Submitted:
30 December 2024
Posted:
31 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
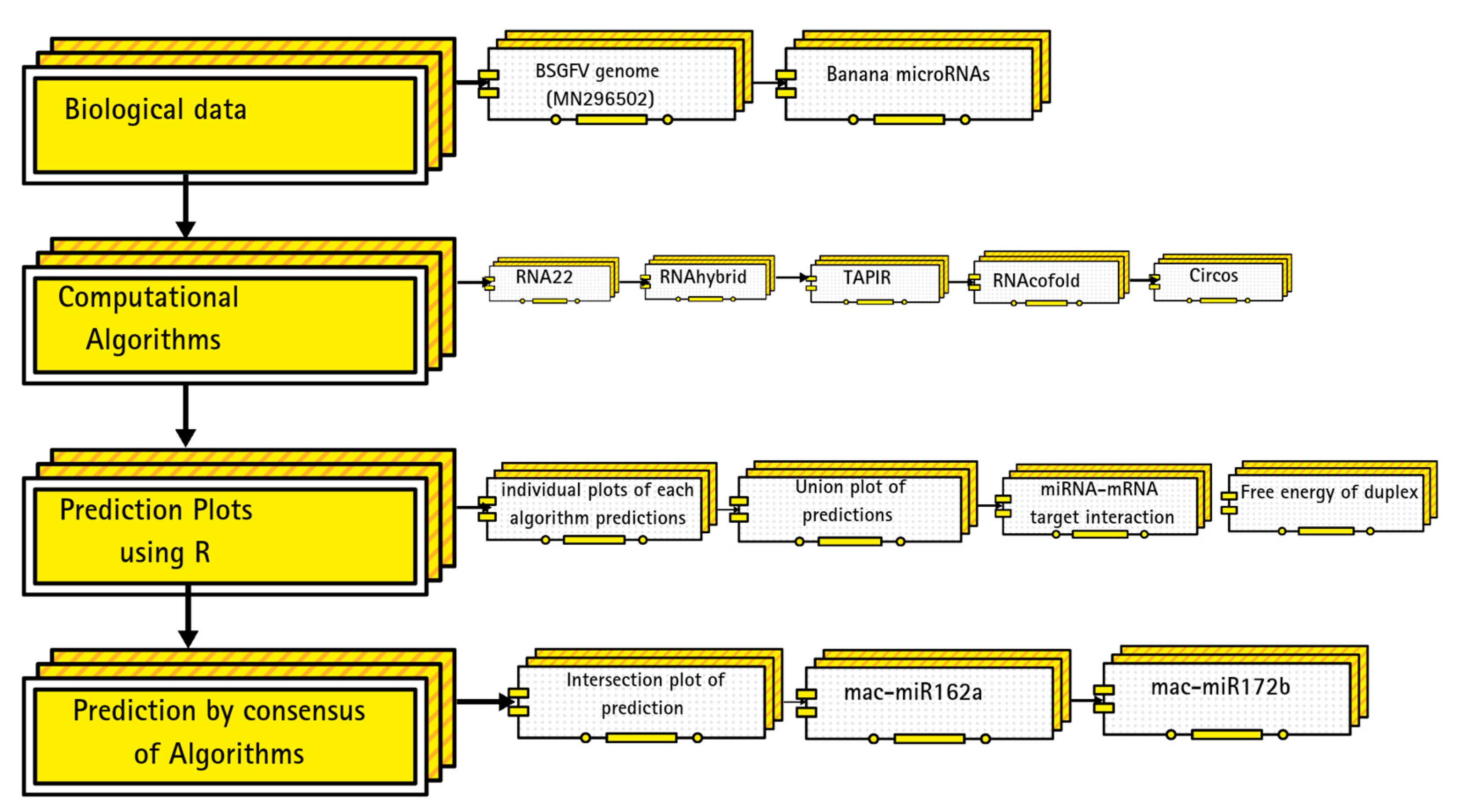
2.1. Biologicl Data
2.2. Target Prediction in BSGFV Genome
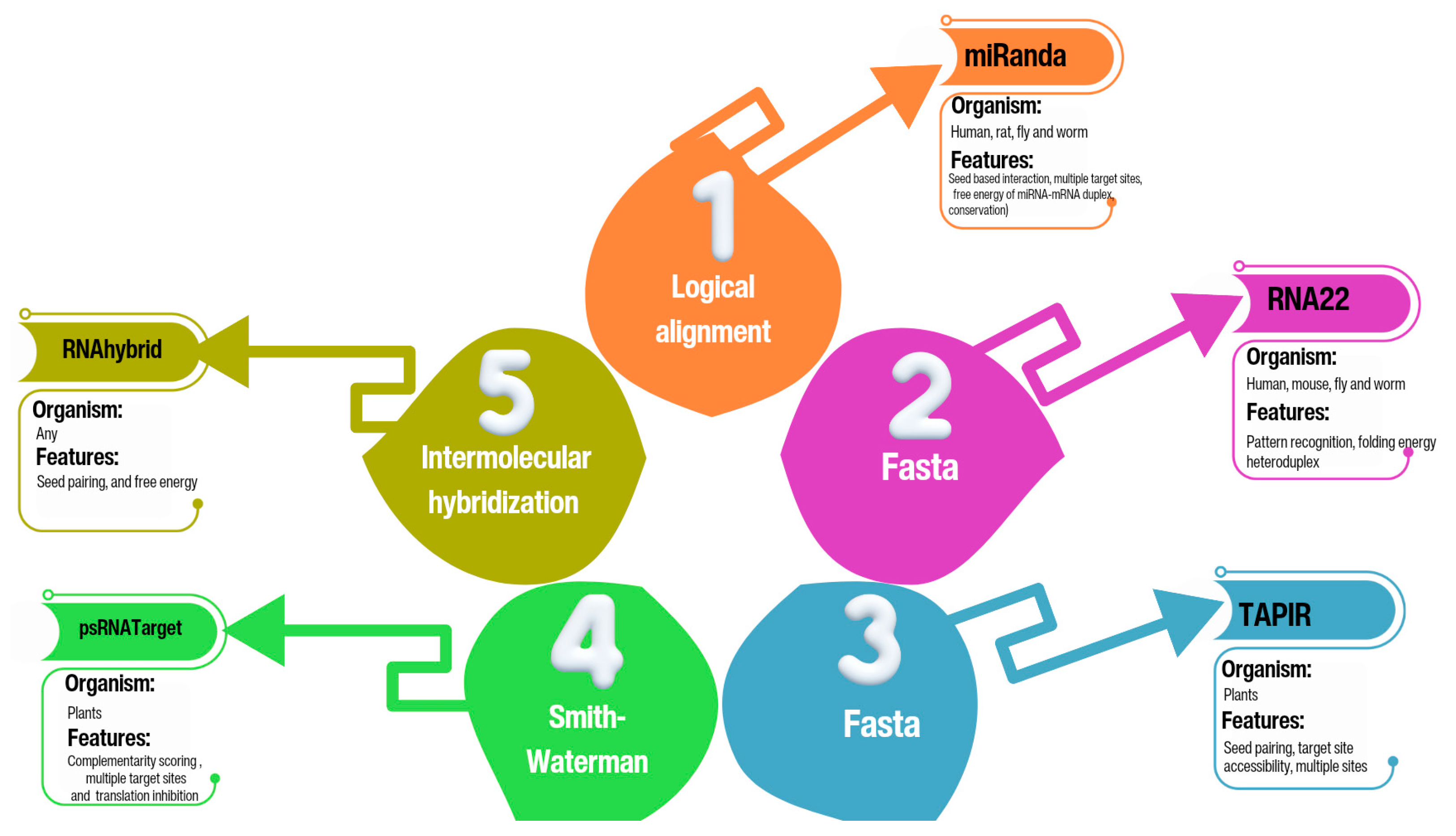
2.3. RNA22 Algorithm
2.4. RNAhybrid Algorithm
2.5. TAPIR Algorithm
2.6. RNAcofold Algorithm
2.7. Discovering Banana Genome-Encoded miRNAs-Target Interaction
2.8. Graphical Representation
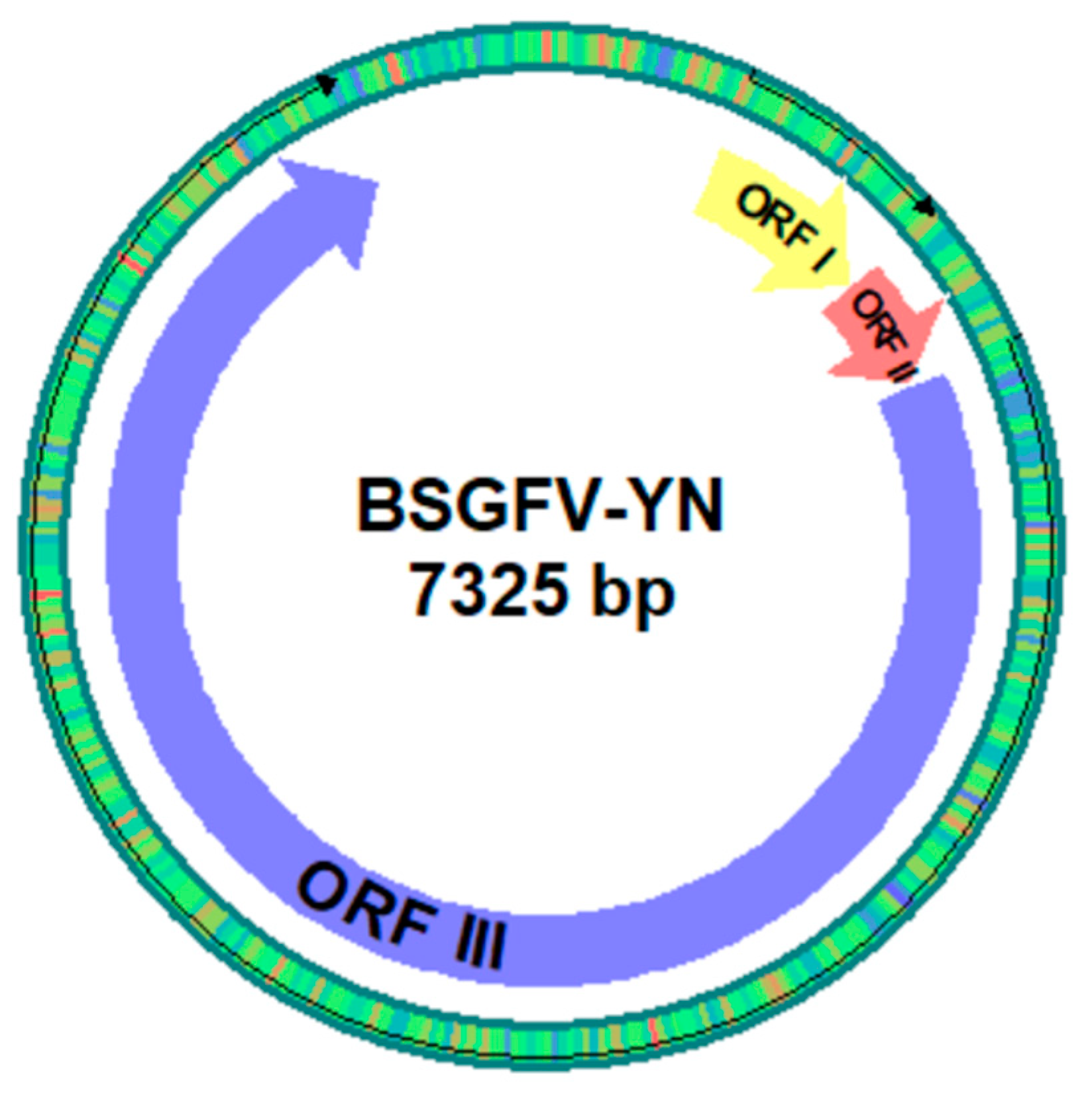
2.9. BSGFV Genome Analysis
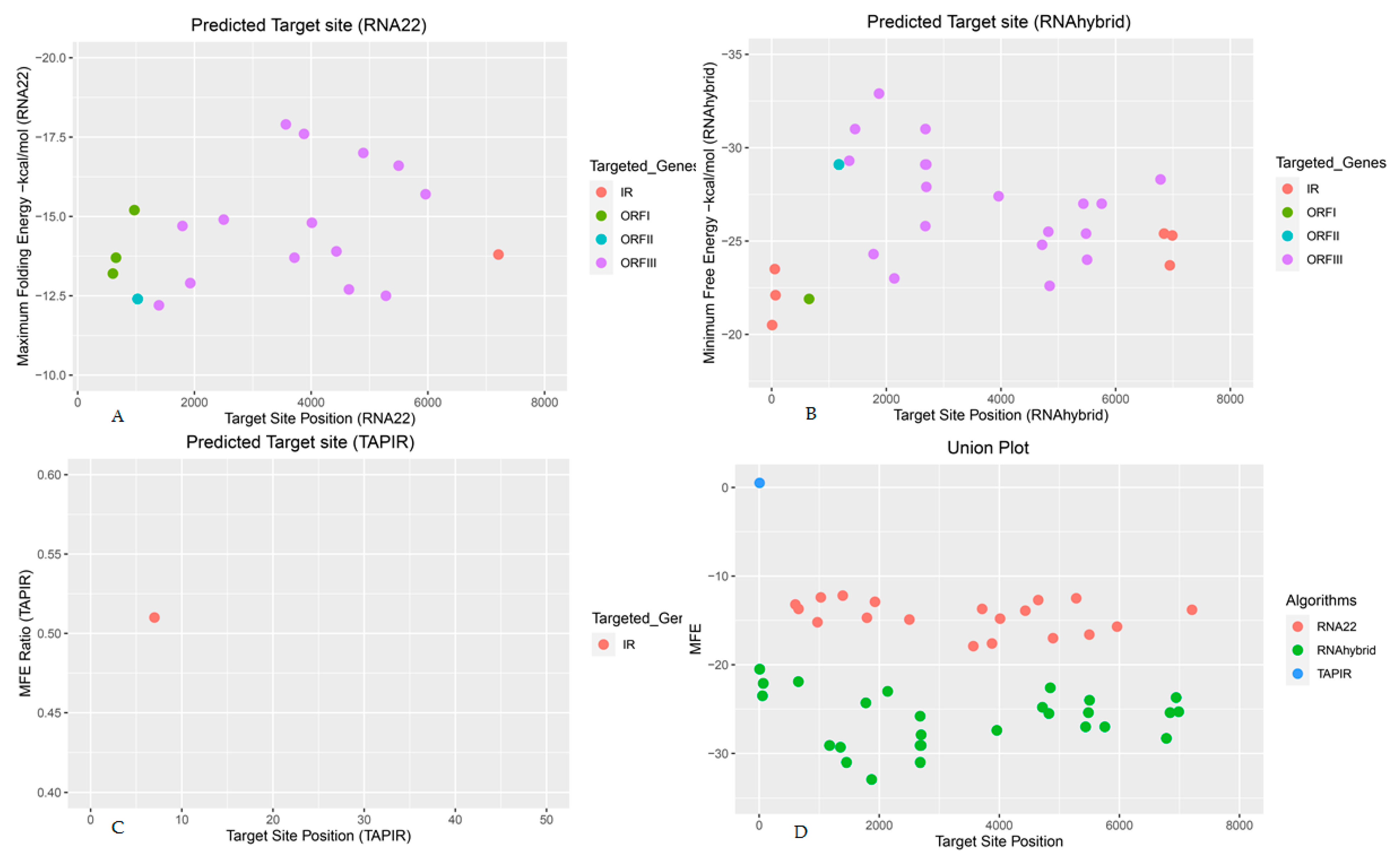
3. Results
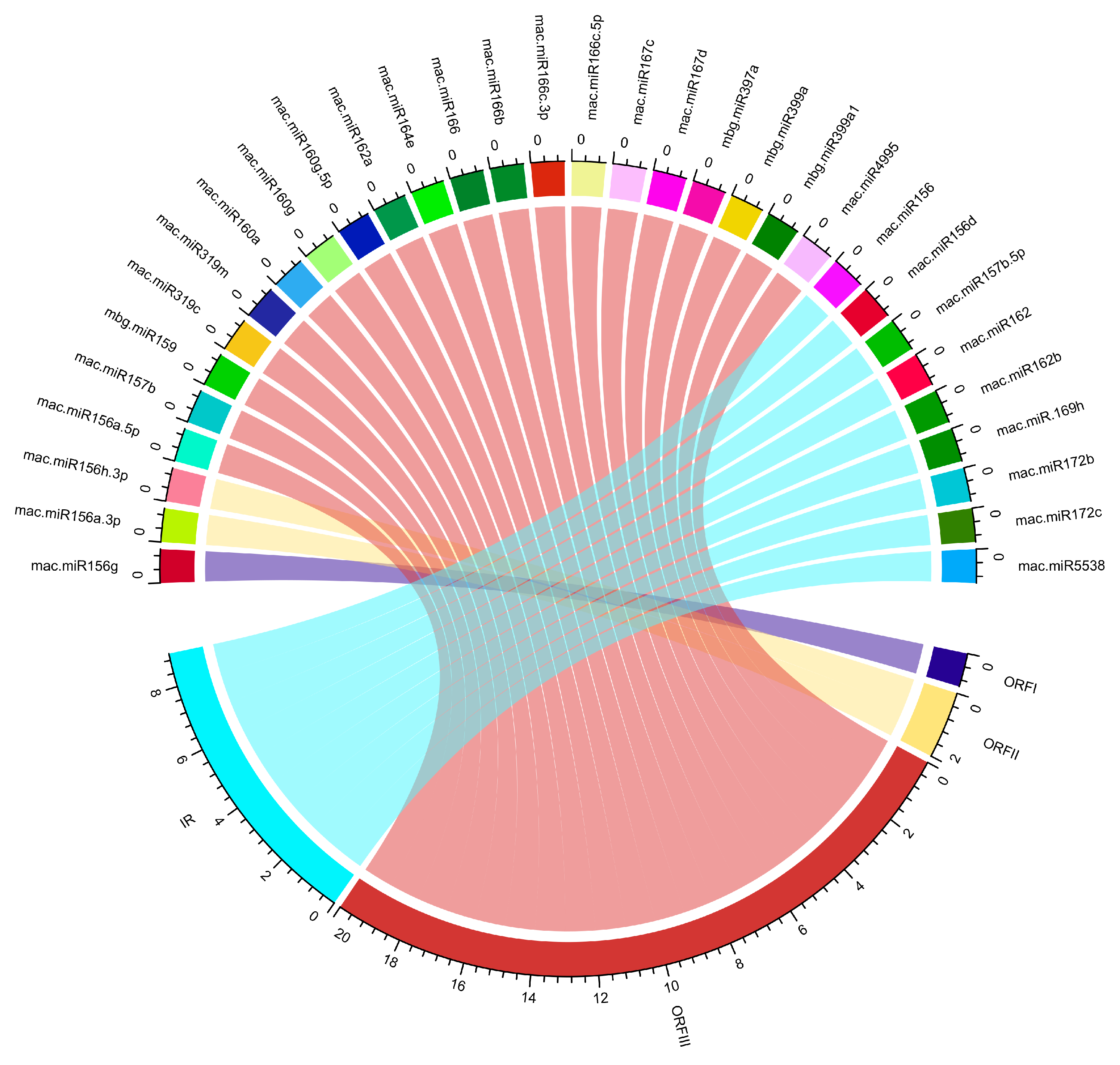
3.1. Banana miRNA’s Loci n BSGFV Genome
3.2. Viral ORF1-Encoding Hypothetical Protein
3.3. Viral ORF11-Encoding DNA Binding Protein
3.4. Viral ORFIII-Encoding Polyprotein
3.5. Banana miRNAs Targetig Intergenic Region of BSGFV genome
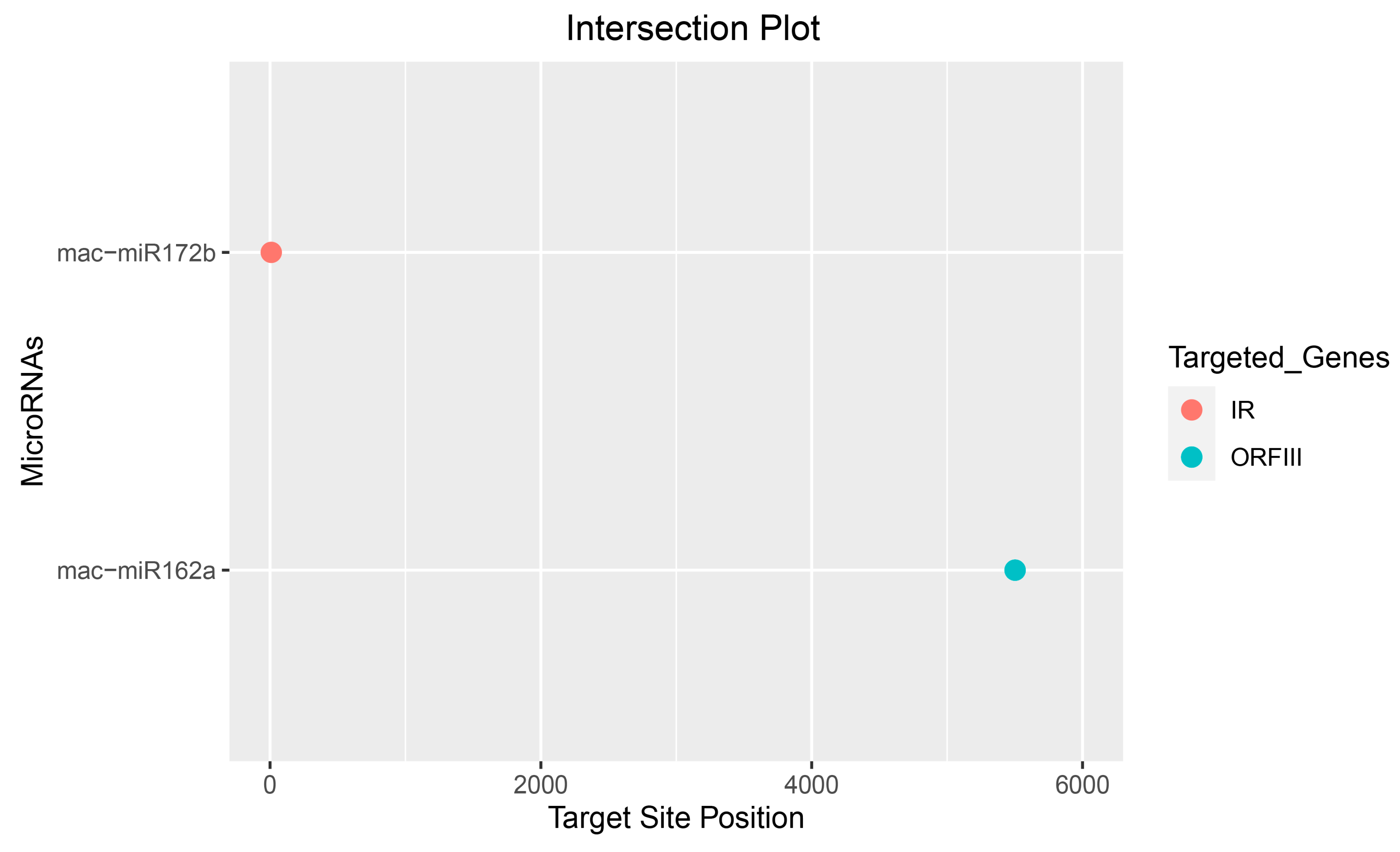
3.6. Identification of Unique Banana-Encoded miRNAs
3.7. Predicting Consensual Banana miRNAs and Silencing BSGFV Genome
3.8. Association of Banana miRNA-Target Interaction
3.9. Evaluation of Free Energy (ΔG)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.-P.; Jenny, C. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of the National Academy of Sciences 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-R.; Liu, X.; Arshad, R.; Wang, X.; Li, W.-M.; Zhou, Y.; Ge, X.-J. Telomere-to-telomere haplotype-resolved reference genome reveals subgenome divergence and disease resistance in triploid Cavendish banana. Horticulture Research 2023, 10, uhad153. [Google Scholar] [CrossRef]
- Teycheney, P.-Y.; Geering, A.D.; Dasgupta, I.; Hull, R.; Kreuze, J.F.; Lockhart, B.; Muller, E.; Olszewski, N.; Pappu, H.; Pooggin, M.M. ICTV virus taxonomy profile: Caulimoviridae. Journal of General Virology 2020, 101, 1025–1026. [Google Scholar] [CrossRef]
