1. Introduction
The limited antiviral drug pharmacopeia together with the rapid selection of drug-resistant viruses present a significant challenge to the medical community [1,2,3,4,5]. The recent outbreak of mpox in Central Africa illustrates this issue as TPOXX, a FDA-approved antiviral for the treatment of smallpox and under the CDC-held Expanded Access-Investigational New Drug (EA-IND) protocol certain mpox patients, failed to improve outcomes in individuals with clade I MPXV infection in a recent clinical trial in the Democratic Republic of the Congo (DRC) [6,7,8]. This lack of efficacy may have been due to sub-optimal potency and de novo drug resistance as TPOXX resistant MPXV variants are readily found in clinical cases of mpox, especially in immunosuppressed patients on long-term treatment [9]. MPXV is classified within the Orthopoxvirus genus, which includes smallpox and vaccinia virus (VACV), in the family Poxviridae. Members of this genus possess linear double-stranded DNA (dsDNA) genomes and a life cycle restricted to the cytoplasm of infected cells [10], involving four distinct infectious virus stages: the intracellular mature virion (IMV), the intracellular enveloped virion (IEV), the cell-associated enveloped virion (CEV), and the extracellular enveloped virion (EEV) [11]. The small molecule antiviral TPOXX targets the viral F13 protein resulting in inhibition of the IMV wrapping in both VACV and MPXV [12,13]. F13L, a conserved gene within the Poxviridae family, is essential for virus envelopment and release [12,14], and likely for efficient viral entry [15,16]. VACV can be used as an accurate surrogate for the investigation of MPXV antivirals, as both share functionally conserved proteins in their genomes [17,18]. Both viruses exhibit similar resistance mechanisms to TPOXX associated with mutations in the F13L gene. VACV resistance to TPOXX has been linked to several mutations including G277C in the F13L gene [19], whereas T289A and A290V mutations in F13L have been confirmed to confer MPXV with resistance to TPOXX [9]. Several additional mutations including R291K, S215F, and P243S in the F13L gene have been linked, but yet to be confirmed, to MPXV resistance to TPOXX [9]. MPXV mutations associated with increased resistance to TPOXX have been identified among severely immunocompromised mpox patients who required prolonged courses of TPOXX [20,21,22]. However, spread of TPOXX-resistant MPXV has been also reported among persons with no previous TPOXX treatment [20]. The genome of MPXV Clade Ib contains large deletions in gene OPG032 [23,24], but these deletions do not contribute to TPOXX resistance. Likewise, none of the multiple mutations consistent with APOBEC3-mediated cytosine deamination present in the genome of Clade Ib MPXV has been directly implicated in TPOXX resistance [9,23]. However, the recent outbreak of clade Ib MPXV [23] in DRC [24] and Sweden [25,26] together with the recently released results of the international clinical trial STOMP (NCT05597735), highlight the challenges posed by the limited efficacy of TPOXX against both clades of MPXV, and the urgent need for alternative antivirals [7,8].
We have documented that mycophenolate mofetil (MMF), an FDA-approved drug, has potent antiviral activity against both VACV and MPXV in cell-based assays [18]. N-myristoylation has been shown to be crucial for the propagation of several viruses [27,28], and VACV multiplication was effectively inhibited by the NMT inhibitor IMP-1088 [29]. In this study, we used VACV as a surrogate model for MPXV to assess the efficacy of combination therapy based on the combination of direct-acting (TPOXX) and a host-targeting (MMF or IMP-1088) antivirals as a strategy to counteract MPXV TPOXX resistance. This approach targets multiple steps of the virus life cycle, which via synergistic effects can result in more effective control of viral burden and associated improved clinical outcomes of mpox, as well as posing a high genetic barrier to the emergence of drug-resistant MPXV.
2. Materials and Methods
Cell Lines and Viruses
Homo sapiens A549 (ATCC CCL-185) cell line was maintained in Dulbecco’s modified Eagle medium (DMEM) (ThermoFisher Scientific, Waltham, MA, USA) containing 10% heat- inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 100 μg/mL streptomycin, and 100 U/mL penicillin. The bi-reporter recombinant VACV expressing fluorescent protein (GFP, downstream of the A27L locus) and nanoluciferase (Nluc, downstream of the F13L locus), rVACV Nluc/GFP was previously described and characterized [30,31]. MPXV 2003 (NR-2500) was obtained from BEI Resources.
Compounds
The compounds previously identified to have antiviral activity against RNA viruses in the ReFRAME library were previously described [18,32] were purchased from MedChem Exp and include mycophenolate mofetil, MMF (cat. No. HY-B0199), buparvaquone (cat. No. HY-17581), OSU-03012 (cat. No. HY-10547), AVN-944 (cat. No. HY-13560), brequinar (cat. No. HY-108325), Valinomycin (cat. No. HY-N6693). The NMT inhibitors IMP-1088 was purchased from Cayman Chemical (cat. No. 25366-1) and DDD85646 (Synonyms: IMP-366) was purchased from MedChem Exp (Cat. No.: HY-103056). Tecovirimat (Synonyms: ST-246), referred to as TPOXX here, was purchased from MedChem Exp (Cat. No.: HY- HY-14805).
Determination of Compound EC50
A549 cells were seeded on 96-well clear-bottom black plates (4.0 × 104 cells/well) and infected (MOI 0.01) 20 h later with rVACV Nluc/GFP using 4 technical replicates. After a 60 min adsorption, the virus inoculum was aspirated off and compound-containing media was added to the cells. At 48 h pi, the cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes, then washed once with PBS, and GFP expression was determined by fluorescence using a fluorescent plate reader BioTek Cytation 5 (Agilent, Santa Clara, CA, USA). The mean relative fluorescence units were normalized to vehicle control (DMSO)-treated cells, which were assigned a value of 100%. The half-maximal effective concentrations (EC50) were determined using GraphPad Prism, v10 (Prism10).
Determination of Compound CC50
Cell viability was determined using the 4′,6-diamidino-2-phenylindole (DAPI) staining, which is a blue-fluorescent DNA stain that exhibits about 20-fold enhancement of fluorescence upon binding to AT regions of double stranded DNA. A549 cells were plated on 96-well clear-bottom plates (4.0 × 104 cells/well) using four technical replicates. Serial dilutions of each compound were added to the cells, and at 48 h after the drug treatment, cells were fixed with 8% PFA (final 4%) and incubated for 30 minutes, then washed once with DPBS, signals were quantified using the BioTek Cytation 5. The resulting optical densities were normalized to the vehicle-treated (DMSO) control samples, which were assigned a value of 100%. The half-maximal cytotoxic concentrations (CC50) were determined using Prism10.
Drug Combination Assays in VACV-Infected Cells
A549 cells were seeded on 96-well clear-bottom black plates (4.0 × 104 cells/well) and infected (MOI 0.01) 20 h later with rVACV Nluc/GFP. After a 60 min adsorption, the virus inoculum was aspirated, and compound-containing media were added to the cells. The compounds were prepared in serial dilution, 2x concentration, then added to predetermined matrix of compounds mixture, to bring final concentration into 1x. At 48 h pi, cell culture supernatants (CCS) were collected, then cells were fixed with 4% paraformaldehyde, and GFP expression levels were determined using a fluorescent plate reader (BioTek Cytation 5). Mean relative fluorescence units were normalized to vehicle-treated (DMSO) control cells, which were assigned a value of 100%. Compounds half-maximal effective concentrations (EC50) were determined using Prism10. To assess the effect of drug combination treatments in production of rVACV Nluc/GFP infectious progeny, CCS were diluted (1:10) and 50 µL used to infect A549 cells seeded on 96-well clear-bottom black plates (4.0 × 104 cells/well). After a 60 min adsorption, the virus inoculum was aspirated, and media were added to the cells. At 48 h pi, CCS were collected, and cells fixed with 4% paraformaldehyde, and GFP expression levels determined using a fluorescent plate reader (BioTek Cytation 5). Mean relative fluorescence units were normalized to vehicle control (DMSO)-treated cells, which were assigned a value of 100%. The half-maximal effective concentrations (EC50) were determined using Prism10.
