Submitted:
24 December 2024
Posted:
25 December 2024
You are already at the latest version
Abstract
Keywords:
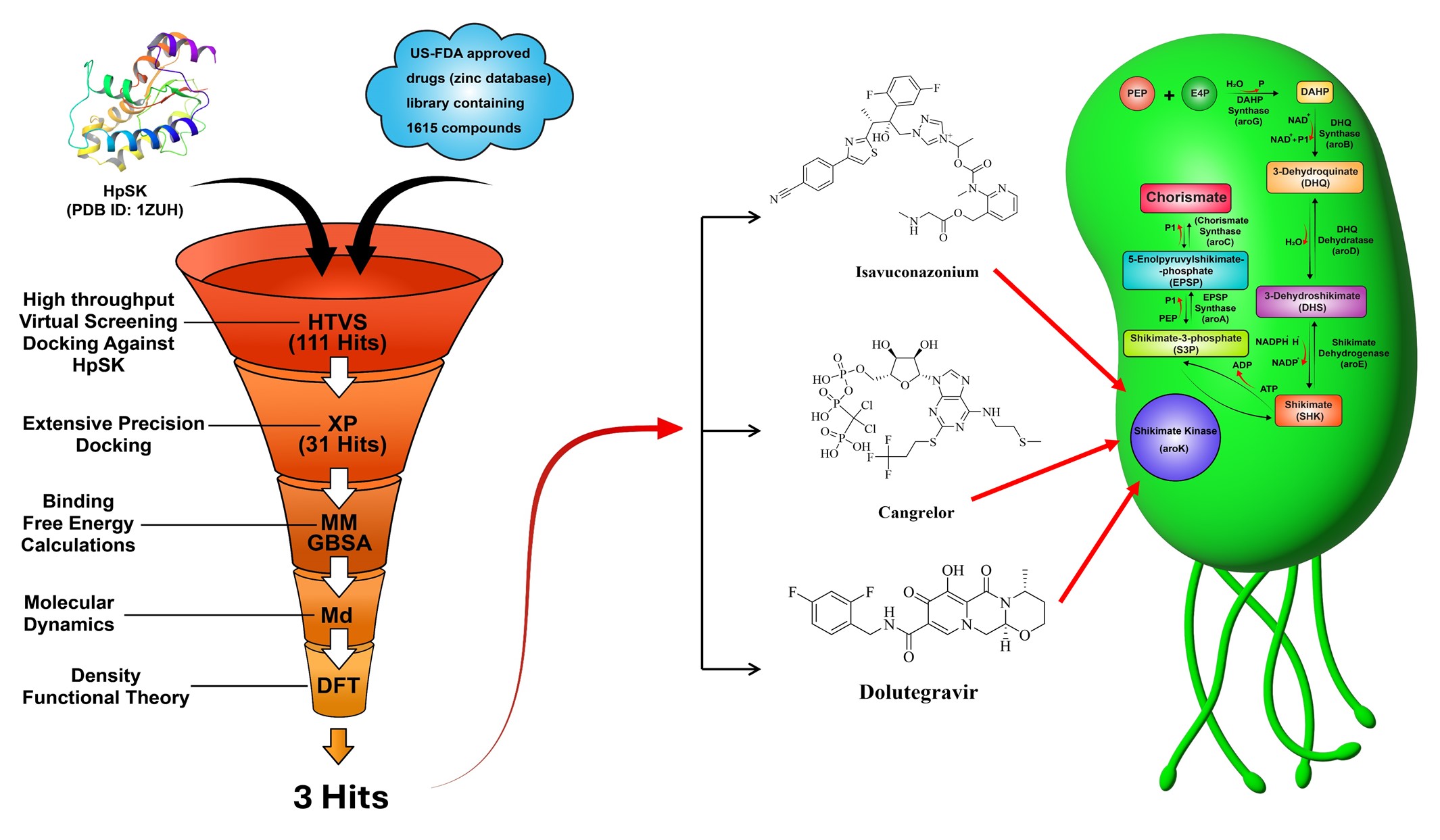
1. Introduction
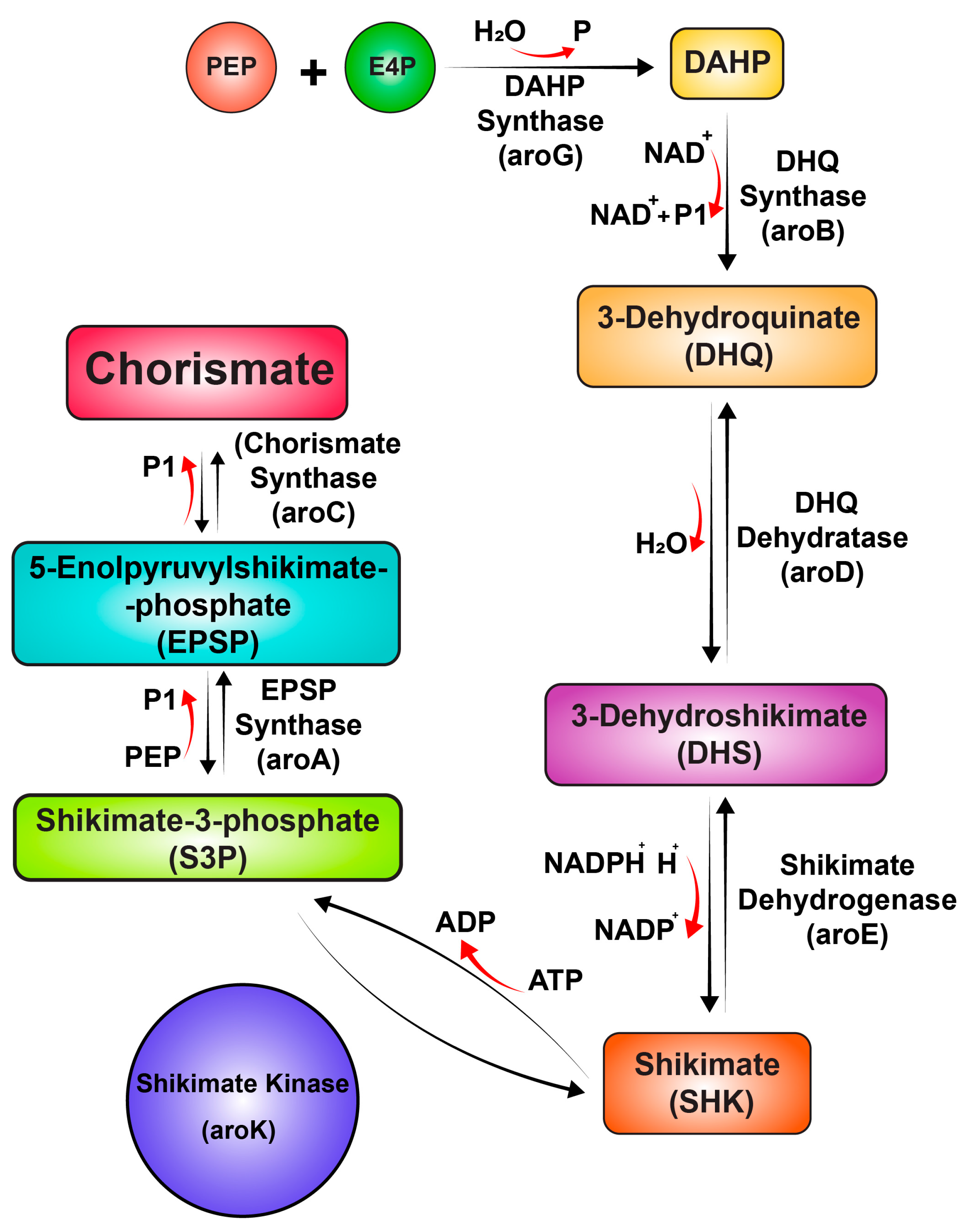
