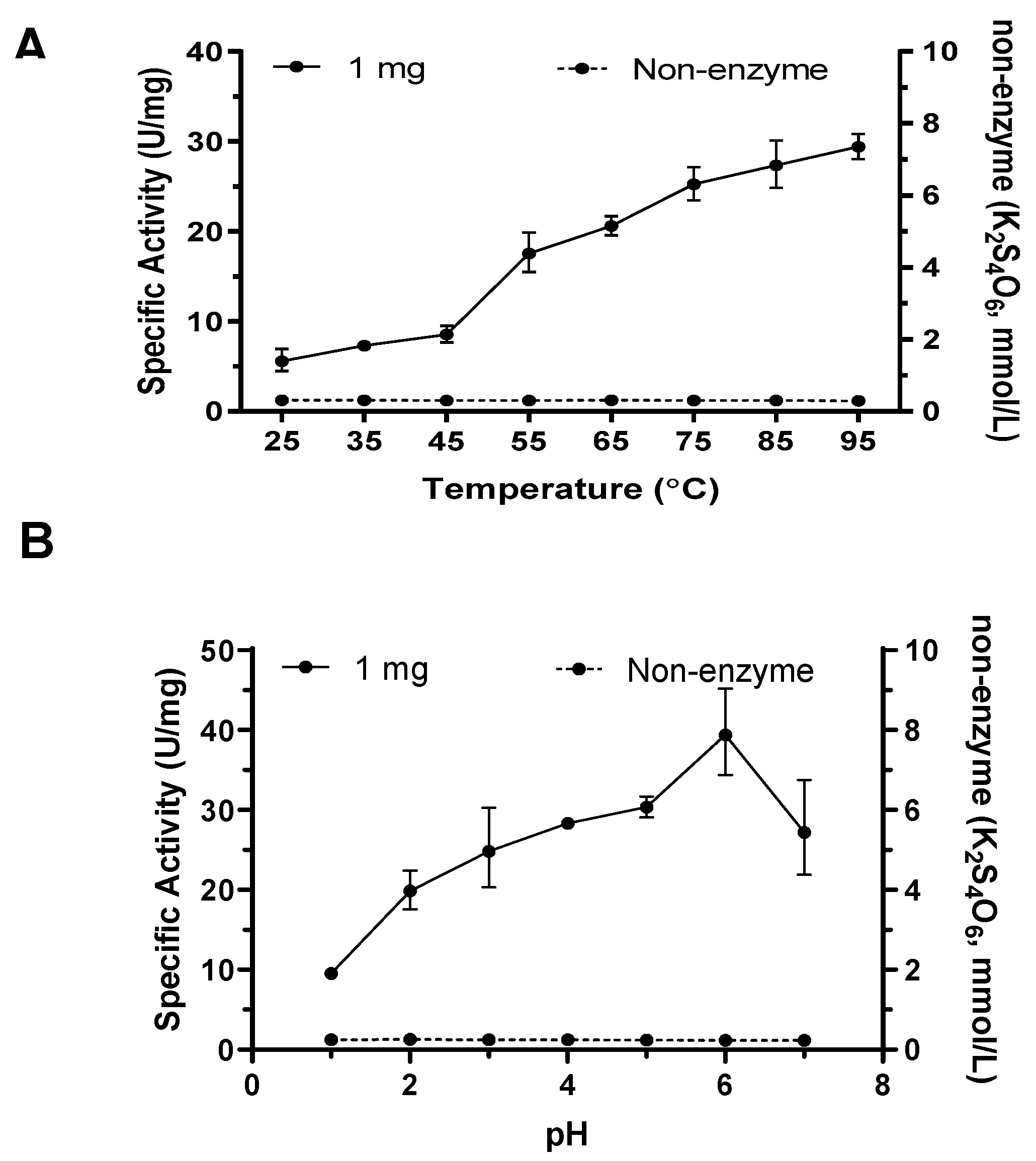
The enzyme activity of TTH
Mc was tested under different temperature and pH conditions. As the temperature increased, the enzymatic efficiency increased almost linearly. The specific activity remained at approximately 30 U/mg at 95 ºC (
Figure 4A). However, the maximal and optimum temperature of TTH
Mc could not be determined due to the inability to reach a higher temperature range. The pH experiment showed that the maximum activity occurred at pH 6.0; specific activity of TTH
Mc increased from 9.54 ± 0.17 U/mg at pH 1.0 to 39.44 ± 1.75 U/mg at pH 6.0, then decreased at pH 7.0 (
Figure 4B). 0.01 mol/L Magnesium greatly stimulated the enzyme activity, while 0.01 mol/L calcium seriously inhibited the enzyme activity. Other divalent metal ions such as Fe
2+, Zn
2+, Mn
2+, and Ni
2+ had less effect on the specific activity of the enzyme (
Table 2 &
Figure S4).
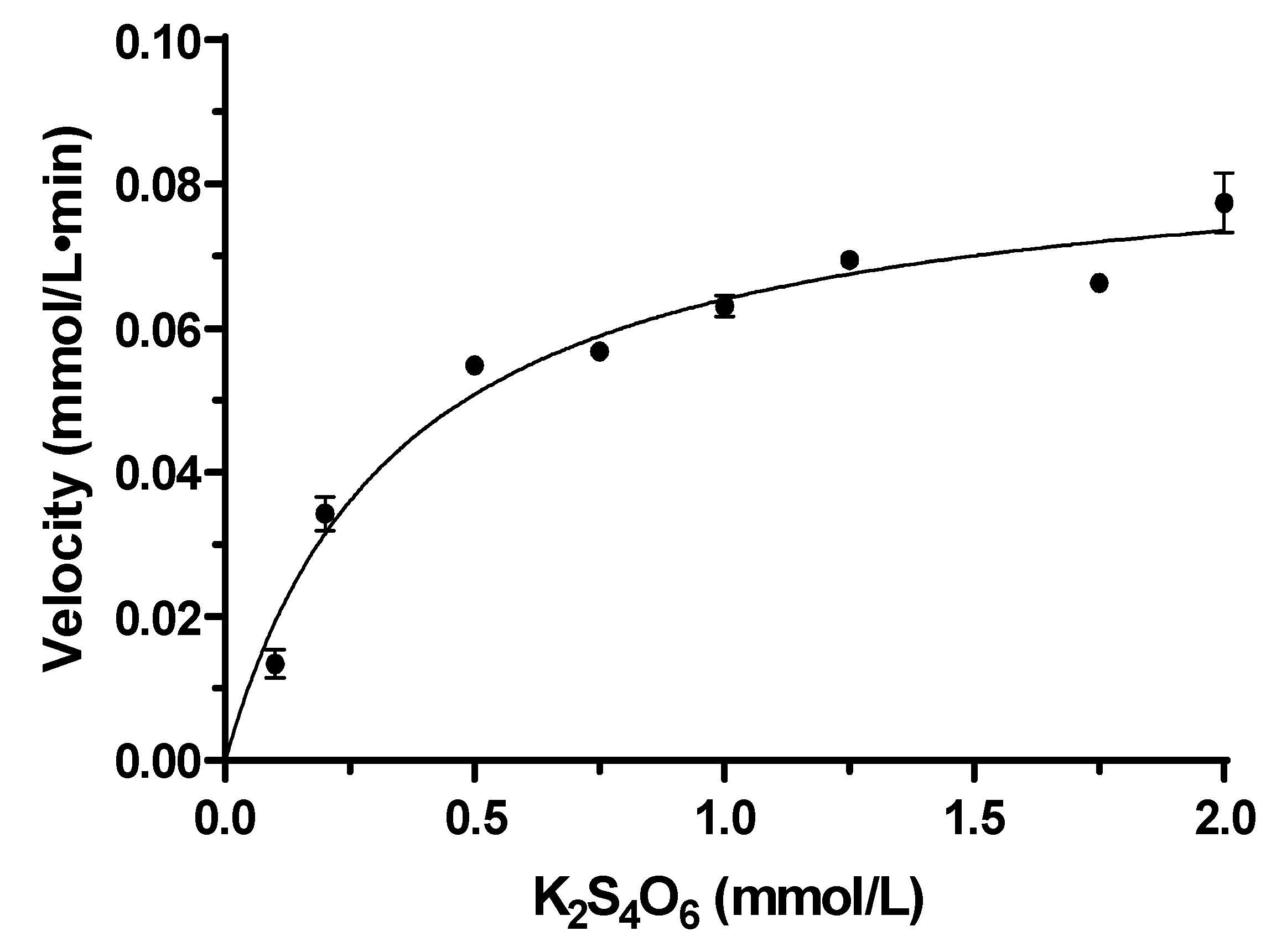
The reaction process of tetrathionate catalyzed by TTH