For synergetic studies, we prepared an 8-by-8 matrix (i.e., two drugs were tested at all possible 64 combinations of the two drugs). A549 cells, 4 x 104/well, were seeded in 96-well, black, clear-bottom plates in a final volume of 100µl/well. The plates were incubated at 37°C with 5% CO2 overnight. Stock solutions of each drug (A and B) were made at 2x in 3000 µL. U-bottom 96 well plates were added 125µL of media, then 2x drug stock was added into first row or column, and 1:2 serial dilutions were performed. Samples in the 2x plated were mixed into a 1:1 mixture to obtain 1x final concentration ready to be added to wells of the plate containing infected cells after 60 minutes adsorption period (SF1). Normalized values from matrix data were analyzed using SynergyFinder+ software, an open-access platform for multidrug combination synergies [33]. For the combination synergy model, we used the four models: ZIP, HSA, Loewe, and Bliss.; however, we sorted synergetic score based on ZIP’s logistic growth fit and scores because both ZIP and Loewe can interpolate from logistic growth fit, as opposed to HSA and Bliss where they align with linear fit. Also, ZIP model assumed independent mechanistic pathways involved in the synergy. In the synergy matrix, the 100% value of GFP signal indicates a total inhibition, while 0% means no inhibition. The initial combination therapy assessment was done using one biological replicate, while the rest of the combination therapy assays were done using three independent biological replicates. Complete baseline correction was conducted on all values in SynergyFinder+.
Drug Combination Assays in MPXV-Infected Cells
A549 cells (4x104 cells/96-well, triplicate) were infected with MPXV 2003 (BEI Resources NR-2500) (MOI 0.01-0.02, 50 µL/well in DMEM/2% FBS, 1% PSG) for 1 h at 37°C degrees with occasional shaking. After the adsorption time, viral inoculum was aspirated off and 100 µL of serial dilutions from combination plate TPOXX and MMF or IMP-1088 and MMF was added to cells. At 24 h pi, CCS was aspirated off and cells were fixed in 10% neutral buffered formalin at 4°C. After 24 h, cells were washed with ddH2O 3x and overlayed with 50 µL of 1x DAPI in PBS for 30 min at room temperature (RT). Cells were washed with PBS and imaged for DAPI fluorescence using a Synergy H1 Hybrid Multi-Mode Reader. Cells were then washed and permeabilized with 0.5% Triton X -100 in 1x PBS for 15 mins at RT, followed by 2 h incubation at 37°C in PBS containing 2.5% BSA to block non-specific antibody binding. Cells were incubated with a rabbit polyclonal serum to VACV-A33R (BEI Resources NR-628) (1:5,000) in PBS/2.5% BSA for 2 h at 37°C. After washing, cells were incubated with a secondary anti-rabbit antibody, washed and incubated with ABC reagent for 45 min at 37°C, followed by incubation with DAB reagent for 15 mins at RT (VectorLabs ABC HRP and DAB). Cells were allowed to dry and read using the Bioreader 7000 Fz machine.
3. Results
Dose-Response Inhibitory Effect of Selected Drugs on VACV Multiplication
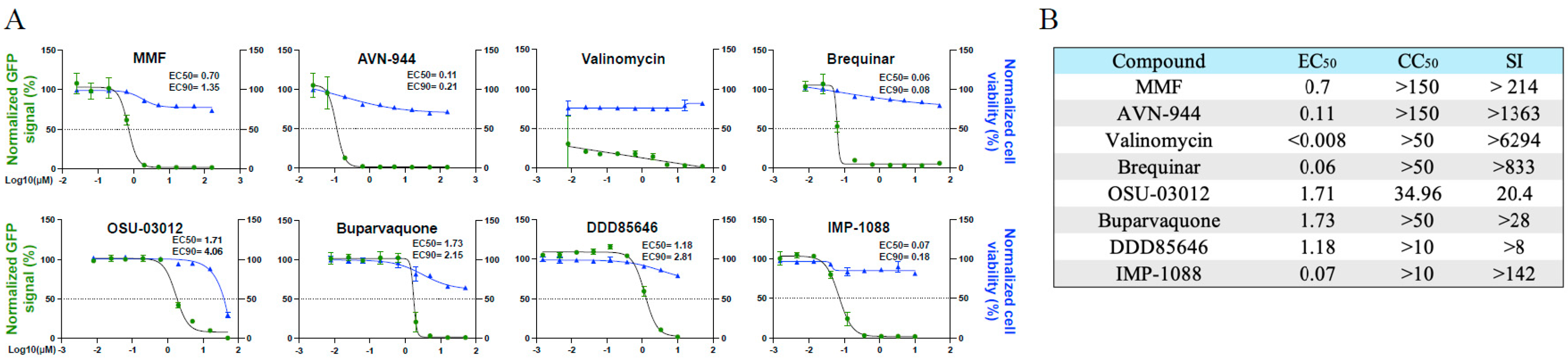
To select compounds for combination therapy with TPOXX, we conducted dose-response assays to confirm the previously shown anti-VACV activity of MMF, AVN-944, brequinar, valinomycin, OSU-03012, and buparvaquone [18]. In addition, we examined the dose-response of the NMT inhibitors DDD85646 and IMP-1088 on VACV multiplication. All eight tested compounds exhibited potent dose-dependent inhibitory effect on VACV multiplication at 48 h pi (
Figure 1A) and had robust selectivity indices (SI) (
Figure 1B), similar to described findings that were measured at 24 h pi [18].