- Meyer, J.; Kasdorf, G.; Nel, L.; Pietersen, G. Transmission of activated-episomal banana streak OL (badna) virus (BSOLV) to cv. Williams banana (Musa sp.) by three mealybug species. Plant Disease 2008, 92, 1158–1163. [Google Scholar] [CrossRef]
- Li, W.-l.; Yu, N.-t.; Wang, J.-h.; Li, J.-c.; Liu, Z.-x. The complete genome of Banana streak GF virus Yunnan isolate infecting Cavendish Musa AAA group in China. PeerJ 2020, 8, e8459. [Google Scholar] [CrossRef]
- Rao, X.; Chen, H.; Lu, Y.; Liu, R.; Li, H. Distribution and location of BEVs in different genotypes of bananas reveal the coevolution of BSVs and bananas. International Journal of Molecular Sciences 2023, 24, 17064. [Google Scholar] [CrossRef]
- Rao, X.-Q.; Wu, Z.-L.; Wang, W.; Zhou, L.; Sun, J.; Li, H.-P. Genetic diversity analysis reveals new badnaviruses infecting banana in South China. Journal of Plant Pathology 2020, 102, 1065–1075. [Google Scholar] [CrossRef]
- Medberry, S.L.; Lockhart, B.; Olszewski, N.E. Properties of Commelina yellow mottle virus's complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic acids research 1990, 18, 5505–5513. [Google Scholar] [CrossRef]
- Bouhida, M.; Lockhart, B.; Olszewski, N.E. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. Journal of General Virology 1993, 74, 15–22. [Google Scholar] [CrossRef]
- Hagen, L.S.; Jacquemond, M.; Lepingle, A.; Lot, H.; Tepfer, M. Nucleotide sequence and genomic organization of cacao swollen shoot virus. Virology 1993, 196, 619–628. [Google Scholar] [CrossRef]
- Ishwara Bhat, A.; Selvarajan, R.; Balasubramanian, V. Emerging and re-emerging diseases caused by Badnaviruses. Pathogens 2023, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The current global scenario. Viruses 2016, 8, 177. [Google Scholar] [CrossRef]