2. Results and Discussion
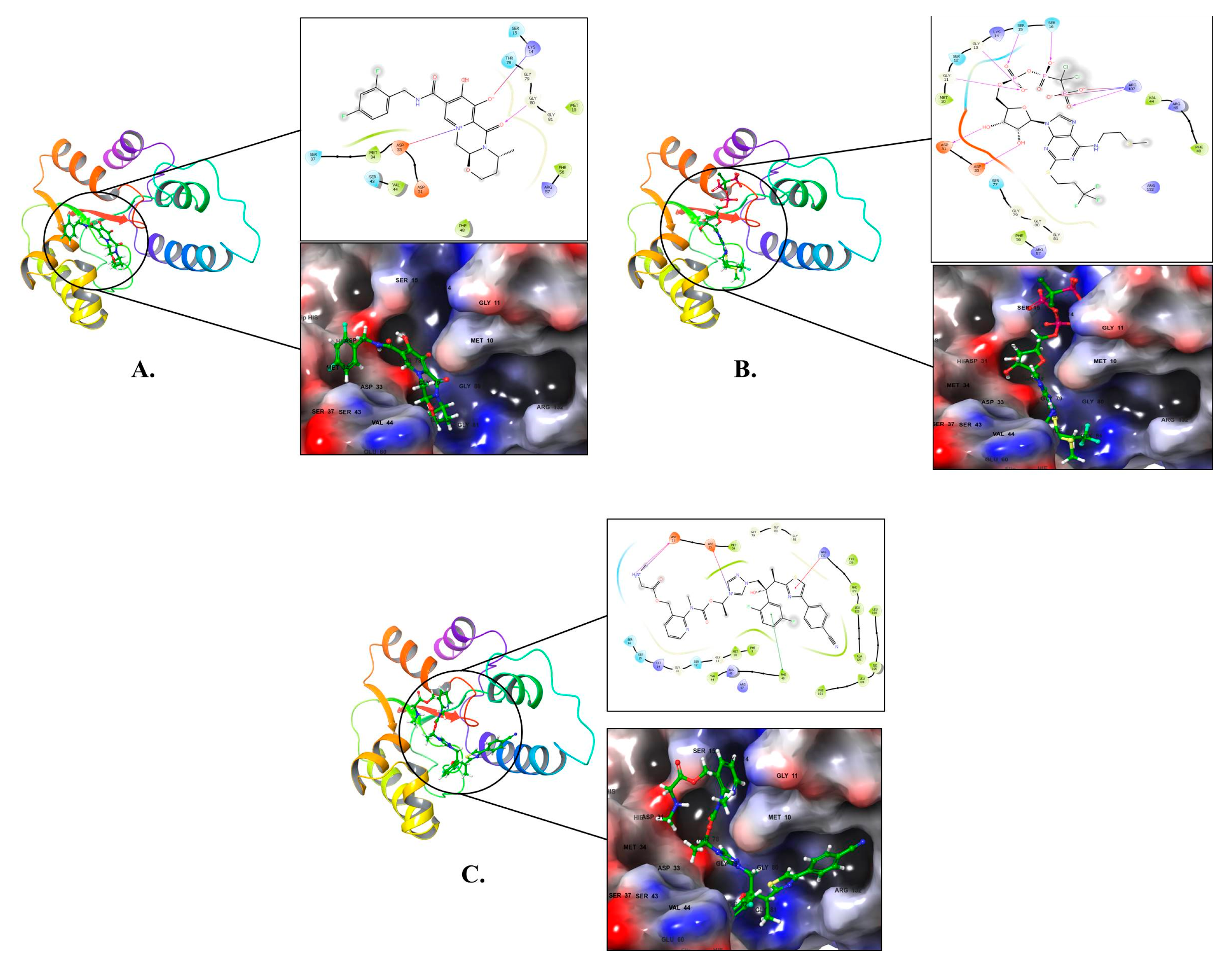
2.1. Molecular Docking and MM-GBSA Calculations
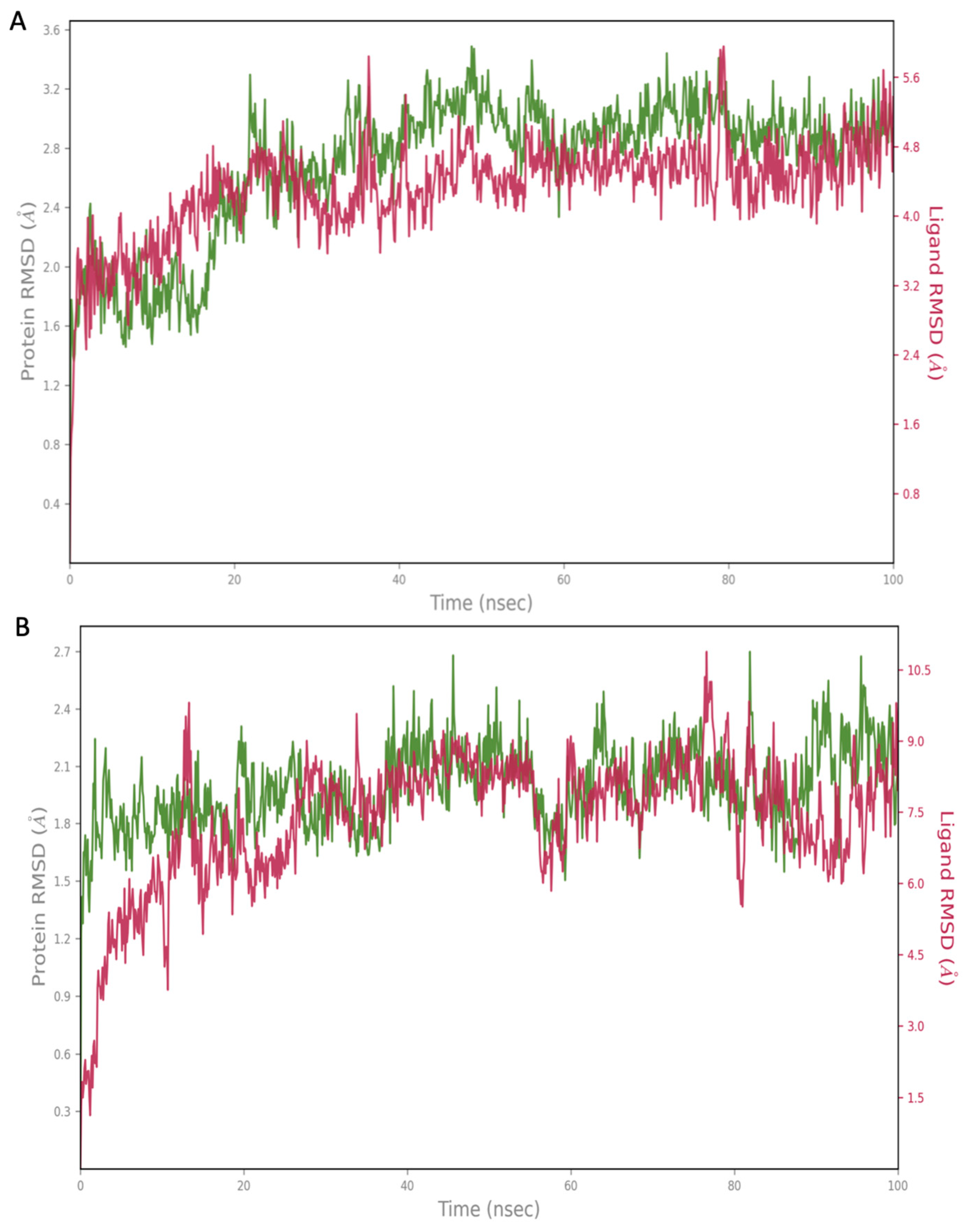
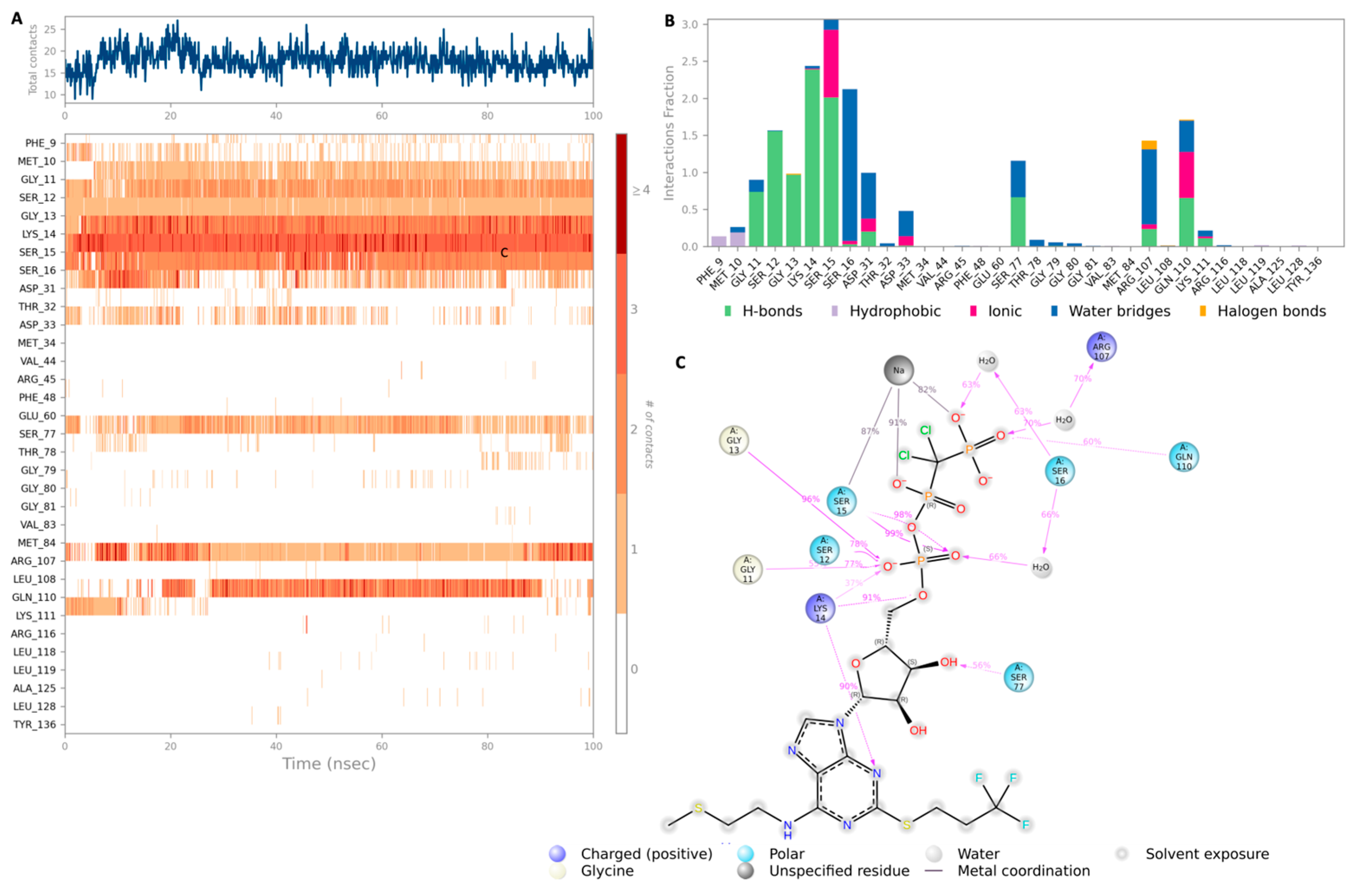