Assessment of Synergistic Anti-VACV Activity of Selected Drugs in Combination with TPOXX
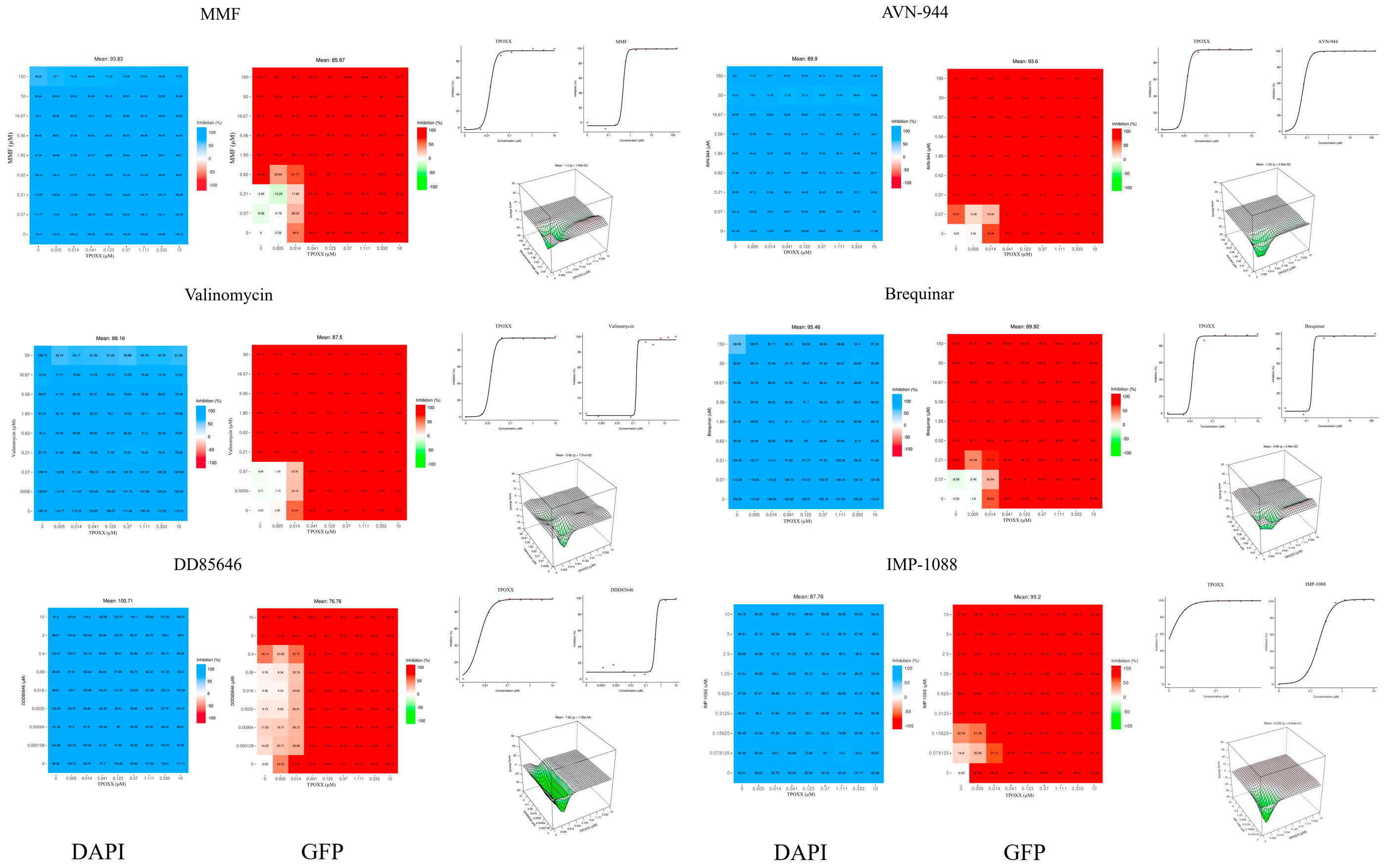
To investigate the synergistic activity of the different compounds with TPOXX, we designed a matrix to screen for synergy using a 96-well plate format. All eight tested compounds showed strong anti-VACV activity in combination therapy (
Figure 2). We measured their synergistic effect with TPOXX based on four models: ZIP, Bliss, Loewe, and HSA, only the three-dimensional plot of ZIP synergy depiction is shown. All tested combinations demonstrated significant inhibition. Nevertheless, the high dose applied in this experiment exceeded the detection threshold of the synergy software, preventing us from obtaining accurate measurements of the synergistic activity.
MMF is a prodrug that serves as a potent irreversible inhibitor of inosine-5'-monophosphate dehydrogenase (IMPDH), leading to the depletion of guanosine nucleotides; it is selective for isoform II of IMPDH [34]. MMF has been shown to exhibit antiviral activity against different viruses [35,36,37], including VACV [38,39]. AVN-944 is a specific and potent pan-inhibitor of IMPDH [40]. Brequinar, an inhibitor of dihydroorotate dehydrogenase (DHODH), has demonstrated potential in synergistic studies [41]. Valinomycin acts as an ionophore, disrupting the potassium ion gradient across the cell membrane [42]. OSU-03012 inhibits phosphoinositide-dependent kinase 1, thereby interfering with Akt signaling, with a preference for the GRP78 chaperone [43,44,45]. Buparvaquone disrupts cytochrome b in eukaryotic cells [46,47]. Both DDD85646 and its analog IMP-1088 are highly selective pan-NMT inhibitors known to exhibit antiviral activity against viruses, including VACV, which rely on myristoylated proteins to complete their life cycle [27,28,29]. Importantly, the IMP-1088 derivative NMT inhibitor PCLX-001 has advanced toward phase II clinical trial drug and has exhibited a good safety profile in phase I, where it completed six dose escalations without dose-limiting toxicities, reaching clinically active exposure levels [48,49,50]. Based on the compounds known features, including their clinical profiles, we selected MMF and IMP-1088 for further detailed investigation of their use in combination therapy with TPOXX.
Detailed Assessment of the Synergistic Anti-VACV Activity of MMF and IMP-1088 in Combination with TPOXX
To test MMF and IMP-1088 synergistic effects with TPOXX, we treated VACV-infected cells with the indicated drug combinations, starting at 1 μM for TPOXX and IMP-1088, and 5 μM for MMF, and using two-fold serial dilutions. MMF and IMP-1088 showed strong dose response synergistic effect in combination with TPOXX with ZIP-synergy scores reaching values > of 43 and 70 for IMP-1088 and MMF, respectively (
Figure 3). To assess the effect of the combination therapy on production of extracellular infectious viral progeny (EEV), we collected the CCS from the drug combination treatments and used them to infect fresh A549 cells. We found that combination therapy potently inhibited production of EEV progeny (
Figure 3).
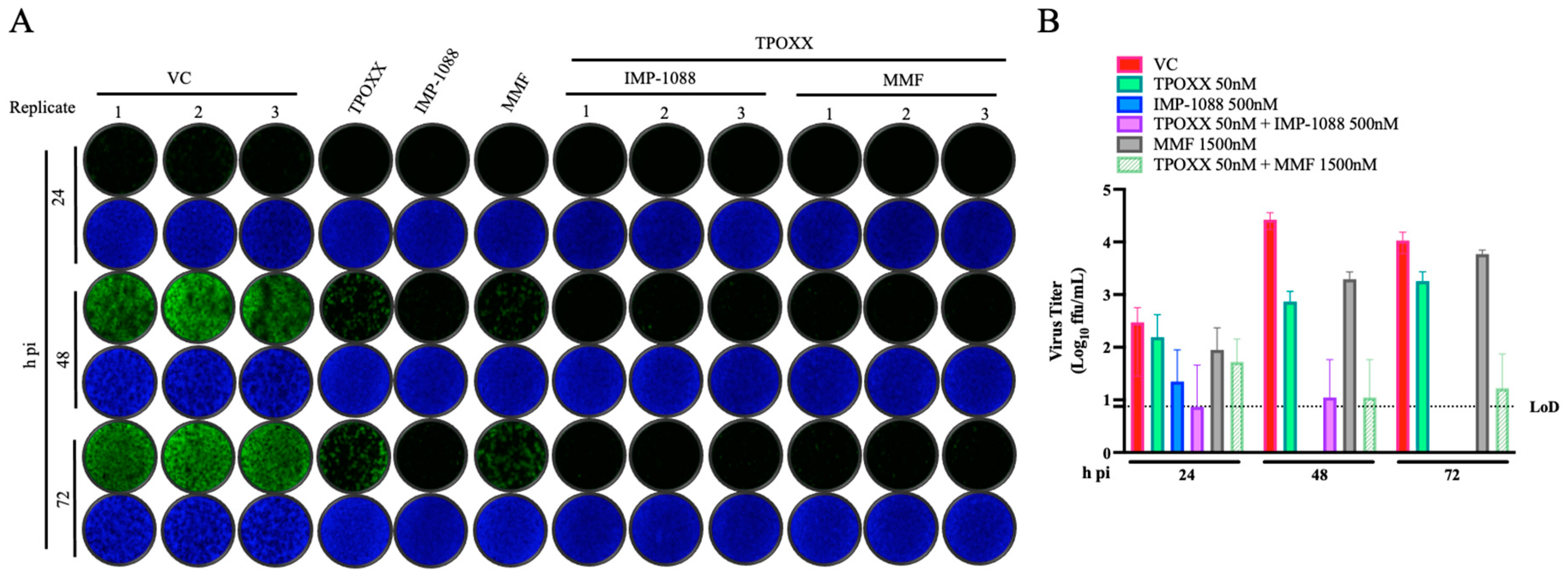
Effect of Combination Therapy on VACV Multi-Step Growth Kinetics
To assess the effect combination therapy on VACV growth kinetic during multiple rounds of infection, we infected A549 cells with rVACV Nluc/GFP at MOI of 0.01 and treated VACV-infected A549 cells with the indicated drug combinations. At the indicated hpi, we determined titers of infectious rVACV Nluc/GFP in CCS (
Figure 4).