- Koeppe, S.; Kawchuk, L.; Kalischuk, M. RNA interference past and future applications in plants. International Journal of Molecular Sciences 2023, 24, 9755. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.-Q. RNA interference: Promising approach to combat plant viruses. International Journal of Molecular Sciences 2022, 23, 5312. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zheng, B.; Yu, Y.; Won, S.Y.; Mo, B.; Chen, X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. The EMBO journal 2011, 30, 814–822. [Google Scholar] [CrossRef]
- Fang, X.; Cui, Y.; Li, Y.; Qi, Y. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nature Plants 2015, 1, 1–9. [Google Scholar] [CrossRef]
- Manavella, P.A.; Koenig, D.; Weigel, D. Plant secondary siRNA production determined by microRNA-duplex structure. Proceedings of the National Academy of Sciences 2012, 109, 2461–2466. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes & development 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Liu, W.-w.; Meng, J.; Cui, J.; Luan, Y.-s. Characterization and function of microRNA∗ s in plants. Frontiers in plant science 2017, 8, 2200. [Google Scholar] [CrossRef]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. Journal of cellular physiology 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annual review of microbiology 2010, 64, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Zhang, X.; Ji, H.; Yasir, M.; Farooq, T.; Dai, X.; Li, F. Large Artificial microRNA Cluster Genes Confer Effective Resistance against Multiple Tomato Yellow Leaf Curl Viruses in Transgenic Tomato. Plants 2023, 12, 2179. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, J.; Li, C. Research Progress on miRNAs and Artificial miRNAs in Insect and Disease Resistance and Breeding in Plants. Genes 2024, 15, 1200. [Google Scholar] [CrossRef]