Mc followed a Michaelis-Menten curve; the substrate hydrolysis velocity (V) was influenced by the enzyme as well as substrate concentration. When the enzyme concentrations were 1.15 μg/L and 2.30 μg/L, the half-lives of the substrates were 55.12 min and 27.36 min respectively. The reaction rate constants corresponding to the half-life were K
1.15μg=0.01258 mmol/L·min and K
2.30μg=0.02533 mmol/L·min respectively and the enzyme concentration showed inversely proportional to the half-life (
Figure S5). The kinetic parameters of TTH activity were measured at a temperature of 95°C and pH of 6.0 with 1.15 µg TTH
Mc. The apparent K
m value for tetrathionate was determined to be 0.35 mmol/L while the observed V
max was measured as 86.3 μmol/min (
Figure 5).
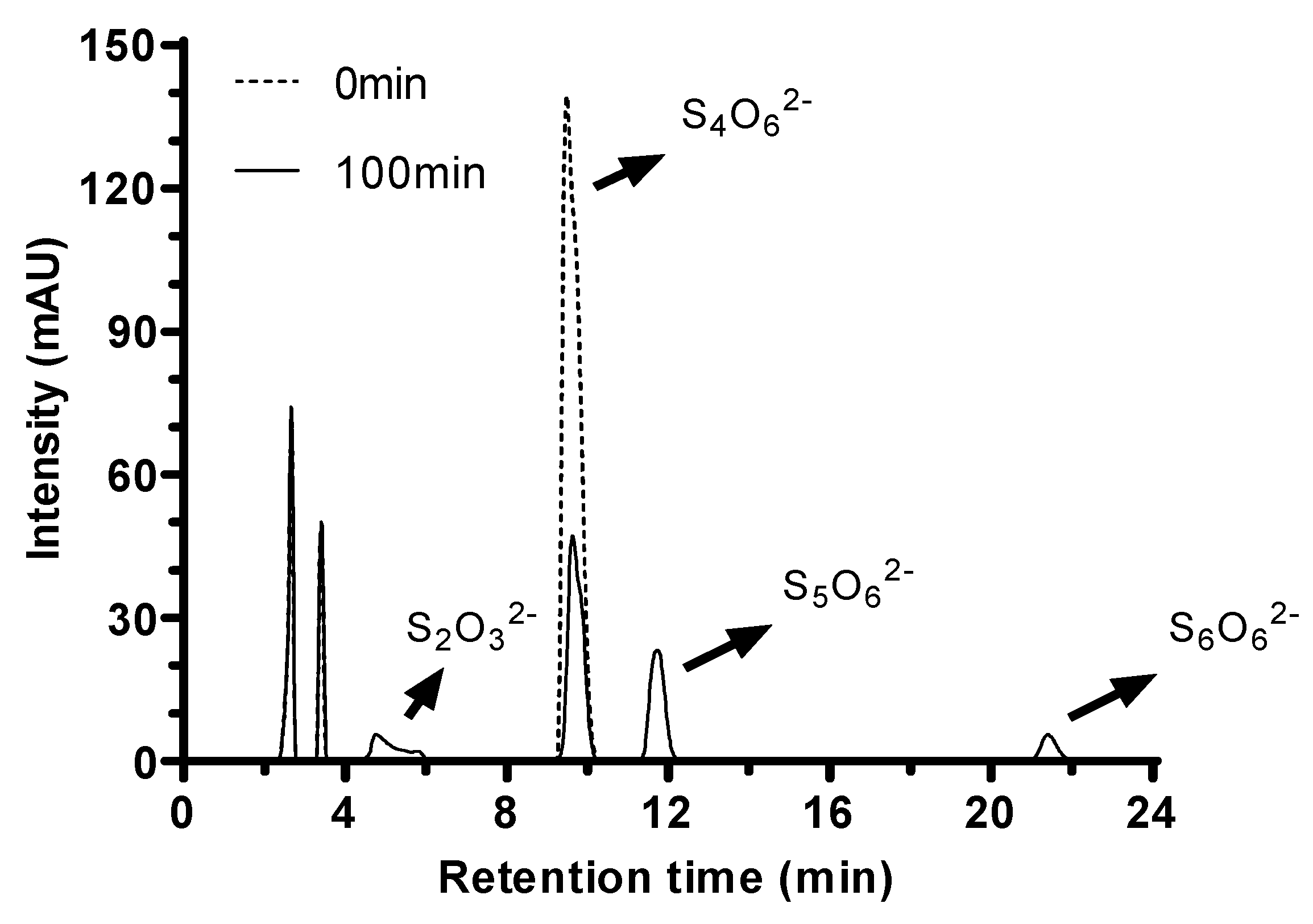
2.7. Hydrolysis Products of Tetrathionate by Tetrathionate Hydrolase
Analysis of the enzymatic reaction mixture by HPLC revealed the presence of four peaks. Besides the peak of tetrathionate at a retention time of 9.5 min, thiosulfate presented at a retention time of 5 min (
Figure 6. Retention times of pentathionate and hexathionate were 11.8 min and 21.6 min respectively, which were also identified using standards described by Miura & Kawaoi [
29].