Effect of IMP-1088 and MMF Combination Therapy on VACV Infection
To determine whether IMP-1088 and MMF exhibit synergistic antiviral activity against VACV, we treated VACV-infected cells with the indicated combinations of IMP-1088 and MMF. We found that IMP-1088 and MMF exhibited a strong dose-dependent synergistic response with ZIP-synergy score values > 66 (
Figure 5). Importantly, the combination of MMF and IMP-1088 at drug concentrations associated with high synergy score values lacked noticeable effects on cell viability (
Table S1).
Effect of Combination Therapy on MPXV Infection
We next examined whether our findings on the efficacy of antiviral combination therapy to inhibit VACV multiplication in cultured cells were also applicable to infection with bona fide MPXV. Combination therapy of TPOXX and MMF exhibited a strong dose-dependent synergistic antiviral activity against MPXV, with ZIP-synergy score values > 37 (
Figure 6A and B). Likewise, combination therapy of IMP-1088 and MMF also exhibited a strong dose-dependent synergistic response with ZIP-synergy score values > 64 (
Figure 6A and B).
Triple Therapy in VACV
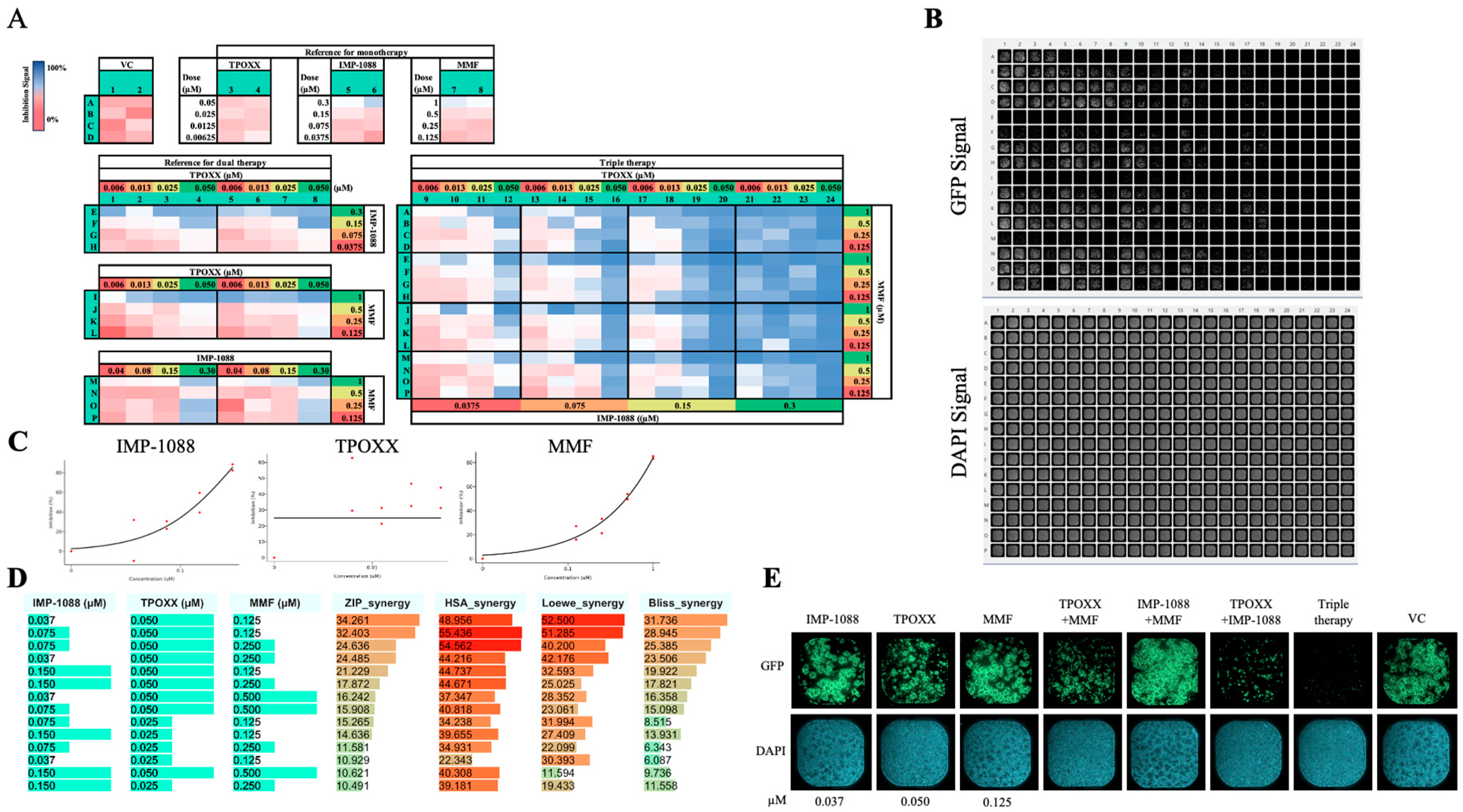
To explore whether triple combination therapy could result in a stronger inhibition at lower doses of each drug, we designed an experiment to investigate the synergistic effect of IMP-1088, TPOXX and MMF at doses starting at 0.3 µM, 0.05 µM and 1 µM, respectively, and serially diluted (1:2). We observed a strong dose response synergistic effect among the three drugs reaching over 34, 48, 52 and 31 score of ZIP, HAS, Leowe, and BLISS models, respectively (
Figure 7). This strong synergistic antiviral effect occurred in the absence of noticeable effects on cell viability (
Figure 7b). Consistent with its potent antiviral activity, this triple combination therapy protected A549 cells from MPXV induced cytopathic effect (
Figure 7E).
Discussion
The mpox disease continues to spread in Africa and other parts of the world, posing a threat to global health security. This situation is exacerbated by the emergence of more recent virulent forms of MPXV, limited orthopoxvirus vaccines repurposed for prevention of mpox, very narrow arsenal of antivirals effective against MPXV, and the emergence of viral variants with increased resistance to TPOXX, currently the only drug available for use to treat mpox. Hence, the development of novel therapeutics to treat mpox represents an important unmet clinical need. In this study we have presented evidence supporting the use of two safe drugs, MMF and the NMT inhibitor IMP-1088, either alone or in combination with TPOXX, as potential therapeutic strategies against MPXV. We [18] and others [51] have shown that MMF inhibits multiplication of VACV and MPXV, and the use of MMF as an antiviral has been successful in treating other viral infections [35,36]. Evidence indicates that this antiviral activity of MMF is mediated by the ability of its metabolite mycophenolic acid to inhibit IMPDH, a key enzyme in purine biosynthesis and DNA replication [34]. MMF is an immunosuppressant that is indicated for transplant recipients as prophylactic therapy [52], however, other indications in dermatological and rheumatological disease are well established [53,54]. The immunosuppressive effect of MMF is caused by its cytostatic effect on lymphocytes due to prolonged use, which is related to the high dependence of activated lymphocytes on robust IMPDH activity to generate the high purine levels required for active lymphocyte proliferation. Accordingly, no differences in lymphocyte cell count were observed between MMF-treated and control groups at day 3 post-treatment [47,48], which provides a therapeutic window for the antiviral use of MMF with limited cytostatic effect on lymphocytes. The use of MMF in combination therapy allows effective lower doses of MMF, and short treatment duration, which will further contribute to minimize MMF immunosuppressive activity while maintaining its potent antiviral activity. Using the myristoylator bioinformatic tool that predicts potential myristoylation signals in proteins [56,57] we identified four proteins in VACV and MPXV containing predicted myristoylation sites, including MPXVgp079, the counterpart of VACV G9R; MPXVgp080, the IMV membrane protein homologous to VACV L1R; MPXVgp127, the counterpart of VACV A16L. However, in contrast to the strong myristoylation signal exhibited by VACV E7R, its MPXV counterpart gp055, also known as F6R, is not predicted to be myristoylated. Likewise, MPXVgp039, but not its VACV counterpart F7L, shows probability of myristoylation. (
Table S2). The findings from our bioinformatics analysis are consistent with published data on acylation of VACV proteins [58]. G9R [29,59], A16L [60], and L1R [61,62] proteins are components of the VACV entry-fusion complex and essential for virus multiplication, and the NMT inhibitor IMP-1088 potently inhibited VACV multiplication via targeting mainly myristoylation of the L1R protein [29,59]. Multiplication of VACV may require myristoylation of specific host cell proteins, and IMP-1088 mediated inhibition of this process may contribute to the anti-VACV activity of IMP-1088. The PCLX-001 NMT-inhibitor has successfully completed phase I clinical trial and advanced toward phase II trial [48,49,50], supporting the safety of NMT-inhibitors.