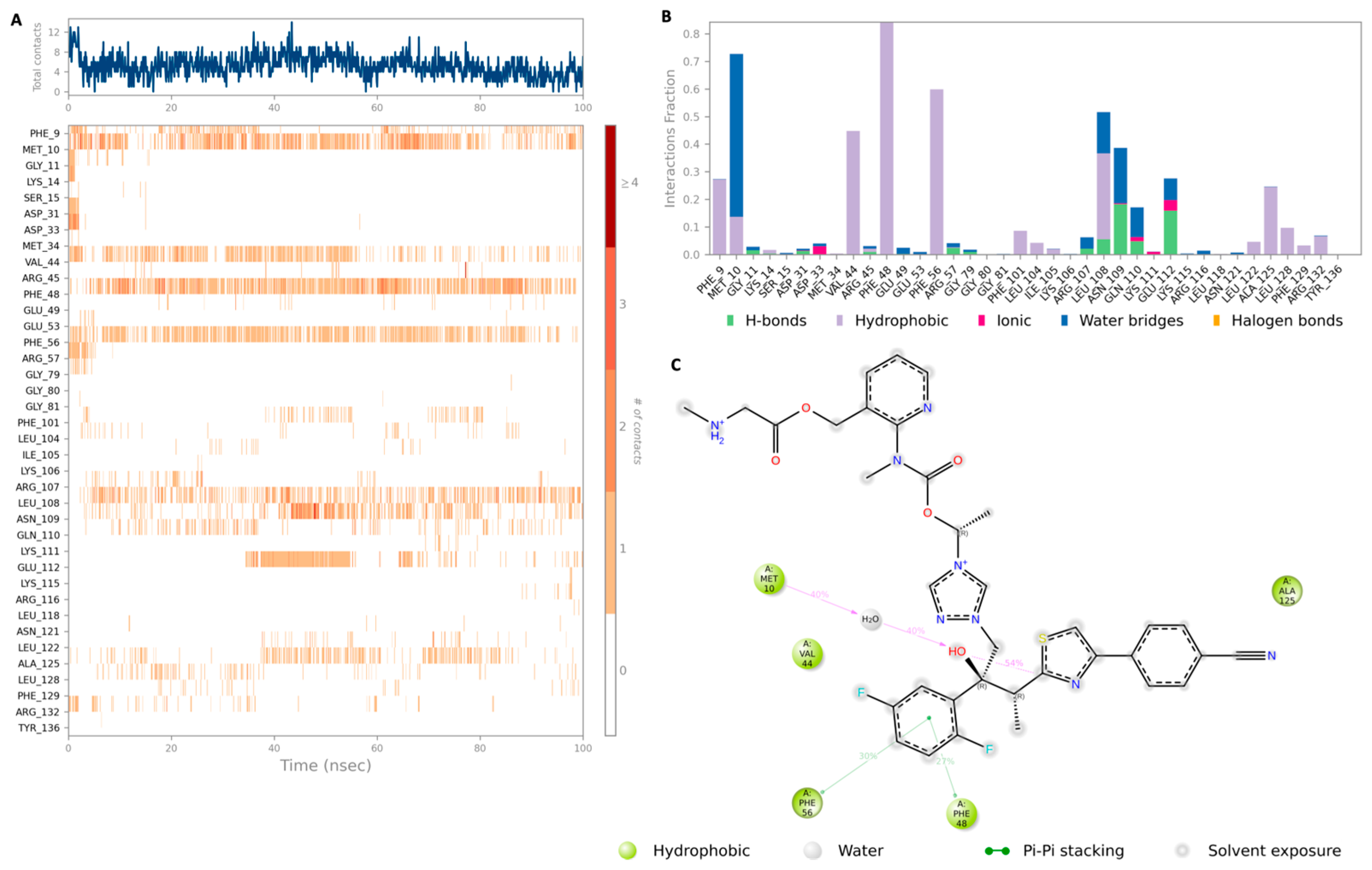
2.2. Molecular Dynamics (MD) Simulation
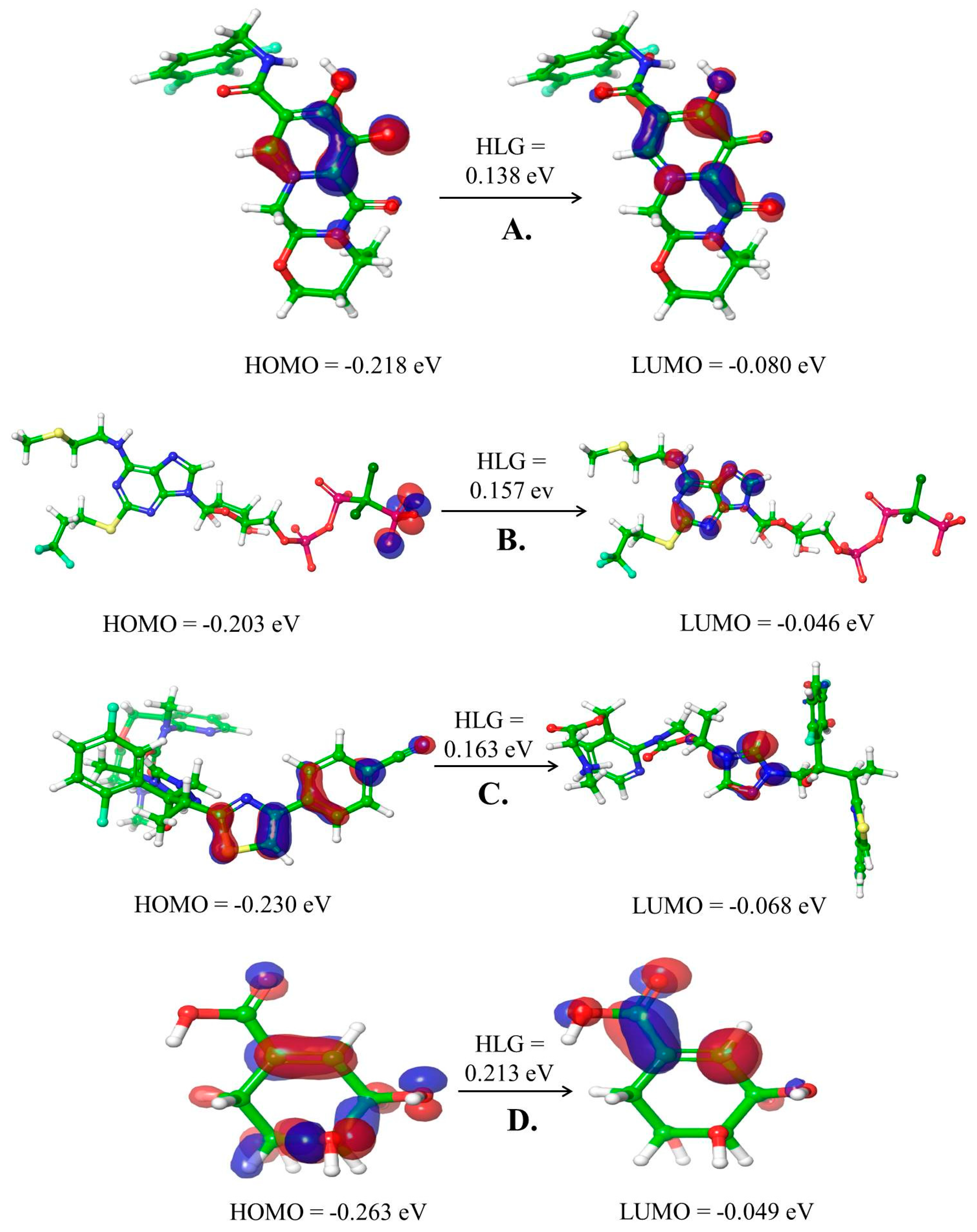
2.3. Density Functional Theory (DFT) Calculations
3. Materials and Methods
3.1. Software
3.2. Preparation of Ligands
3.3. Protein Target Retrieval and Preparation
3.4. Site Mapping and Generation of Receptor Gride
3.5. Molecular Docking
3.6. Free Binding Energy Calculation (MM/GBSA)
3.7. Molecular Dynamic (MD) Simulations
3.8. Density Functional Theory (DFT) Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parikh, N.S.; Ahlawat, R. Helicobacter Pylori. StatPearls 2023.