3. Discussion
Tetrathionate plays a significant role in the metabolism of acidophilic microorganisms, particularly in the RISC pathway as it serves both as a substrate and as a mesostate [
31]. The location in cells and characters of the enzyme responsible for tetrathionate metabolism varies greatly among different species [12, 15, 22]. There is a lack of systematic studies on tetrathionate hydrolase of acidothermophilic sulfur-oxidizing archaeon
Metallosphaera genus, another member of the
Sulfolobaceae family. In our study, a tetrathionate hydrolase was purified from
Metallosphaera cuprina Ar-4, , the main objective of this investigation was to gain a comprehensive understanding of the tetrathionate hydrolase, determine its cellular localization, and characterize its biological properties.
3.1. TTH in Archaea and Bacteria Species
The TTH
Mc encoded by Mcup_1281 (PQQ-binding-like beta-propeller repeat protein) of
M. cuprina is clustered to that of other
Metallophaera sp. However, it exhibits differences with that of
Sulfolobus spp.,
Acidianus spp., and
Sulfurisphaera spp. It has been observed that the TTH cluster is conserved in
Metallophaera [
12]. Additionally, the region surrounding TTH contains an ABC transporter, a 4Fe-4S cluster domain-containing protein, and a vitamin epoxide reductase family protein (
Figure S6). Tetrathionate hydrolases are widely found in sulfur-oxidizing archaea and bacteria, but the number of gene copies and protein location are not consistent across microorganisms [
15]. There is no information available on the regulatory system that controls the tetrathionate intermediate pathway in sulfur-oxidizing archaea, such as the two-component system RsrS-RsrR found in
Acidithiobacillus species [
32]. We hypothesize that sulfur-oxidizing archaea have developed a specialized regulatory system to adapt to challenging environmental conditions.
The top-scoring BLAST entries of TTH
Mc at the Uniprot and NCBI databases (pairwise identity >30%) were highly similar homologs found in acidophilic archaea and bacteria (
Figure S7). These homologs belong to the PQQ family proteins and are likely to be authentic tetrathionate hydrolases. The proteins formed separate branches according to their respective species, indicating conservation of tetrathionate hydrolases within the same species. However, there were some variations observed in tetrathionate hydrolases among different species. In addition, it was noted that the
Metallosphaera spp. lacked
tth2 gene copies of tetrathionate hydrolases found in
Acidianus spp.
Unlike the tetrathionate hydrolase of
Ac. ambivalens, TTH
Mc can also be detected in cells of
M. cuprina Ar-4 grown on S
0 as the sole energy medium. The transcription level of TTH
Mc is higher in cells grown on K
2S
4O
6 than in cells grown on S
0. This suggests that the expression of tetrathionate hydrolase varies among different species and that tetrathionate metabolism in
M. cuprina Ar-4 is TTH-dependent, similar to sulfur-oxidizing bacteria [
33]. We also found the presence of Fe (II) in the broth from tetrathionate- and sulfur-grown
M. cuprina Ar-4 cells, through the addition of Fe (III) to the medium, there is a possibility that it came from the contact of tetrathionate hydrolase with ferric reductase, although this was not shown [
34].
3.2. TTH is Found in the Cytoplasm and Periplasm of K2S4O6 and S0-Grown Cells
It has been suggested that TTH (TTH1) of
Ac. ambivlaens is located in the pseudo-periplasmic space [
15]. Additionally, it has been stated that TTH of
Ac. hospitalis was a secreted protein with a zipper-like shape [
24]. However, we found that TTH
Mc is a soluble protein and it has the highest specific activity in periplasm and cytoplasm compared to other fractions. Moreover, the optimal pH of TTH
Mc is 6.0, unlike TTH of other microorganisms. These results indicate that TTH
Mc is located in both the periplasm and the cytoplasm.
A series of purification strategies are based on the chemical properties of protein, such as its hydrophobicity, charge property and molecular weight. In a recent study, tetrathionate hydrolase was purified using four chromatographies [
15]. The ion exchange column chromatography process with NaCl concentrations of 150 mM-300 mM resulted in similar charge properties of TTH
Mc as
Ac. ambivalens TTH1 [
15]. However, the hydrophobicity character differed between TTH1 and TTH
Mc. TTH
Mc precipitated in 2 mol/L (NH
4)
2SO
4. The specific activity consistently increased with each high number of purification steps, but the recovery yield was only 1.24%. This low yield may be attributed to purification steps, as increased steps often lead to a higher protein loss rate. Additionally, metal ions or cofactors like calcium may play a crucial role in stabilizing the protein structure [
35].
3.3. Biochemical Properties of Tetrathionate Hydrolase
Some biochemical properties of TTH
Mc of
M. cuprina are similar to those of other acidophilic archaea [
15]. The optimal temperature for this enzyme is above 95°C, which is much higher than the enzymes from mesophilic species (optimum at 40–65°C), even higher than the optimal growth temperature of
M. cuprina [
6]. By the protein sequence of the TTH
Mc, we calculated that the isoelectric point (pI) of TTH
Mc is 4.70 by ExPASy ProtParam (
https://web.expasy.org/protparam/), different from that of TTH1 of
Ac. ambivlaens which is 6.48 [
15]. Nevertheless, TTH
Mc exhibited an optimal pH of 6.0, which is higher than the typical pH ranges [1.0-4.0] observed for TTHs from other sulfur-oxidizing bacteria and archaea [
15]. This suggests that TTH
Mc is more effective in the intracellular environment. The molecular mass of the enzyme is approximately 57 kDa, however, no homodimer formation was observed. At pH 6.0 and 95°C, the K
m value of TTH
Mc for tetrathionate is 0.35 mmol/L. Reported K
m values for TTH vary widely, suggesting differing substrate affinities across species. Among them, the TTH from
Thiobacillus ferrooxidans (
Ac. ferrooxidans) stands out with a K
m value of 50 μmol/L, appearing a higher substrate affinity [
23].