Combination therapy of antivirals with synergistic effects can pose a high genetic barrier to the emergence of drug-resistant viral variants that often jeopardize monotherapy approaches, as well as facilitate the use of reduced drug doses within therapeutic range and alleviate, or prevent, side effects associated with high drug doses using in monotherapy. Combination antiviral therapy can also help to counteract the ability of many viruses, including orthopoxviruses [63,64,65], to evade host cell antiviral pathways such as interferon (IFN), RNAse L, and PKR [66,67,68].
The success of combination therapy is illustrated by the current therapeutic approaches to treat HIV and HCV infections involving combinations of two to four antivirals targeting different steps of the virus life cycle [69,70,71,72,73], which promotes synergistic antiviral effects [74,75] that raise a genetic barrier to the emergence of drug-resistant viruses [76,77]. This situation is particularly relevant to MPXV infections of immunocompromised individuals, where prolonged virus multiplication can favor selection and human-to-human propagation of drug-resistant MPXV variants.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, H.W. and J.C.T.; methodology, H.W., B.C.; and E.C. software, H.W.; validation, H.W., B.C, and E.C.; formal analysis, H.W. and J.C.T.; investigation, H.W. and J.C.T; resources, J.C.T. and L.S.; writing—original draft preparation, H.W. and R.K.; writing—review and editing, H.W., R.K., B.C., E.C., M.G., M.L., R.B., L.S., J.C.T.; visualization, H.W..; supervision, H.W. and J.C.T.; project administration, H.W..; funding acquisition, L.S. and J.C.T. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This work was supported by grant UM1 TR004407 Pilot Award 2023. We want to thank BEI Resources for providing MPXV USA-2003 (NR-2500) and the rabbit polyclonal antibody against VACV A33R (NR-628) protein. EMC was supported by the University of Texas Health San Antonio Initiative to Maximize Student Development (IMSD) T32 Program.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Günthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2022 Update of the Drug Resistance Mutations in HIV-1. Top Antivir Med 2022, 30, 559–574. [Google Scholar]
- Boden, D.; Hurley, A.; Zhang, L.; Cao, Y.; Guo, Y.; Jones, E.; Tsay, J.; Ip, J.; Farthing, C.; Limoli, K.; et al. HIV-1 Drug Resistance in Newly Infected Individuals. JAMA 1999, 282, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, H.; Liu, X.; Iketani, S.; Lin, M.; Zhang, X.; Bian, Q.; Wang, H.; Sun, H.; Hong, S.J.; et al. Molecular Mechanisms of SARS-CoV-2 Resistance to Nirmatrelvir. Nature 2023, 622, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Wyles, D.L.; Luetkemeyer, A.F. Understanding Hepatitis C Virus Drug Resistance: Clinical Implications for Current and Future Regimens. Top Antivir Med 2017, 25, 103–109. [Google Scholar]
- Duan, S.; Govorkova, E.A.; Bahl, J.; Zaraket, H.; Baranovich, T.; Seiler, P.; Prevost, K.; Webster, R.G.; Webby, R.J. Epistatic Interactions between Neuraminidase Mutations Facilitated the Emergence of the Oseltamivir-Resistant H1N1 Influenza Viruses. Nat Commun 2014, 5, 5029. [Google Scholar] [CrossRef]
- Staff, G.B. Disappointing Results for Tecovirimat Mpox Trial in the DRC. Global Biodefense 2024.
- Lenharo, M. Hopes Dashed for Drug Aimed at Monkeypox Virus Spreading in Africa. Nature 2024, 632, 965–965. [Google Scholar] [CrossRef]
- NIH Study Finds Tecovirimat Was Safe but Did Not Improve Mpox Resolution or Pain Available online:. Available online: https://www.nih.gov/news-events/news-releases/nih-study-finds-tecovirimat-was-safe-did-not-improve-mpox-resolution-or-pain (accessed on 10 December 2024).
- Smith, T.G.; Gigante, C.M.; Wynn, N.T.; Matheny, A.; Davidson, W.; Yang, Y.; Condori, R.E.; O’Connell, K.; Kovar, L.; Williams, T.L.; et al. Tecovirimat Resistance in Mpox Patients, United States, 2022–2023 - Volume 29, Number 12—23 - Emerging Infectious Diseases Journal - CDC. 20 December. [CrossRef]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb Perspect Biol 2013, 5, a010199. [Google Scholar] [CrossRef]
- Smith, G.L.; Vanderplasschen, A.; Law, M. The Formation and Function of Extracellular Enveloped Vaccinia Virus. J Gen Virol 2002, 83, 2915–2931. [Google Scholar] [CrossRef] [PubMed]
- Honeychurch, K.M.; Yang, G.; Jordan, R.; Hruby, D.E. The Vaccinia Virus F13L YPPL Motif Is Required for Efficient Release of Extracellular Enveloped Virus. Journal of Virology 2007, 81, 7310–7315. [Google Scholar] [CrossRef]
- Yang, G.; Pevear, D.C.; Davies, M.H.; Collett, M.S.; Bailey, T.; Rippen, S.; Barone, L.; Burns, C.; Rhodes, G.; Tohan, S.; et al. An Orally Bioavailable Antipoxvirus Compound (ST-246) Inhibits Extracellular Virus Formation and Protects Mice from Lethal Orthopoxvirus Challenge. J Virol 2005, 79, 13139–13149. [Google Scholar] [CrossRef] [PubMed]
- Blasco, R.; Moss, B. Role of Cell-Associated Enveloped Vaccinia Virus in Cell-to-Cell Spread. J Virol 1992, 66, 4170–4179. [Google Scholar] [CrossRef]
- Bryk, P.; Brewer, M.G.; Ward, B.M. Vaccinia Virus Phospholipase Protein F13 Promotes Rapid Entry of Extracellular Virions into Cells. J Virol 2018, 92, e02154–17. [Google Scholar] [CrossRef]
- Blasco, R.; Moss, B. Extracellular Vaccinia Virus Formation and Cell-to-Cell Virus Transmission Are Prevented by Deletion of the Gene Encoding the 37,000-Dalton Outer Envelope Protein. J Virol 1991, 65, 5910–5920. [Google Scholar] [CrossRef] [PubMed]