- Al-Roshdi, M.R.; Ammara, U.; Khan, J.; Al-Sadi, A.M.; Shahid, M.S. Artificial microRNA-mediated resistance against Oman strain of tomato yellow leaf curl virus. Frontiers in Plant Science 2023, 14, 1150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yuan, Q.; Ai, X.; Chen, J.; Lu, Y.; Yan, F. Transgenic Rice Plants Expressing Artificial miRNA Targeting the Rice Stripe Virus MP Gene Are Highly Resistant to the Virus. Biology 2022, 11, 332. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, N.; Shen, W.; Li, J.-F. Engineered artificial microRNA precursors facilitate cloning and gene silencing in arabidopsis and rice. International Journal of Molecular Sciences 2019, 20, 5620. [Google Scholar] [CrossRef]
- Bi, F.; Meng, X.; Ma, C.; Yi, G. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC genomics 2015, 16, 1–15. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, Y.; Tang, R.; Qu, H.; Duan, X.; Jiang, Y. Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress. BMC genomics 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Xia, Y.; Lai, Z.; Do, Y.-Y.; Huang, P.-L. Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas. Plants 2023, 12, 3414. [Google Scholar] [CrossRef]
- Chai, J.; Feng, R.; Shi, H.; Ren, M.; Zhang, Y.; Wang, J. Bioinformatic identification and expression analysis of banana microRNAs and their targets. PLoS ONE 2015, 10, e0123083. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S. Database resources of the national center for biotechnology information. Nucleic acids research 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Loher, P.; Rigoutsos, I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 2012, 28, 3322–3323. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic acids research 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome research 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Gandrud, C. Reproducible research with R and RStudio; Chapman and Hall/CRC: 2018.
- Cheng, C.-P.; Lockhart, B.; Olszewski, N.E. The ORF I and II proteins ofcommelinayellow mottle virus are virion-associated. Virology 1996, 223, 263–271. [Google Scholar] [CrossRef]