3.4. Evolutionary Insights of TTHs Functionality
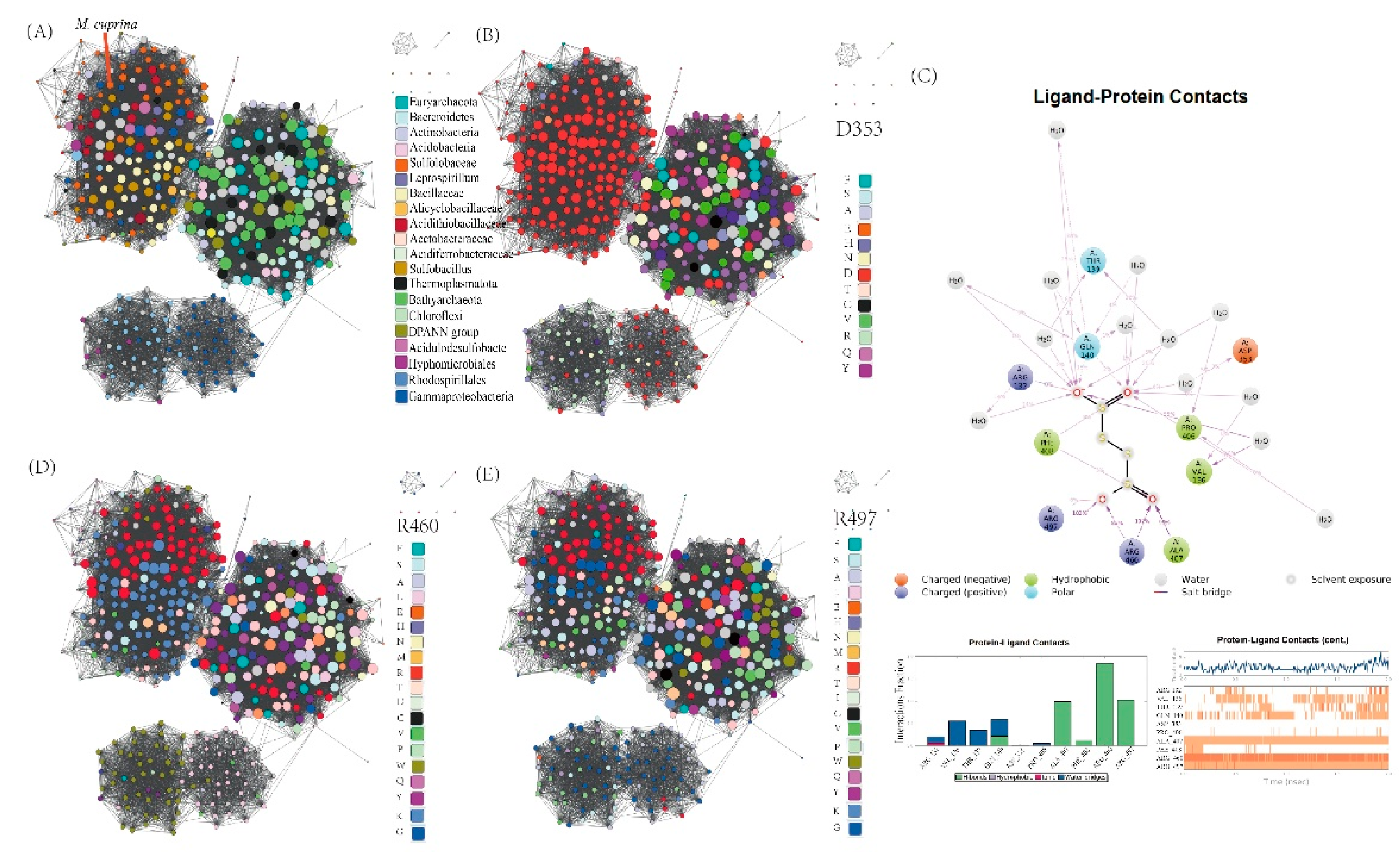
To delve deeper into the evolutionary history of TTHs functionality, we constructed a protein sequence similarity network (SSN) using TTH
Mc as the query [
36]. The analysis reveals that TTHs from the
Sulfolobaceae family cluster closely with homologs predominantly from other acidophilic sulfur/iron-oxidizing bacterial groups such as the
Acidithiobacillaceae, Alicyclobacillaceae, Sulfobacillus, and
Leptospirillum. These homologs form a dense cluster on the periphery of the entire SSN (
Figure 7A) and maintain few connections with distantly related homologs of unknown function. This pattern supports the occurrence of cross-domain horizontal gene transfer (HGT) of TTH, further corroborated by the unconserved genomic contexts within the
Sulfolobaceae strains and the presence of nearby transposase element (
Figure S8). Previous studies have shown that an aspartate residue corresponding to Asp353 in TTH
Mc is crucial for the activity of the TTH from
Acidithiobacillus ferrooxidans [
19]. This aspartate residue is highly conserved within the TTH sub-cluster and is occasionally substituted by another acidic residue, glutamate (
Figure 7B). This suggests that this aspartate residue initiates the reaction through protonating the S (alpha) atom of the tetrathionate [
19].
Classical molecular dynamics were utilized to explore additional key interacting residues in TTH
Mc (
Figure 7C). The analysis identified Arg460, Arg497, and Ala407 as the residues with the highest interaction rates. These residues likely serve as electrostatic stabilizers, forming hydrogen bonds with the terminal oxygen atoms adjacent to the S (alpha) atom of tetrathionate. Furthermore, Gln140, Thr139, Val136, and Arg132 of TTH
Mc, positioned on a helix extending from the narrower propeller side, interact with the substrate via water-mediated bridges.. Within the taxa from the TTH sub-SSN, Arg460 and Arg497 are frequently substituted by another basic residue (lysine) (
Figure 7D-E). Additionally, other interacting residues lack aligned counterparts in remote sub-SSNs at equivalent positions, indicating the absence of corresponding structural regions (marked in grey) (
Figure S9A-G). Moreover, three methionine residues—Met172, Met238, and Met279—situated near the tetrathionate binding pocket have been identified as putative metal ligands in TTH
Mc (
Figure S9H-J). These residues are conserved within the sub-SSN of TTHs, though variations exist such as histidine replacing methionine in some instances. These findings suggest that the presence of these interacting residues likely enhances substrate binding and TTH activity across the TTH protein superfamily.
To further validate the functional significance of TTH-like proteins, future research should focus on functional assays and detailed experimental investigations. Such experiments will aid in confirming the role of these identified residues and their contributions to enzymatic activity. Overall, this study offers significant insights into evolutionary dynamics of TTHMc, setting the stage for a deeper understanding of tetrathionate hydrolase activity in related proteins.
4. Materials and Methods
4.1. Strains and Growth Conditions
Metallosphaera cuprina Ar-4T was cultured at 65 °C in three different mediums described from modifying Allen medium [6, 14]. The BSM (Basic Salts Medium) contained (NH4)2SO4 1.3 g, K2HPO4 0.28 g, MgSO4·7H2O 0.25 g, CaCl2·2H2O 0.07 g, FeCl3·6H2O 0.02 g, MnCl2·4H2O 1.8 mg, Na2B4O7·10H2O 4.5 mg, ZnSO4·7H2O 0.22 mg, CuCl2·2H2O 0.05 mg, Na2MoO4·2H2O 0.03 mg, CoSO4·7H2O 0.01 mg, VoSO4·2H2O 0.03 mg. The initial pH was adjusted to 3.5 with sterile 50% H2SO4. About 5 g sterile S0 or 5 mmol/L K2S4O6 as sulfur source and 0.2% yeast extract were added in BSM to promote cells growing. 2 g/L yeast extract was added in BSM for heterotrophic culture strains. Cell growth was monitored by optical density at 600 nm.