- Verardi, P.H.; Titong, A.; Hagen, C.J. A Vaccinia Virus Renaissance. Hum Vaccin Immunother 2012, 8, 961–970. [Google Scholar] [CrossRef]
- Chiem, K.; Nogales, A.; Lorenzo, M.; Morales Vasquez, D.; Xiang, Y.; Gupta, Y.K.; Blasco, R.; de la Torre, J.C.; Martínez-Sobrido, L. Identification of In Vitro Inhibitors of Monkeypox Replication. Microbiology Spectrum 2023, 11, e04745–22. [Google Scholar] [CrossRef] [PubMed]
- Duraffour, S.; Lorenzo, M.M.; Zöller, G.; Topalis, D.; Grosenbach, D.; Hruby, D.E.; Andrei, G.; Blasco, R.; Meyer, H.; Snoeck, R. ST-246 Is a Key Antiviral to Inhibit the Viral F13L Phospholipase, One of the Essential Proteins for Orthopoxvirus Wrapping. J Antimicrob Chemother 2015, 70, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, J.M.; Hemarajata, P.; Espinosa, A.; Hacker, J.K.; Wynn, N.T.; Smith, T.G.; Gigante, C.M.; Davidson, W.; Vega, J.; Edmondson, H.; et al. Community Spread of a Human Monkeypox Virus Variant with a Tecovirimat Resistance-Associated Mutation. Antimicrobial Agents and Chemotherapy 2023, 67, e00972–23. [Google Scholar] [CrossRef]
- Garrigues, J.M.; Hemarajata, P.; Karan, A.; Shah, N.K.; Alarcón, J.; Marutani, A.N.; Finn, L.; Smith, T.G.; Gigante, C.M.; Davidson, W.; et al. Identification of Tecovirimat Resistance-Associated Mutations in Human Monkeypox Virus - Los Angeles County. Antimicrobial Agents and Chemotherapy 2023, 67, e00568–23. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.G.; Gigante, C.M.; Wynn, N.T.; Matheny, A.; Davidson, W.; Yang, Y.; Condori, R.E.; O’Connell, K.; Kovar, L.; Williams, T.L.; et al. Tecovirimat Resistance in Mpox Patients, United States, 2022–2023 - Volume 29, Number 12—23 - Emerging Infectious Diseases Journal - CDC. 20 December. [CrossRef]
- Vakaniaki, E.H.; Kacita, C.; Kinganda-Lusamaki, E.; O’Toole, Á.; Wawina-Bokalanga, T.; Mukadi-Bamuleka, D.; Amuri-Aziza, A.; Malyamungu-Bubala, N.; Mweshi-Kumbana, F.; Mutimbwa-Mambo, L.; et al. Sustained Human Outbreak of a New MPXV Clade I Lineage in Eastern Democratic Republic of the Congo. Nat Med 2024, 1–5. [Google Scholar] [CrossRef]
- Masirika, L.M.; Udahemuka, J.C.; Schuele, L.; Ndishimye, P.; Otani, S.; Mbiribindi, J.B.; Marekani, J.M.; Mambo, L.M.; Bubala, N.M.; Boter, M.; et al. Ongoing Mpox Outbreak in Kamituga, South Kivu Province, Associated with Monkeypox Virus of a Novel Clade I Sub-Lineage, Democratic Republic of the Congo, 2024. Euro Surveill 2024, 29, 2400106. [Google Scholar] [CrossRef] [PubMed]
- Mpox - Sweden Available online:. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON531 (accessed on 31 August 2024).
- Ett fall av mpox klad 1 rapporterat i Sverige Available online:. Available online: https://www.folkhalsomyndigheten.se/nyheter-och-press/nyhetsarkiv/2024/augusti/ett-fall-av-mpox-klad-i-rapporterat-i-sverige/ (accessed on 31 August 2024).
- Ramljak, I.C.; Stanger, J.; Real-Hohn, A.; Dreier, D.; Wimmer, L.; Redlberger-Fritz, M.; Fischl, W.; Klingel, K.; Mihovilovic, M.D.; Blaas, D.; et al. Cellular N-Myristoyltransferases Play a Crucial Picornavirus Genus-Specific Role in Viral Assembly, Virion Maturation, and Infectivity. PLOS Pathogens 2018, 14, e1007203. [Google Scholar] [CrossRef]
- Witwit, H.; Betancourt, C.A.; Cubitt, B.; Khafaji, R.; Kowalski, H.; Jackson, N.; Ye, C.; Martinez-Sobrido, L.; de la Torre, J.C. Cellular N-Myristoyl Transferases Are Required for Mammarenavirus Multiplication. Viruses 2024, 16, 1362. [Google Scholar] [CrossRef] [PubMed]
- Priyamvada, L.; Kallemeijn, W.W.; Faronato, M.; Wilkins, K.; Goldsmith, C.S.; Cotter, C.A.; Ojeda, S.; Solari, R.; Moss, B.; Tate, E.W.; et al. Inhibition of Vaccinia Virus L1 N-Myristoylation by the Host N-Myristoyltransferase Inhibitor IMP-1088 Generates Non-Infectious Virions Defective in Cell Entry. PLoS Pathog 2022, 18, e1010662. [Google Scholar] [CrossRef] [PubMed]
- Chiem, K.; Lorenzo, M.M.; Rangel-Moreno, J.; Garcia-Hernandez, M.D.L.L.; Park, J.-G.; Nogales, A.; Blasco, R.; Martínez-Sobrido, L. Bi-Reporter Vaccinia Virus for Tracking Viral Infections In Vitro and In Vivo. Microbiol Spectr 9, -21. [CrossRef]
- Lorenzo, M.M.; Sánchez-Puig, J.M.; Blasco, R. Genes A27L and F13L as Genetic Markers for the Isolation of Recombinant Vaccinia Virus. Sci Rep 2019, 9, 15684. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Cubitt, B.; Chen, E.; Hull, M.V.; Chatterjee, A.K.; Cai, Y.; Kuhn, J.H.; de la Torre, J.C. The ReFRAME Library as a Comprehensive Drug Repurposing Library to Identify Mammarenavirus Inhibitors. Antiviral Research 2019, 169, 104558. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genomics Proteomics Bioinformatics 2022, 20, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C.; Eugui, E.M. Mechanisms of Action of Mycophenolate Mofetil in Preventing Acute and Chronic Allograft Rejection. Transplantation 2005, 80, S181. [Google Scholar] [CrossRef] [PubMed]
- Sajgure, A.; Kulkarni, A.; Joshi, A.; Sajgure, V.; Pathak, V.; Melinkeri, R.; Pathak, S.; Agrawal, S.; Naik, M.; Rajurkar, M.; et al. Safety and Efficacy of Mycophenolate in COVID-19: A Nonrandomised Prospective Study in Western India. The Lancet Regional Health - Southeast Asia 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Neyts, J.; Andrei, G.; De Clercq, E. The Novel Immunosuppressive Agent Mycophenolate Mofetil Markedly Potentiates the Antiherpesvirus Activities of Acyclovir, Ganciclovir, and Penciclovir In Vitro and In Vivo. Antimicrobial Agents and Chemotherapy 1998, 42, 216–222. [Google Scholar] [CrossRef]