- Jaufeerally-Fakim, Y.; Khorugdharry, A.; Harper, G. Genetic variants of Banana streak virus in Mauritius. Virus Research 2006, 115, 91–98. [Google Scholar] [CrossRef]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef]
- Tripathi, J.; Ntui, V.; Ron, M.; Muiruri, S.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun Biol 2019, 2, 46. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Brown, J.K.; Iqbal, M.S.; Yu, N. Genome-Wide Identification of Cotton MicroRNAs Predicted for Targeting Cotton Leaf Curl Kokhran Virus-Lucknow. Microbiology Research 2023, 15, 1–19. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Ali, B.; Brown, J.K.; Shahid, I.; Yu, N. In silico identification of cassava genome-encoded MicroRNAs with predicted potential for targeting the ICMV-Kerala begomoviral pathogen of cassava. Viruses 2023, 15, 486. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Murtaza, N.; Brown, J.K.; Yu, N. In silico apple genome-encoded microRNA target binding sites targeting apple chlorotic leaf spot virus. Horticulturae 2023, 9, 808. [Google Scholar] [CrossRef]
- Chipman, L.B.; Pasquinelli, A.E. miRNA targeting: Growing beyond the seed. Trends in genetics 2019, 35, 215–222. [Google Scholar] [CrossRef]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for sequence-based miRNA target prediction: What to choose? International journal of molecular sciences 2016, 17, 1987. [Google Scholar] [CrossRef]
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common features of microRNA target prediction tools. Frontiers in genetics 2014, 5, 23. [Google Scholar] [CrossRef]
- Wenzhi, W.; Ashraf, M.A.; Ghaffar, H.; Ijaz, Z.; Zaman, W.u.; Mazhar, H.; Zulfqar, M.; Zhang, S. In Silico Identification of Sugarcane Genome-Encoded MicroRNAs Targeting Sugarcane Mosaic Virus. Microbiology Research 2024, 15, 273–289. [Google Scholar] [CrossRef]