4.2. Sequence Analysis
The homologous amino acid sequences were searched in Uniprot and NCBI databases with Mcup_1281 as a probe. The multiple alignment (the homologs amino acid sequences from
Metallaspahera spp.,
Acidianus spp., and
Acidithiobacillus spp., etc) and a phylogenetic dendrogram were constructed using MAFFT version 7 with the L-INS-I algorithm, the BLOSUM 30 similarity matrix, a gap opening penalty of 1.53, and the offset value of 0.1. Using the ITOL (
http://itol.embl) aimed to modifying the phylogenetic tree. The amino sequences of TTH from
Metallaspahera,
Acidianus,
Acidithiobacillus were compared and analyzed by ESPript 3.0 (
http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Second structure of Mcup_1281 was predicted by SOPMA (
https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html). Prediction of signal sequences and transmembrane domain were done with TMHMM and SignalP (
http://www.cbs.dtu.dk/services/). Three-dimensional models of Mcup_1281 were predicted by AlphaFold2 [
37] and the template model was quinoprotein alcohol dehydrogenase of
Pseudomonas putida (PDB accession number 1kv9). The image of prediction model was shown by Viewer Lite. MD simulations for the protein-ligand complex were performed using the Desmond Molecular Dynamics System version 3.6 (D. E. Shaw Research, New York, NY, 2008) with OPLS_2005 force field.
4.3. RNA Extraction from Cells and Transcription Analysis for Mcup_1281
Cells grown on three different mediums containing 5 g/L S0, 5 mmol/L K2S4O6 and 2 g/L yeast extract respectively were harvested by 10000×g centrifugation for 10 min. Total RNA of cells was extracted with E.Z.N.A Total RNA Kit I (Omega). The quantity and quality of the RNA was measured by UV absorption (A260/A280=1.8-2.0) using NanoDrop One (Thermo Fisher Scientific, USA). The RNA was reverse transcribed by HiFi-Script cDNA Synthesis Kit (CWBIO, China). 16S rRNA gene of Ar-4T was set as the reference gene and the mcup_1281 as target gene. The target DNA fragments used for synthesis were amplified with the primers Real-TTH-F (GCGGGGTTCACAAGTTTGAC) and Real-TTH-R (GCCTGTGCTGGATGATGT), while the reference gene fragments primers were Real-16S rRNA-F (TTGGGATCGAGGGCTGAAAC) and Real-16S rRNA-R (TCCCCTACGGCTACCTTGTT). LightCycler® 480 System (Roche, Switzerland) was used for quantitative PCR and program included: Pre-incubation 95℃ 1 cycle; Amplification; degeneration at 95℃ for 10 s, annealing at 55℃ for 20 s, extension at 72℃ for 40 cycles; Melting curve: 95℃ for 5 s, 55℃ for 1 min, 1 cycle.
4.4. Tetrathionate Hydrolases Purification from Whole Cells
Purification of tetrathionate hydrolases from whole cells was carried out by modifying the method described by Protze (15). 5 g of wet cell was collected from the TT medium or S0 medium. Cells were resuspended in 50 mL 40 mmol/L K3PO3 buffer (pH 7.0) with 10 mmol/L MgCl2, 25 μg/mL RNase, 10 μg/mL DNase I and 1 mmol/L phenylmethanesulfonyl fluoride (PMSF). After adding 1 mm glass beads, cells were disrupted three times through a homogenizer (Fast-prep-24, MP, USA) with 6.0 m/s, 30 s. Cell debris and unbroken cells were discarded by centrifugation at 10000×g and 4 °C for 15 min. The soluble fraction and membrane were separated by centrifugation at 140000×g and 4°C for 60 min. The supernatant was dialyzed against 40 mmol K3PO3 and 10 mmol/L MgCl2 (pH 7.0) at 4 °C for 16 h and the membrane was stored at -80 °C with phosphate buffer saline (PBS) (pH 7.2). The dialyzed soluble fraction containing crude enzyme was applied on the DEAE Sepharose fast-flow column (4×5 mL column volume (CV); GE Healthcare) equilibrated with 40 mmol/L K3PO3 and 10 mmol/L MgCl2, pH 7.0. Proteins were eluted by NaCl solution with a linear concentration gradient of 0-0.2 mol/L, 0.2-0.4 mol/L, 0.4-0.6 mol/L, 0.6-0.8 mol/L, and 0.8-1.0 mol/L, each for 5-10 CV. The eluent fraction showing activity of tetrathionate hydrolases were pooled and dialyzed against 50 mmol/L Tris/HCl (pH 8.0) supplemented 1 mol/L (NH4)2SO4 for 16 h at 4 °C. After dialysis, the fraction was centrifugated at 40000×g and 4℃. The supernatant was next applied onto Phenyl-Sepharose Fast-Flow column [5mL column volume (CV); GE Healthcare]. Protein was eluted by 10 CV (NH4)2SO4 with a linear gradient from 1 mol/L to 0 mol/L. Fractions showing tetrathionate hydrolases activity were pooled and concentrated with Centriprep YM-30 centrifugal ultrafiltration tube (15 mL Millipore, USA) and then eluted by 40 mmol/L PBS (pH 7.5) with flow rate of 0.5 mL/min through Superdex 200 size exclusion column (30 mm×600 mm; GE Healthcare). The peak showing the tetrathionate hydrolases activity was applied on the Q-Sepharose-HP column [5 mL column; GE Healthcare]. Protein was eluted by NaCl with a linear gradient from 0.15 mol/L to 0.6 mol/L. Every purification fraction was detected with Silver-stained SDS-PAGE and HPLC enzyme assay to ensure tetrathionate hydrolases activity.
4.5. Cell Fractionation Separation
Cells of M. cuprina Ar-4 were grown to stationary phases and centrifugated at 10000×g for 30 min. The cell-free supernatant was in turn filtered through filters with hole sizes of 2.2 μm, 0.8 μm, and 0.22 μm. The filtrate then was concentrated with Centriprep YM-30 centrifugal ultrafiltration tube (15 mL Millipore, USA) as extracellular protein.
1 g wet cells were washed twice with 10 mmol/L Tris-HCl buffer solution and resuspended with 50 mL 10 mmol/L Tris-HCl buffer solution (pH 7.3), then equivalent volume of 10 mmol/L Tris-HCl buffer containing 40% sucrose was slowly added into and rocked slowly for 30 min at room temperature followed by centrifugation with 10000×g at 4 °C. Then the cells were quickly resuspended with 50 mL 5 mmol/L MgCl
2 at 4 °C and remove spheroplast by centrifugation at 10000×g for 30 min. The supernatant was periplasmic protein [
38].
Spheroplast were washed three times with BSM and resuspended with 100 mmol/L Tris-HCl and disrupted by sonication using Vibra Cell, cell debris was removed by 10000×g for 15 min and the soluble fraction was centrifuged by 100000×g for 60 min at 4 °C to separate the membrane (precipitate) and cytoplasmic (supernatant) fraction.