- Dunne, C.; Dolan, B.; Clyne, M. Factors That Mediate Colonization of the Human Stomach by Helicobacter Pylori. World journal of gastroenterology 2014, 20, 5610–5624. [Google Scholar] [CrossRef] [PubMed]
- Jearth, V.; Rath, M.M.; Chatterjee, A.; Kale, A.; Panigrahi, M.K. Drug-Resistant Helicobacter Pylori: Diagnosis and Evidence-Based Approach. Diagnostics 2023, 13. [Google Scholar] [CrossRef]
- Asaka, M.; Sepulveda, A.R.; Sugiyama, T.; Graham, D.Y. Gastric Cancer. Helicobacter pylori: Physiology and Genetics 2001.
- Stages of Stomach Cancer - NCI.
- Ilic, M.; Ilic, I. Epidemiology of Stomach Cancer. World journal of gastroenterology 2022, 28, 1187–1203. [Google Scholar] [CrossRef]
- Sukri, A.; Lopes, B.S.; Hanafiah, A. The Emergence of Multidrug-Resistant Helicobacter Pylori in Southeast Asia: A Systematic Review on the Trends and Intervention Strategies Using Antimicrobial Peptides. Antibiotics (Basel, Switzerland) 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Liyen Cartelle, A.; Uy, P.P.; Koehler, T.E.; Yap, J.E.L. Persistent Helicobacter Pylori Infection: An Insight to the Limitations of Current Clinical Practice. Cureus 2020, 12, e12309. [Google Scholar] [CrossRef] [PubMed]
- Al-Fakhrany, O.M.; Elekhnawy, E. Helicobacter Pylori in the Post-Antibiotics Era: From Virulence Factors to New Drug Targets and Therapeutic Agents. Archives of microbiology 2023, 205, 301. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. The Lancet. Infectious diseases 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Al-Fakhrany, O.M.; Elekhnawy, E. Helicobacter Pylori in the Post-Antibiotics Era: From Virulence Factors to New Drug Targets and Therapeutic Agents. Archives of microbiology 2023, 205, 301. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.M.; Marchiosi, R.; Abrahão, J.; Constantin, R.P.; dos Santos, W.D.; Ferrarese-Filho, O. Revisiting the Shikimate Pathway and Highlighting Their Enzyme Inhibitors; 2024; Vol. 23; ISBN 0123456789.
- Uchil, R.R.; Kohli, G.S.; Katekhaye, V.M.; Swami, O.C. Strategies to Combat Antimicrobial Resistance. Journal of clinical and diagnostic research : JCDR 2014, 8, ME01-4. [Google Scholar] [CrossRef]
- Herrmann, K.M.; Weaver, L.M. The Shikimate Pathway. Annu. Rev. Plant. Physiol. Plant. Mol. Biol 1999, 50, 473–503. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Krömer, J.O. Metabolic Engineering of the Shikimate Pathway for Production of Aromatics and Derived Compounds—Present and Future Strain Construction Strategies. Frontiers in Bioengineering and Biotechnology 2018, 6. [Google Scholar] [CrossRef]
- Cheng, W.-C.; Chen, Y.-F.; Wang, H.-J.; Hsu, K.-C.; Lin, S.-C.; Chen, T.-J.; Yang, J.-M.; Wang, W.-C. Structures of Helicobacter Pylori Shikimate Kinase Reveal a Selective Inhibitor-Induced-Fit Mechanism. PLOS ONE 2012, 7, e33481. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.M.; Marchiosi, R.; Abrahão, J.; Constantin, R.P.; dos Santos, W.D.; Ferrarese-Filho, O. Revisiting the Shikimate Pathway and Highlighting Their Enzyme Inhibitors; 2024; Vol. 23; ISBN 0123456789.
- Yuan, J.; Zhong, S.; Long, Y.; Guo, J.; Yu, Y.; Liu, J. Shikimate Kinase Plays Important Roles in Anthocyanin Synthesis in Petunia. International journal of molecular sciences 2022, 23. [Google Scholar] [CrossRef]
- Cheng, W.-C.; Chen, Y.-F.; Wang, H.-J.; Hsu, K.-C.; Lin, S.-C.; Chen, T.-J.; Yang, J.-M.; Wang, W.-C. Structures of Helicobacter Pylori Shikimate Kinase Reveal a Selective Inhibitor-Induced-Fit Mechanism. PLOS ONE 2012, 7, e33481. [Google Scholar] [CrossRef] [PubMed]
- Parish, T.; Stoker, N.G. The Common Aromatic Amino Acid Biosynthesis Pathway Is Essential in Mycobacterium Tuberculosis. Microbiology 2002, 148, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nature Reviews Drug Discovery 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Schrödinger Schrödinger | Schrödinger Is the Scientific Leader in Developing State-of-the-Art Chemical Simulation Software for Use in Pharmaceutical, Biotechnology, and Materials Research. Available online: https://www.schrodinger.com/ (accessed on March 2024).