- Cho, J.; Yi, H.; Jang, E.Y.; Lee, M.-S.; Lee, J.-Y.; Kang, C.; Lee, C.H.; Kim, K. Mycophenolic Mofetil, an Alternative Antiviral and Immunomodulator for the Highly Pathogenic Avian Influenza H5N1 Virus Infection. Biochem Biophys Res Commun 2017, 494, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Boyle, D.B.; Coupar, B.E.H. A Dominant Selectable Marker for the Construction of Recombinant Poxviruses. Gene 1988, 65, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Falkner, F.G.; Moss, B. Escherichia Coli Gpt Gene Provides Dominant Selection for Vaccinia Virus Open Reading Frame Expression Vectors. J Virol 1988, 62, 1849–1854. [Google Scholar] [CrossRef]
- Strovel, J.W.; Jain, J.; Natarajan, P.; Lawrence, T.; Castaneda, J.; Chakiath, M.; Zuck, K.; Harding, M.W.; Kelliher, K.; Shames, B.; et al. Global Gene Expression Effects of AVN-944, a Novel Small Molecule Inhibitor of Inosine Monophosphate Dehydrogenase (IMPDH). Cancer Research 2006, 66, 736. [Google Scholar]
- Cuthbertson, C.R.; Guo, H.; Kyani, A.; Madak, J.T.; Arabzada, Z.; Neamati, N. The Dihydroorotate Dehydrogenase Inhibitor Brequinar Is Synergistic with ENT1/2 Inhibitors. ACS Pharmacol Transl Sci 2020, 3, 1242–1252. [Google Scholar] [CrossRef]
- Huang, S.; Liu, Y.; Liu, W.-Q.; Neubauer, P.; Li, J. The Nonribosomal Peptide Valinomycin: From Discovery to Bioactivity and Biosynthesis. Microorganisms 2021, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.X.; Packer, M.D.; Huang, J.; Akhmametyeva, E.M.; Kulp, S.K.; Chen, C.-S.; Giovannini, M.; Jacob, A.; Welling, D.B.; Chang, L.-S. Growth Inhibitory and Anti-Tumour Activities of OSU-03012, a Novel PDK-1 Inhibitor, on Vestibular Schwannoma and Malignant Schwannoma Cells. European Journal of Cancer 2009, 45, 1709–1720. [Google Scholar] [CrossRef]
- Zhang, S.; Suvannasankha, A.; Crean, C.D.; White, V.L.; Johnson, A.; Chen, C.-S.; Farag, S.S. OSU-03012, a Novel Celecoxib Derivative, Is Cytotoxic to Myeloma Cells and Acts through Multiple Mechanisms. Clinical Cancer Research 2007, 13, 4750–4758. [Google Scholar] [CrossRef] [PubMed]
- Rayner, J.O.; Roberts, R.A.; Kim, J.; Poklepovic, A.; Roberts, J.L.; Booth, L.; Dent, P. AR12 (OSU-03012) Suppresses GRP78 Expression and Inhibits SARS-CoV-2 Replication. Biochemical Pharmacology 2020, 182, 114227. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Aguado-Martínez, A.; Manser, V.; Wong, H.N.; Haynes, R.K.; Hemphill, A. Repurposing of Antiparasitic Drugs: The Hydroxy-Naphthoquinone Buparvaquone Inhibits Vertical Transmission in the Pregnant Neosporosis Mouse Model. Veterinary Research 2016, 47, 32. [Google Scholar] [CrossRef]
- Monteiro, L.M.; Löbenberg, R.; Barbosa, E.J.; de Araujo, G.L.B.; Sato, P.K.; Kanashiro, E.; de Araujo Eliodoro, R.H.; Rocha, M.; de Freitas, V.L.T.; Fotaki, N.; et al. Oral Administration of Buparvaquone Nanostructured Lipid Carrier Enables in Vivo Activity against Leishmania Infantum. European Journal of Pharmaceutical Sciences 2022, 169, 106097. [Google Scholar] [CrossRef] [PubMed]
- Pacylex Pharmaceuticals Announces First Patient Dosed in a Phase 2a Study of PCLX-001 in Patients with Relapsed/Refractory Non-Hodgkin Lymphoma Available online:. Available online: https://www.pacylex.com/post/pacylex-pharmaceuticals-announces-first-patient-dosed-in-a-phase-2a-study-of-pclx-001-in-patients-with-relapsed-refractory-non-hodgkin-lymphoma (accessed on 5 November 2024).
- Sangha, R.S.; Jamal, R.; Spratlin, J.L.; Kuruvilla, J.; Sehn, L.H.; Weickert, M.; Berthiaume, L.G.; Mackey, J.R. A First-in-Human, Open-Label, Phase I Trial of Daily Oral PCLX-001, an NMT Inhibitor, in Patients with Relapsed/Refractory B-Cell Lymphomas and Advanced Solid Tumors. JCO 2023, 41, e15094–e15094. [Google Scholar] [CrossRef]
- Sangha, R.; Davies, N.M.; Namdar, A.; Chu, M.; Spratlin, J.; Beauchamp, E.; Berthiaume, L.G.; Mackey, J.R. Novel, First-in-Human, Oral PCLX-001 Treatment in a Patient with Relapsed Diffuse Large B-Cell Lymphoma. Curr Oncol 2022, 29, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Hishiki, T.; Morita, T.; Akazawa, D.; Ohashi, H.; Park, E.-S.; Kataoka, M.; Mifune, J.; Shionoya, K.; Tsuchimoto, K.; Ojima, S.; et al. Identification of IMP Dehydrogenase as a Potential Target for Anti-Mpox Virus Agents. Microbiology Spectrum 2023, 11, e00566–23. [Google Scholar] [CrossRef] [PubMed]
- Cellcept.
- Mycophenolate Mofetil in Severe Atopic Dermatitis: A Review. JDDonline - Journal of Drugs in Dermatology.
- Mycophenolate Available online:. Available online: https://www.versusarthritis.org/about-arthritis/treatments/drugs/mycophenolate/ (accessed on 12 November 2024).
- He, X.; Smeets, R.L.; Koenen, H.J.P.M.; Vink, P.M.; Wagenaars, J.; Boots, A.M.H.; Joosten, I. Mycophenolic Acid-Mediated Suppression of Human CD4+ T Cells: More than Mere Guanine Nucleotide Deprivation. Am J Transplant 2011, 11, 439–449. [Google Scholar] [CrossRef]
- Bologna, G.; Yvon, C.; Duvaud, S.; Veuthey, A.-L. N-Terminal Myristoylation Predictions by Ensembles of Neural Networks. Proteomics 2004, 4, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- NMT - The Predictor Available online:. Available online: https://mendel.imp.ac.at/myristate/SUPLpredictor.htm (accessed on 20 December 2024).