The membrane fraction was washed three times with distilled deionized water, resuspended in an elution buffer, and stayed on ice overnight. The formate buffer (pH 3.0) contained 0.5% (v/v) N-lauroylsarcosine, 1% (v/v) SDS, and 1% (v/v) Triton X-100 or 100 mmol/L Tris-HCl buffer (pH 7.0) contained 0.5% (v/v) Triton X-100, 5 mmol/L EDTA were used as elution buffer [
38]. The fraction was centrifuged by 100000×g for 60 min at 4℃ and the soluble fraction was concentrated with Centriprep YM-30 centrifugal ultrafiltration tube (15 mL Millipore, USA) as the membrane protein fraction that was tested for TTH activity.
4.6. Enzyme Characteristic Assay
Tetrathionate hydrolase activity of different fractions was determined by two methods.
4.6.1. Continuous Method of Enzyme Activity Assay
The continuous assay is based on the principle that tetrathionate hydrolases hydrolyze K
2S
4O
6 to produce a series of sulfur-containing compounds which increase absorbance at 290 nm [
29]. The reaction mixture (1 mL, pH 3.5) contained 50 mmol/L K
3PO
3, 1 mmol/L K
2S
4O
6, and 25-100 μL cell lysate or distillation water as control. Enzyme activity was detected by full-wavelength scan which was found that the absorption value of the mixture increases from 250 nm to 430 nm. This could represent that tetrathionate hydrolases hydrolyzes K
2S
4O
6. In order to ensure the sensitivity and practicality of the measurement, the change rate of absorbance at 290 nm was selected to define the catalytic efficiency of tetrathionate hydrolases [
29]. One unit enzyme activity was defined as the ΔA
290 min
-1.
Pyrophosphatase activity was detected with the method of Richter and Schäfer to assess the purity of each fraction [15, 39]. 1μg protein from each fraction was incubated with reaction buffer containing 40 mM acetate, 40 mM imidazole, 40 mM Tris, 2.5 mM MgC12 and 0.2 mM Ppi (Na4 P2 O7) for 30 mins at 60 °C. The concentration of Ppi was detected by Pyrophosphate Assay Kit (MAK168, Sigma-Aldrich, USA). A unit Pyrophosphatase activity was defined as the amount of protein required for hydrolysis of 1 µmol/L of Ppi in one min. The protein concentration of different fraction of cells was detected by BCA assay.
4.6.2. Discontinuous Method of Enzyme Activity Assay
For understanding the products from tetrathionate hydrolyzation, we used HPLC assays method described by Miura & Kawaoi [
29] to detect the sulfur-containing intermediates and quantify the activity of purified tetrathionate hydrolases. The K
2S
4O
6 used in the study was purchased from the Merck KGaA (Germany) and its purity was over 98%. Na
2S
2O
3 was purchased from Sinopharm (China), its purity was over 99%. 1)The determination of optimal temperature of TTH was proceeded in 1mL 50 mmol/L formate buffer (pH 3.0) containing 1-2 mmol/L K
2S
4O
6 and 1 mol/L (NH
4)
2SO
4 (Sigma-Aldrich, USA). The reaction was started by adding 25 μL protein and hatched in water bath from 25 °C to 95 °C with the interval of 10 °C. 2) The determination of optimal pH of TTH carried in the same solution system of temperature test and set pH at 1-7 with the interval of 1.0, hatched at 65 °C. 3) Vmax and Km were detected and calculated at 1.15 µg TTH with different tetrathionate concentration. To stop the enzyme reaction, the sample was rapidly frozen in liquid nitrogen for 3 min and then boiled in boiling water for 5 min, repeated 3 times. The mixture was filtered by 0.22 μm filter of polyethersulfone (SLGP033R, Millipore, USA). The contents of chemical compounds were determined using an Agilent 1290 infinity II (Agilent, Germany) consisting of a binary pump system and auto-sampler. A chromatography separation was achieved with an Agilent Eclipse Plus C18 column (4.6 mm×150 mm, 5 µm) and maintained at 30 °C during the run. The mobile phase was acetonitrile (Thermo Fisher, USA)/water (20:80 V/V) with 6 mmol/L Tetrapropylammonium hydroxide (TPA, Sigma-Aldrich, USA) adjusted pH as 5.0 by acetic acid. The flow rate was 0.8 mL·min
-1 and the UV-detector was set at 230 nm. A unit enzyme activity of tetrathionate hydrolase was defined as the amount of protein required for hydrolysis of 1 µmol/L of S
4O
62- in one min. The specific enzyme activity was the activity of per mg TTH.
4.7. Effects of Bivalent Ions on Enzyme Activity
The series of bivalent metal ions almost were metal chlorides, including magnesium chloride, zinc chloride, ferric chloride, nickel chloride, manganese chloride, and calcium chloride. The effects of metal ions on TTHMc activity were detected by discontinuous methods of enzyme activity assay with 0.01 mol/L metal ion at 95°C and pH 6.0.
4.8. Statistics
Statistics were performed using Prism 8 (Graphpad). All quantification tests were independently performed for at least three replications. Data were represented as the mean. Significance was determined using one-way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 throughout the manuscript.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Figure S1: AlphaFold2-predicted structure of tetrathionate hydrolase (TTH) from
Metallosphaera cuprina Ar-4 (A), multiple sequence alignments of TTH
Mc and closely related PDB entries (B); Figure S2: Silver-stained SDS gels of TTH
Mc by different purification steps; Figure S3: Specific activity for TTH of
M. cuprina Ar-4 during the purified process; Figure S4: Effect of bivalent metal ion on the activity of the TTH from
M. cuprina Ar-4; Figure S5: The reaction process of tetrathionate catalyzed by various concentrations of TTH
Mc; Figure S6: TTH clusters in
Metallophaera spp.; Figure S7: Phylogenetic relationship of TTHs among acidophilic archaea and bacteria; Figure S8: Genome context comparison of TTHs from
Sulfolobaceae strains; Figure S9: Sequence similarity network (SSN) built with TTH
Mc as the query; Table S1: The TTH total activity and percentages in differential cell fractions of whole cell.
Author Contributions
CY. J and SJ. L conceived the study. P. W and LJ. L prepared the TTH and quantified the TTH activity. LZ. L, HQ. Y, DF. L performed evolutionary insights and structure analysis of TTH functionality. P. W, XT. L and YL. Q contributed to cell collection. All authors contributed to data analysis and manuscript writing.
Funding
This work was funded by the Major Research Plan of National Natural Science Foundation of China (grant 92251307, 91851206), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0830000) and National Nature Science Foundation of China (31670124).
Data availability Statement
All data in this study are available within the article, supporting information, and/or from the corresponding author on reasonable request.