- Watts, K.S.; Dalal, P.; Tebben, A.J.; Cheney, D.L.; Shelley, J.C. Macrocycle Conformational Sampling with Macromodel. Journal of Chemical Information and Modeling 2014, 54, 2680–2696. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, F.; Richards, N.G.J.; Guida, W.C.; Liskamp, R.; Lipton, M.; Caufield, C.; Chang, G.; Hendrickson, T.; Still, W.C. Macromodel—an Integrated Software System for Modeling Organic and Bioorganic Molecules Using Molecular Mechanics. Journal of Computational Chemistry 1990, 11, 440–467. [Google Scholar] [CrossRef]
- Cheng, W.C.; Chang, Y.N.; Wang, W.C. Structural Basis for Shikimate-Binding Specificity of Helicobacter Pylori Shikimate Kinase. Journal of Bacteriology 2005, 187, 8156. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. Journal of Computer-Aided Molecular Design 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. Journal of Chemical Theory and Computation 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Cheng, W.C.; Chang, Y.N.; Wang, W.C. Structural Basis for Shikimate-Binding Specificity of Helicobacter Pylori Shikimate Kinase. Journal of Bacteriology 2005, 187, 8156. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. Journal of Chemical Information and Modeling 2009, 49, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T. New Method for Fast and Accurate Binding-Site Identification and Analysis. Chemical Biology and Drug Design 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. Journal of Medicinal Chemistry 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Alzain, A.A.; Mukhtar, R.M.; Abdelmoniem, N.; Shoaib, T.H.; Osman, W.; Alsulaimany, M.; Aljohani, A.K.B.; Almadani, S.A.; Alsaadi, B.H.; Althubyani, M.M.; et al. Modulation of NRF2/KEAP1-Mediated Oxidative Stress for Cancer Treatment by Natural Products Using Pharmacophore-Based Screening, Molecular Docking, and Molecular Dynamics Studies. Molecules (Basel, Switzerland) 2023, 28. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, T.H.; Ibraheem, W.; Abdelrahman, M.; Osman, W.; Sherif, A.E.; Ashour, A.; Ibrahim, S.R.M.; Ghazawi, K.F.; Miski, S.F.; Almadani, S.A.; et al. Exploring the Potential of Approved Drugs for Triple-Negative Breast Cancer Treatment by Targeting Casein Kinase 2: Insights from Computational Studies. PLOS ONE 2023, 18, e0289887. [Google Scholar] [CrossRef]
- Eltaib, L.; Alzain, A.A. Targeting the Omicron Variant of SARS-CoV-2 with Phytochemicals from Saudi Medicinal Plants: Molecular Docking Combined with Molecular Dynamics Investigations. Journal of Biomolecular Structure and Dynamics 2022, 0, 1–13. [Google Scholar] [CrossRef]
- Alzain, A.A.; Elbadwi, F.A. Identification of Novel TMPRSS2 Inhibitors for COVID-19 Using e-Pharmacophore Modelling, Molecular Docking, Molecular Dynamics and Quantum Mechanics Studies. Informatics in Medicine Unlocked 2021, 26, 100758. [Google Scholar] [CrossRef] [PubMed]
- Lyne, P.D.; Lamb, M.L.; Saeh, J.C. Accurate Prediction of the Relative Potencies of Members of a Series of Kinase Inhibitors Using Molecular Docking and MM-GBSA Scoring. Journal of Medicinal Chemistry 2006, 49, 4805–4808. [Google Scholar] [CrossRef] [PubMed]