- Mohamed, N.A.; Ngah, N.M.F.N.C.; Abas, A.; Talip, N.; Sarian, M.N.; Hamezah, H.S.; Harun, S.; Bunawan, H. Candidate miRNAs from Oryza sativa for Silencing the Rice Tungro Viruses. Agriculture 2023, 13, 651. [Google Scholar] [CrossRef]
- Nivedha, M.; Harish, S.; Angappan, K.; Karthikeyan, G.; Kumar, K.; Murugan, M.; Jayakanthan, M. In silico identification and validation of microRNAs from the genome of Solanum lycopersicum targeting Groundnut bud necrosis orthotospovirus. Physiological and Molecular Plant Pathology 2023, 127, 102086. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Tariq, H.K.; Hu, X.-W.; Khan, J.; Zou, Z. Computational biology and machine learning approaches identify rubber tree (Hevea brasiliensis Muell. Arg.) genome encoded MicroRNAs targeting rubber tree virus 1. Applied Sciences 2022, 12, 12908. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Ashraf, F.; Feng, X.; Hu, X.; Shen, L.; Khan, J.; Zhang, S. Potential targets for evaluation of sugarcane yellow leaf virus resistance in sugarcane cultivars: In silico sugarcane miRNA and target network prediction. Biotechnology & Biotechnological Equipment 2021, 35, 1980–1991. [Google Scholar] [CrossRef]
- Shahid, M.N.; Rashid, S.; Iqbal, M.S.; Jamal, A.; Khalid, S.; Shamim, Z. In silico prediction of potential mirnas to target zymv in cucumis melo. Pak. J. Bot 2022, 54, 1319–1325. [Google Scholar] [CrossRef]
- Gaafar, Y.Z.A.; Ziebell, H. Novel targets for engineering Physostegia chlorotic mottle and tomato brown rugose fruit virus-resistant tomatoes: In silico prediction of tomato microRNA targets. PeerJ 2020, 8, e10096. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Gao, Z.; Wang, F.; Xu, T.; Qi, M.; Liu, Y.; Li, T. MicroRNA162 regulates stomatal conductance in response to low night temperature stress via abscisic acid signaling pathway in tomato. Frontiers in Plant Science 2023, 14, 1045112. [Google Scholar] [CrossRef]