Acknowledgments
We thank the Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences, for their reliable technical support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shi, J. C., Huang, T. L., Wen, G., Liu, F., Qiu, X. P., andWang, B. S. (2016) The Variation Characteristic of Sulfides and VOSc in a Source Water Reservoir and Its Control Using a Water-Lifting Aerator, Int J Environ Res Public Health 13, 427. [CrossRef]
- Vavourakis, C. D., Mehrshad, M., Balkema, C., van Hall, R., Andrei, A. S., Ghai, R. et al. (2019) Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake, BMC Biol 17, 69. [CrossRef]
- Urbieta, M. S., Rascovan, N., Vazquez, M. P., andDonati, E. (2017) Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities, BMC Genomics 18, 445. [CrossRef]
- Zhang, X., Liu, Z., Wei, G., Yang, F., andLiu, X. (2018) In Silico Genome-Wide Analysis Reveals the Potential Links Between Core Genome of Acidithiobacillus thiooxidans and Its Autotrophic Lifestyle, Front Microbiol 9, 1255. [CrossRef]
- Parey, K., Demmer, U., Warkentin, E., Wynen, A., Ermler, U., andDahl, C. (2013) Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum, PLoS One 8, e74707. [CrossRef]
- Liu, L. J., You, X. Y., Guo, X., Liu, S. J., andJiang, C. Y. (2011) Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon, Int J Syst Evol Microbiol 61, 2395-2400. [CrossRef]
- Liu, L. J., Stockdreher, Y., Koch, T., Sun, S. T., Fan, Z., Josten, M. et al. (2014) Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina, J Biol Chem 289, 26949-26959. [CrossRef]
- Peng, T. J., Liu, L. J., Liu, C., Yang, Z. F., Liu, S. J., andJiang, C. Y. (2015) Metallosphaera tengchongensis sp. nov., an acidothermophilic archaeon isolated from a hot spring. Int J Syst Evol Microbiol 65, 537-542. [CrossRef]
- Johnson, D. B. (2014) Biomining - biotechnologies for extracting and recovering metals from ores and waste materials, Current Opinion in Biotechnology 30, 24-31. [CrossRef]
- Batty, J. D., and Rorke, G. V. (2006) Development and commercial demonstration of the BioCOP™ thermophile process, Hydrometallurgy 83, 83-89. [CrossRef]
- Liu, L. J., Jiang, Z., Wang, P., Qin, Y. L., Xu, W., Wang, Y. et al. (2021) Physiology, Taxonomy, and Sulfur Metabolism of the Sulfolobales, an Order of Thermoacidophilic Archaea, Front Microbiol 12, 768283. [CrossRef]
- Auernik, K. S., andKelly, R. M. (2008) Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes, Appl Environ Microbiol 74, 7723-7732. [CrossRef]
- Segerer, A., Neuner, A., Kristjansson, J. K., andStetter, K. O. (1986) Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov.: Facultatively Aerobic, Extremely Acidophilic Thermophilic Sulfur-Metabolizing Archaebacteria,Int J Syst Evol Microbiol 36, 559-564. [CrossRef]
- Brock, T. D., Brock, K. M., Belly, R. T., andWeiss, R. L. (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature, Arch Mikrobiol 84, 54-68. [CrossRef]
- Protze, J., Müller, F., Lauber, K., Naß, B., Mentele, R., Lottspeich, F. et al. (2011) An Extracellular Tetrathionate Hydrolase from the Thermoacidophilic Archaeon Acidianus ambivalens with an Activity Optimum at pH 1, Front Microbiol 2, 68 . [CrossRef]
- Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D. S., andBonnefoy, V. (2009) Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans, BMC Genomics 10, 394 . [CrossRef]
- Rohwerder, T., andSand, W. (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp, Microbiology (Reading) 149, 1699-1710 . [CrossRef]
- Nguyen, P. M., Do, P. T., Pham, Y. B., Doan, T. O., Nguyen, X. C., Lee, W. K. et al. (2022) Roles, mechanism of action, and potential applications of sulfur-oxidizing bacteria for environmental bioremediation, Sci Total Environ 852, 158203 . [CrossRef]
- Kanao, T., Hase, N., Nakayama, H., Yoshida, K., Nishiura, K., Kosaka, M. et al. (2021) Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans, Protein Science 30, 328-338 . [CrossRef]
- Chen, L., Ren, Y., Lin, J., Liu, X., Pang, X., andLin, J. (2012) Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant, PLoS One 7, e39470 . [CrossRef]
- Gumulya, Y., Boxall, N. J., Khaleque, H. N., Santala, V., Carlson, R. P., and Kaksonen, A. H. (2018) In a quest for engineering acidophiles for biomining applications: challenges and opportunities, Genes (Basel) 9, . [CrossRef]
- Bugaytsova, Z., and Lindström, E. B. (2004) Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus, Eur J Biochem 271, 272-280 . [CrossRef]
- Lu, W. P., and Kelly, D. (1988) Cellular Location and Partial Purification of the 'Thiosulphate-oxidizing Enzyme' and 'Trithionate Hydrolyase' from Thiobacillus tepidarius, J Gen Microbiol 134, 877-885 . [CrossRef]
- Krupovic, M., Peixeiro, N., Bettstetter, M., Rachel, R., and Prangishvili, David. (2012) Archaeal Tetrathionate Hydrolase Goes Viral: Secretion of a Sulfur Metabolism Enzyme in the Form of Virus-Like Particles, Appl Environ Microbiol 78, 5463 - 5465 . [CrossRef]
- You, X.-Y., Liu, C., Wang, S.-Y., Jiang, C.-Y., Shah, S. A., Prangishvili, D. et al. (2011) Genomic analysis of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles, Extremophiles 15, 487-497 . [CrossRef]
- Kanao, T., Onishi, M., Kajitani, Y., Hashimoto, Y., Toge, T., Kikukawa, H. et al. (2018) Characterization of tetrathionate hydrolase from the marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH, Biosci Biotechnol Biochem 82, 152-160 . [CrossRef]
- Kanao, T., Kamimura, K., andSugio, T. (2007) Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans, J Biotechnol 132, 16-22 . [CrossRef]
- Rzhepishevska, O. I., Valdés, J., Marcinkeviciene, L., Gallardo, C. A., Meskys, R., Bonnefoy, V., Holmes, D. S. et al. (2007). Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Applied and environmental microbiology, 73(22), 7367–7372. [CrossRef]
- Govardus A. H. de Jong, W. H., Piet Bos and J. Gijs Kuenen (1997) Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans, Microbiology 143, 499-504 . [CrossRef]
- Miura, Y., and Kawaoi, A. (2000) Determination of thiosulfate, thiocyanate and polythionates in a mixture by ion-pair chromatography with ultraviolet absorbance detection, J Chromatogr A 884, 81-87 . [CrossRef]
- Kanao, T., Matsumoto, C., Shiraga, K., Yoshida, K., Takada, J., andKamimura, K. (2010) Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding, FEMS Microbiol Lett 309, 43-47 . [CrossRef]
- Wang, Z. B., Li, Y. Q., Lin, J. Q., Pang, X., Liu, X. M., Liu, B. Q. et al. (2016) The Two-Component System RsrS-RsrR Regulates the Tetrathionate Intermediate Pathway for Thiosulfate Oxidation in Acidithiobacillus caldus, Front Microbiol 7, 1755 . [CrossRef]
- Wang, R., Lin, J. Q., Liu, X. M., Pang, X., Zhang, C. J., Yang, C. L. et al. (2018) Sulfur Oxidation in the Acidophilic Autotrophic Acidithiobacillus spp, Front Microbiol 9, 3290 . [CrossRef]
- Sugio, T., Taha, T. M., andTakeuchi, F. (2009) Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells, Biosci Biotechnol Biochem 73, 1381-1386 . [CrossRef]
- Cao, X., Hu, X., Zhang, X., Gao, S., Ding, C., Feng, Y. et al. (2017) Identification of metal ion binding sites based on amino acid sequences, PLoS One 12, e0183756 . [CrossRef]
- Atkinson, H. J., Morris, J. H., Ferrin, T. E., andBabbitt, P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies, PLoS One 4, e4345 . [CrossRef]
- Senior, A. W., Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., Green, T. et al. (2020) Improved protein structure prediction using potentials from deep learning, Nature 577, 706-710 . [CrossRef]
- Kalckar, H. M. (1971) The periplasmic galactose binding protein of Escherichia coli, Science 174, 557-565 . [CrossRef]
- Richter, O. M., and Schäfer, G. (1992). Purification and enzymic characterization of the cytoplasmic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Eur. J. Biochem. 209, 343–349.