- Hussein, D. In Silico Investigation of the Human GTP Cyclohydrolase 1 Enzyme Reveals the Potential of Drug Repurposing Approaches towards the Discovery of Effective BH4 Therapeutics. Int J Mol Sci 2023, 24, 1210. [Google Scholar] [CrossRef] [PubMed]
- Suryanarayanan, V.; Singh, S.K. Assessment of Dual Inhibition Property of Newly Discovered Inhibitors against PCAF and GCN5 through in Silico Screening, Molecular Dynamics Simulation and DFT Approach. Journal of Receptors and Signal Transduction 2015, 35, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Current Computer Aided-Drug Design 2012, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Romano, T. Kroemer Structure-Based Drug Design: Docking and Scoring. Current Protein & Peptide Science 2007, 8, 312–328. [Google Scholar] [CrossRef]
- Pattar, S.V.; Adhoni, S.A.; Kamanavalli, C.M.; Kumbar, S.S. In Silico Molecular Docking Studies and MM/GBSA Analysis of Coumarin-Carbonodithioate Hybrid Derivatives Divulge the Anticancer Potential against Breast Cancer. Beni-Suef University Journal of Basic and Applied Sciences 2020, 9. [Google Scholar] [CrossRef]
- Cada, D.; Levien, T.; Baker, D. Dolutegravir. Hospital Pharmacy 2014, 49, 184–195. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Steg, P.G.; Bhatt, D.L.; Capodanno, D.; Angiolillo, D.J. Cangrelor: Clinical Data, Contemporary Use, and Future Perspectives. Journal of the American Heart Association 2021, 10, e022125. [Google Scholar] [CrossRef] [PubMed]
- Shirae, S.; Ose, A.; Kumagai, Y. Pharmacokinetics, Safety, and Tolerability of Single and Multiple Doses of Isavuconazonium Sulfate in Healthy Adult Japanese Subjects. Clinical Pharmacology in Drug Development 2022, 11, 744–753. [Google Scholar] [CrossRef]
- Kavitha, R.; Karunagaran, S.; Chandrabose, S.S.; Lee, K.W.; Meganathan, C. Pharmacophore Modeling, Virtual Screening, Molecular Docking Studies and Density Functional Theory Approaches to Identify Novel Ketohexokinase (KHK) Inhibitors. BioSystems 2015, 138, 39–52. [Google Scholar] [CrossRef]
- Zothantluanga, J.H.; Abdalla, M.; Rudrapal, M.; Tian, Q.; Chetia, D.; Li, J. Computational Investigations for Identification of Bioactive Molecules from Baccaurea Ramiflora and Bergenia Ciliata as Inhibitors of SARS-CoV-2 Mpro. Polycyclic Aromatic Compounds 2023, 43, 2459–2487. [Google Scholar] [CrossRef]
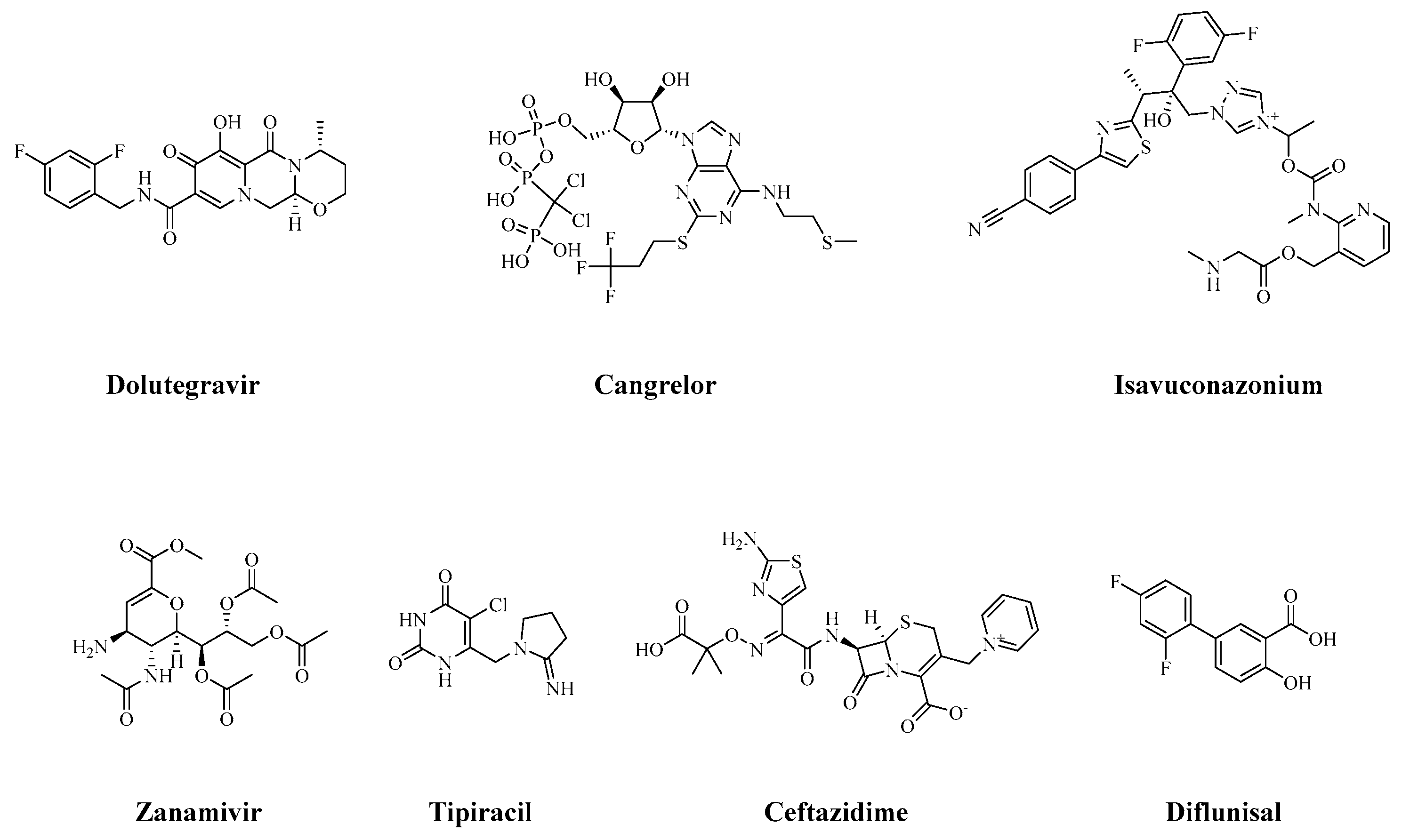
| No.* | ZINC_ID | Medication | Docking Score (XP) | MM-GBSA dG Bind** | |
| 1 | ZINC000058581064 | Dolutegravir | -7.947 | -35.05 | |
| 2 | ZINC000003918138 | Zanamivir | -7.727 | -32.43 | |
| 3 | ZINC000085537017 | Cangrelor | -7.645 | -35.73 | |
| 4 | ZINC000100032379 | Tipiracil | -7.285 | -31.56 | |
| 5 | ZINC000003871960 | Ceftazidime | -6.120 | -33.03 | |
| 6 | ZINC000029571072 | Isavuconazonium | -6.069 | -53.07 | |
| 7 | ZINC000000020243 | Diflunisal | -5.981 | -33.35 | |
| 8 | Shikimate | - | -5.867 | -34.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