- Li, X.-P.; Ma, X.-C.; Wang, H.; Zhu, Y.; Liu, X.-X.; Li, T.-T.; Zheng, Y.-P.; Zhao, J.-Q.; Zhang, J.-W.; Huang, Y.-Y. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and yield. Rice 2020, 13, 1–13. [Google Scholar] [CrossRef]
- Shekhawat, U.K.; Ganapathi, T.R.; Hadapad, A.B. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. Journal of general virology 2012, 93, 1804–1813. [Google Scholar] [CrossRef]
- Ding, T.; Tomes, S.; Gleave, A.P.; Zhang, H.; Dare, A.P.; Plunkett, B.; Espley, R.V.; Luo, Z.; Zhang, R.; Allan, A.C. microRNA172 targets APETALA2 to regulate flavonoid biosynthesis in apple (Malus domestica). Horticulture Research 2022, 9, uhab007. [Google Scholar] [CrossRef]
- Liu, W.; Cheng, C.; Chen, F.; Ni, S.; Lin, Y.; Lai, Z. High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC plant biology 2018, 18, 1–26. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytologist 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Chung, M.-Y.; Nath, U.K.; Vrebalov, J.; Gapper, N.; Lee, J.M.; Lee, D.-J.; Kim, C.K.; Giovannoni, J. Ectopic expression of miRNA172 in tomato (Solanum lycopersicum) reveals novel function in fruit development through regulation of an AP2 transcription factor. BMC plant biology 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Kim, B.H.; Kwon, Y.; Lee, B.-h.; Nam, K.H. Overexpression of miR172 suppresses the brassinosteroid signaling defects of bak1 in Arabidopsis. Biochemical and Biophysical Research Communications 2014, 447, 479–484. [Google Scholar] [CrossRef]
- Tang, M.; Bai, X.; Niu, L.-J.; Chai, X.; Chen, M.-S.; Xu, Z.-F. miR172 regulates both vegetative and reproductive development in the perennial woody plant Jatropha curcas. Plant and Cell Physiology 2018, 59, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal 2008, 53, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wang, W.; Bai, X.; Qi, Y. Gene silencing by artificial microRNAs in Chlamydomonas. The Plant Journal 2009, 58, 157–164. [Google Scholar] [CrossRef]