Figure 1.
The transcriptional expression of mcup_1281 in different cultures. YE, S and K2S4O6 represent that the M. cuprina Ar-4 was cultivated with yeast extraction (YE), sulfur powder (S0) or K2S4O6 as the energy source, respectively. 16S rRNA as the reference gene and mcup_1281 as target gene. Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Figure 1.
The transcriptional expression of mcup_1281 in different cultures. YE, S and K2S4O6 represent that the M. cuprina Ar-4 was cultivated with yeast extraction (YE), sulfur powder (S0) or K2S4O6 as the energy source, respectively. 16S rRNA as the reference gene and mcup_1281 as target gene. Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Figure 2.
Activity of TTHs for cell lysis solution in different cultures analyzed by continuous method. YE, S and K2S4O6 represent that the M. cuprina Ar-4 was cultivated with yeast extraction (YE), sulfur powder (S0) or K2S4O6 as the energy source, respectively. Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Figure 2.
Activity of TTHs for cell lysis solution in different cultures analyzed by continuous method. YE, S and K2S4O6 represent that the M. cuprina Ar-4 was cultivated with yeast extraction (YE), sulfur powder (S0) or K2S4O6 as the energy source, respectively. Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Figure 3.
The TTH specific activity (A) and relative activity (B) in differential cell fractions of whole cell. Per, Periplasmic space; Cyt, cytoplasm; M -3, Membrane (pH 3.0); M -7, Membrane (pH 7.0). Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. TTH activity was measured by HPLC.
Figure 3.
The TTH specific activity (A) and relative activity (B) in differential cell fractions of whole cell. Per, Periplasmic space; Cyt, cytoplasm; M -3, Membrane (pH 3.0); M -7, Membrane (pH 7.0). Error bars represent SD of n=3 independent measurements. Significance was determined using one –way ANOVA analysis, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. TTH activity was measured by HPLC.
Figure 4.
Temperature and pH profiles of TTHMc activity. A. Temperature profile of TTH activity from 25 to 95 ℃; B. pH profile of TTH activity from 1 to 7. Error bars represent SD of n=3 independent measurements. TTH activity was measured by HPLC.
Figure 4.
Temperature and pH profiles of TTHMc activity. A. Temperature profile of TTH activity from 25 to 95 ℃; B. pH profile of TTH activity from 1 to 7. Error bars represent SD of n=3 independent measurements. TTH activity was measured by HPLC.
Figure 5.
Relationship between TTHMc enzymatic reaction rate and K2S4O6 concentration. Error bars represent SD of n=3 independent measurements. TTH activity was measured by HPLC.
Figure 5.
Relationship between TTHMc enzymatic reaction rate and K2S4O6 concentration. Error bars represent SD of n=3 independent measurements. TTH activity was measured by HPLC.
Figure 6.
TTHMc products from M. cuprina Ar-4 analyzed by HPLC method. Products were analyzed following the enzyme addition (Dotted line) and after 100 min incubation (Solid line).
Figure 6.
TTHMc products from M. cuprina Ar-4 analyzed by HPLC method. Products were analyzed following the enzyme addition (Dotted line) and after 100 min incubation (Solid line).
Figure 7.
Sequence similarity network (SSN) built with TTHMc as the query. (A) SSN built with TTHMc as the query colored by taxonomy; (B) SSN built with TTHMc as the query colored by the type of the residues on the position of TTHMc Asp353; (C) Schematic of detailed ligand atom interactions with the protein residues. SSN built with TTHMc as the query colored by the type of the residues on the position of TTHMc (D) R460; (E) R497.
Figure 7.
Sequence similarity network (SSN) built with TTHMc as the query. (A) SSN built with TTHMc as the query colored by taxonomy; (B) SSN built with TTHMc as the query colored by the type of the residues on the position of TTHMc Asp353; (C) Schematic of detailed ligand atom interactions with the protein residues. SSN built with TTHMc as the query colored by the type of the residues on the position of TTHMc (D) R460; (E) R497.
Table 1.
Purification of the TTH from M. cuprina Ar-4. TTH activity was measured by HPLC.
Table 1.
Purification of the TTH from M. cuprina Ar-4. TTH activity was measured by HPLC.
| Fraction |
Total protein (mg)a
|
Total activity (U)b
|
Specific activity (U/mg)c
|
Yield
(%) |
Fold |
| Cell extract |
79.000 ± 5.132 |
56.750 ± 1.303 |
0.454 ± 0.010 |
100.00 |
1.00 |
| DEAE |
35.880 ± 1.322 |
50.232 ± 6.331 |
1.400 ± 0.100 |
88.50 |
3.08 |
| HIC |
1.203 ± 0.043 |
2.634 ± 0.167 |
2.190 ± 0.095 |
4.63 |
4.82 |
| Size exclusion |
0.325 ± 0.007 |
1.221 ± 0.156 |
4.070 ± 0.096 |
2.15 |
8.96 |
| Q-sepharose |
0.058 ± 0.001 |
0.704 ± 0.043 |
12.140 ± 1.02 |
1.24 |
26.7 |
Table 2.
Effects of bivalent ions on enzyme activity. TTH activity was measured by HPLC.
Table 2.
Effects of bivalent ions on enzyme activity. TTH activity was measured by HPLC.
| Metal ions |
Concentration (mol/L) |
Specific activity(U/mg) |
Residual activity(%) |
| Control |
- |
6.57 ± 0.12 |
- |
| Mg2+
|
0.01 |
7.95 ± 0.04 |
121.01 |
| Zn2+
|
0.01 |
6.55 ± 0.12 |
94.79 |
| Fe2+
|
0.01 |
6.23 ± 0.09 |
94.47 |
| Ni2+
|
0.01 |
6.10 ± 0.00 |
92.77 |
| Mn2+
|
0.01 |
6.23 ± 0.09 |
87.72 |
| Ca2+
|
0.01 |
3.93 ± 0.05 |
59.81 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).