- Ashraf, F.; Ashraf, M.A.; Hu, X.; Zhang, S. A novel computational approach to the silencing of Sugarcane Bacilliform Guadeloupe A Virus determines potential host-derived MicroRNAs in sugarcane (Saccharum officinarum L.). PeerJ 2020, 8, e8359. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Feng, X.; Hu, X.; Ashraf, F.; Shen, L.; Iqbal, M.S.; Zhang, S. In silico identification of sugarcane (Saccharum officinarum L.) genome encoded microRNAs targeting sugarcane bacilliform virus. PLoS ONE 2022, 17, e0261807. [Google Scholar] [CrossRef]
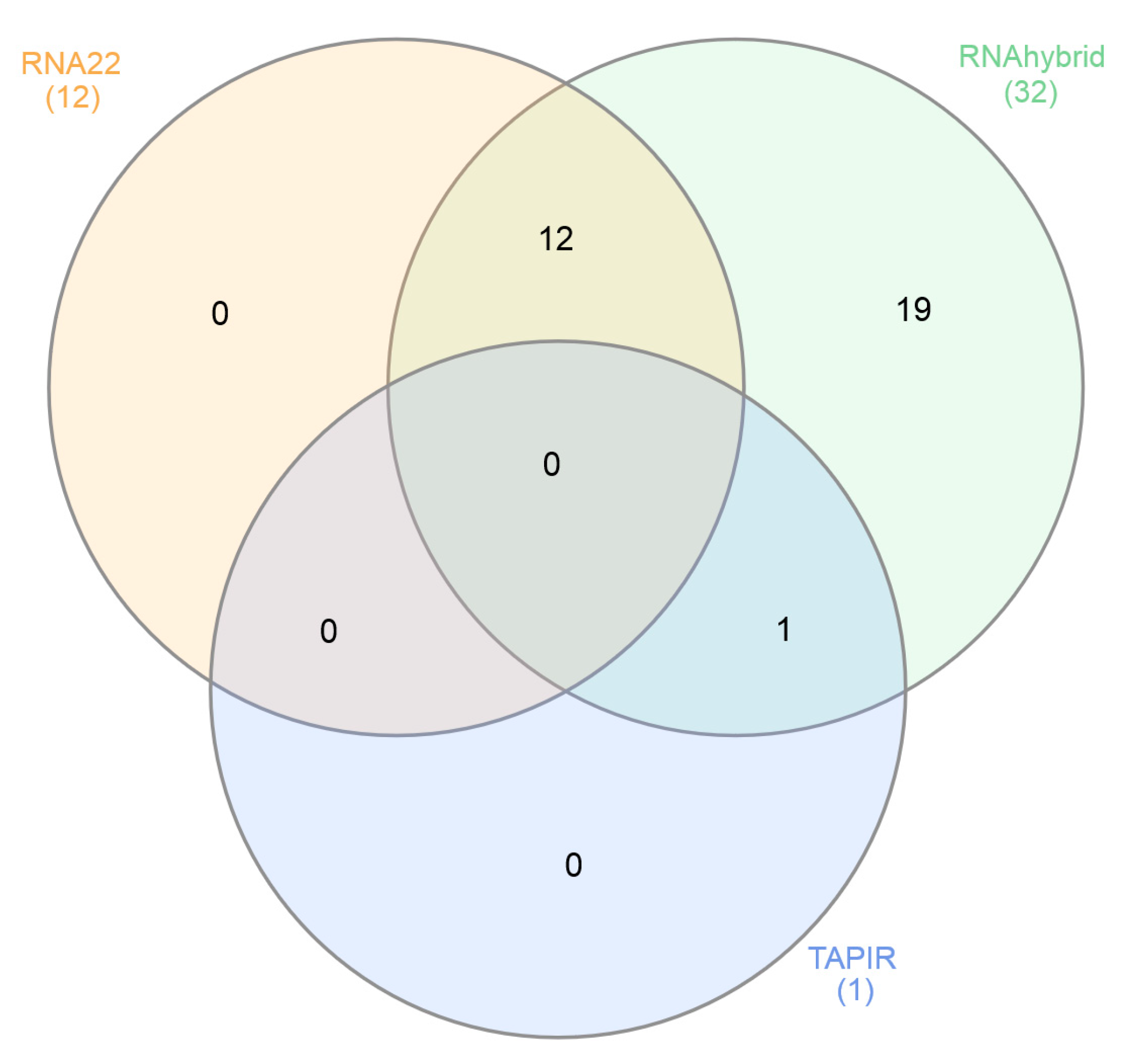
| BSGFV Genes |
RNA22 | RNAhybrid | TAPIR |
|---|---|---|---|
| ORF1 | mbg-miR159, mbg-miR399a, mac-miR4995 | mac-miR156g | |
| ORFII | mbg-miR159 | mac-miR156a-3p, mac-miR156h-3p | |
| ORFIII | mac-miR156a-3p, mac-miR156h-3p, mac-miR319m, mac-miR160a, mac-miR160g-5p, mac-miR162a, mac-miR164e, mac-miR166b, mac-miR399a1, mac-miR4995 |
mac-miR156a-5p, mac-miR157b, mac-miR159, mac-miR319m, mac-miR160a, mac-miR, mac-miR |
|
| IR | mac-miR4995 | mac-miR172b |
| miRNA ID | miRNA–mRNA Pairing | MFE of Binding (Kcal/mol) |
Binding Position/Genes |
|---|---|---|---|
| mac-miR162a | Target 5' A AA U G 3' GAUGGAC CUGC UCC CUAUUUG GACG AGG miRNA 3' CCAG GA U 5' |
−24.00 | 5502 (ORFIII) |
| mac-miR172b | Target 5' G AG G 3' AGCA GUUAAGAUU UCGU UAAUUCUAA miRNA 3' ACA AG GU 5' |
−20.50 | 9 (IR) |
| miRNA ID | miRNA–mRNA Sequence (5′–3′) |
ΔG Duplex (Kcal/mol) |
ΔG Binding (Kcal/mol) |
|---|---|---|---|
| mac-miR162a | 5′ GGAUGCAGAGGUUUAUCGACC 3′ 5′ AGATGGACAACTGCTTCCGAG 3′ |
−20.05 | −15.92 |
| mac-miR172b | 5′ UGAAUCUUAAUGAUGCUACA 3′ 5′ GAGCAAGGTTAAGATTGATGG 3′ |
−14.30 | −13.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