- Martin, K.H.; Grosenbach, D.W.; Franke, C.A.; Hruby, D.E. Identification and Analysis of Three Myristylated Vaccinia Virus Late Proteins. J Virol 1997, 71, 5218–5226. [Google Scholar] [CrossRef]
- Ojeda, S.; Domi, A.; Moss, B. Vaccinia Virus G9 Protein Is an Essential Component of the Poxvirus Entry-Fusion Complex. J Virol 2006, 80, 9822–9830. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, S.; Senkevich, T.G.; Moss, B. Entry of Vaccinia Virus and Cell-Cell Fusion Require a Highly Conserved Cysteine-Rich Membrane Protein Encoded by the A16L Gene. J Virol 2006, 80, 51–61. [Google Scholar] [CrossRef]
- Ravanello, M.P.; Hruby, D.E. Conditional Lethal Expression of the Vaccinia Virus L1R Myristylated Protein Reveals a Role in Virion Assembly. J Virol 1994, 68, 6401–6410. [Google Scholar] [CrossRef]
- Bisht, H.; Weisberg, A.S.; Moss, B. Vaccinia Virus L1 Protein Is Required for Cell Entry and Membrane Fusion. J Virol 2008, 82, 8687–8694. [Google Scholar] [CrossRef] [PubMed]
- Rivas, C.; Gil, J.; Mĕlková, Z.; Esteban, M.; Díaz-Guerra, M. Vaccinia Virus E3L Protein Is an Inhibitor of the Interferon (i.f.n.)-Induced 2-5A Synthetase Enzyme. Virology 1998, 243, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Watson, J.C.; Jacobs, B.L. The E3L Gene of Vaccinia Virus Encodes an Inhibitor of the Interferon-Induced, Double-Stranded RNA-Dependent Protein Kinase. Proc Natl Acad Sci U S A 1992, 89, 4825–4829. [Google Scholar] [CrossRef]
- Liu, S.-W.; Katsafanas, G.C.; Liu, R.; Wyatt, L.S.; Moss, B. Poxvirus Decapping Enzymes Enhance Virulence by Preventing the Accumulation of dsRNA and the Induction of Innate Antiviral Responses. Cell Host Microbe 2015, 17, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Gargan, S.; Ahmed, S.; Mahony, R.; Bannan, C.; Napoletano, S.; O’Farrelly, C.; Borrow, P.; Bergin, C.; Stevenson, N.J. HIV-1 Promotes the Degradation of Components of the Type 1 IFN JAK/STAT Pathway and Blocks Anti-Viral ISG Induction. EBioMedicine 2018, 30, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Garaigorta, U.; Chisari, F.V. Hepatitis C Virus Blocks Interferon Effector Function by Inducing Protein Kinase R Phosphorylation. Cell Host Microbe 2009, 6, 513–522. [Google Scholar] [CrossRef]
- Witwit, H.; Khafaji, R.; Salaniwal, A.; Kim, A.S.; Cubitt, B.; Jackson, N.; Ye, C.; Weiss, S.R.; Martinez-Sobrido, L.; de la Torre, J.C. Activation of Protein Kinase Receptor (PKR) Plays a pro-Viral Role in Mammarenavirus-Infected Cells. J Virol 2024, 98, e0188323. [Google Scholar] [CrossRef] [PubMed]
- FDA-Approved HIV Medicines | NIH Available online:. Available online: https://hivinfo.nih.gov/understanding-hiv/fact-sheets/fda-approved-hiv-medicines (accessed on 10 November 2024).
- Bernstein, D.E.; Aron, J.S.; Kerr, C.A.; Flanigan, C.; Hoffmann, C.J.; Gonzalez, C.J. Treatment of Chronic Hepatitis C Virus Infection in Adults; New York State Department of Health AIDS Institute Clinical Guidelines; Johns Hopkins University: Baltimore (MD), 2023. [Google Scholar]
- Gulick, R.M.; Mellors, J.W.; Havlir, D.; Eron, J.J.; Gonzalez, C.; McMahon, D.; Richman, D.D.; Valentine, F.T.; Jonas, L.; Meibohm, A.; et al. Treatment with Indinavir, Zidovudine, and Lamivudine in Adults with Human Immunodeficiency Virus Infection and Prior Antiretroviral Therapy. N Engl J Med 1997, 337, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Jilek, B.L.; Zarr, M.; Sampah, M.E.; Rabi, S.A.; Bullen, C.K.; Lai, J.; Shen, L.; Siliciano, R.F. A Quantitative Basis for Antiretroviral Therapy for HIV-1 Infection. Nat Med 2012, 18, 446–451. [Google Scholar] [CrossRef]
- Shafran, S.D.; Shaw, D.; Charafeddine, M.; Agarwal, K.; Foster, G.R.; Abunimeh, M.; Pilot-Matias, T.; Pothacamury, R.K.; Fu, B.; Cohen, E.; et al. Efficacy and Safety Results of Patients with HCV Genotype 2 or 3 Infection Treated with Ombitasvir/Paritaprevir/Ritonavir and Sofosbuvir with or without Ribavirin (QUARTZ II-III). J Viral Hepat 2018, 25, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Rabi, S.A.; Sedaghat, A.R.; Shan, L.; Lai, J.; Xing, S.; Siliciano, R.F. A Critical Subset Model Provides a Conceptual Basis for the High Antiviral Activity of Major HIV Drugs. Sci Transl Med 2011, 3, 91ra63. [Google Scholar] [CrossRef]
- Shen, L.; Peterson, S.; Sedaghat, A.R.; McMahon, M.A.; Callender, M.; Zhang, H.; Zhou, Y.; Pitt, E.; Anderson, K.S.; Acosta, E.P.; et al. Dose-Response Curve Slope Sets Class-Specific Limits on Inhibitory Potential of Anti-HIV Drugs. Nat Med 2008, 14, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Lehár, J.; Krueger, A.S.; Avery, W.; Heilbut, A.M.; Johansen, L.M.; Price, E.R.; Rickles, R.J.; Short III, G.F.; Staunton, J.E.; Jin, X.; et al. Synergistic Drug Combinations Tend to Improve Therapeutically Relevant Selectivity. Nat Biotechnol 2009, 27, 659–666. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Williamson, P.R.; Zheng, W. Improving Therapy of Severe Infections through Drug Repurposing of Synergistic Combinations. Curr Opin Pharmacol 2019, 48, 92–98. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Dose-response of selected drugs in A549 cells infected with VACV: A) A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption the virus inoculum was aspirated and media containing serial dilutions (1:3) of the indicated drugs added to cells. At 48 hpi cells were fixed and inhibition of rVACV Nluc/GFP replication was assessed by quantifying levels of GFP expression (green line). Cell viability was determined by DAPI staining (blue line) using BioTek Cytation 5 reader. The EC50 and EC90 values for each compound were calculated using sigmoidal dose-response four parameter and log(agonist) vs response model with F value set to 10 curves, respectively, with GraphPad Prism. The horizontal dotted lines indicate 50% inhibition. Data are means and SD of viral inhibition from quadruplicate wells (n = 4). B) EC50, CC50, and Selectivity index (SI) of each compound.
Figure 1.
Dose-response of selected drugs in A549 cells infected with VACV: A) A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption the virus inoculum was aspirated and media containing serial dilutions (1:3) of the indicated drugs added to cells. At 48 hpi cells were fixed and inhibition of rVACV Nluc/GFP replication was assessed by quantifying levels of GFP expression (green line). Cell viability was determined by DAPI staining (blue line) using BioTek Cytation 5 reader. The EC50 and EC90 values for each compound were calculated using sigmoidal dose-response four parameter and log(agonist) vs response model with F value set to 10 curves, respectively, with GraphPad Prism. The horizontal dotted lines indicate 50% inhibition. Data are means and SD of viral inhibition from quadruplicate wells (n = 4). B) EC50, CC50, and Selectivity index (SI) of each compound.
Figure 2.
Synergistic effects of selected drugs in combination with TPOXX: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and medium containing TPOXX or the indicated drug combinations and concentrations added to cells, and infection allowed to proceed. At 48 h post-infection (hpi), inhibition of rVACV Nluc/GFP multiplication was assessed based on levels of GFP expression and cell viability determined by DAPI staining using the BioTek Cytation 5 plate reader. The heat maps of synergy matrices, dose-response, and three-dimensional synergy plot and scores were determined using SynergyFinder+ software, the table template was used to input the data, then the data was uploaded to the software, in the specify response step of the software, the % inhibition option for GFP signal or % viability option for DAPI were used respectively.
Figure 2.
Synergistic effects of selected drugs in combination with TPOXX: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and medium containing TPOXX or the indicated drug combinations and concentrations added to cells, and infection allowed to proceed. At 48 h post-infection (hpi), inhibition of rVACV Nluc/GFP multiplication was assessed based on levels of GFP expression and cell viability determined by DAPI staining using the BioTek Cytation 5 plate reader. The heat maps of synergy matrices, dose-response, and three-dimensional synergy plot and scores were determined using SynergyFinder+ software, the table template was used to input the data, then the data was uploaded to the software, in the specify response step of the software, the % inhibition option for GFP signal or % viability option for DAPI were used respectively.
Figure 3.
A. Synergistic anti-VACV activity of MMF or IMP-1088 in combination with TPOXX: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and the indicated drug combinations and concentrations added to the cells. At 48 hpi, inhibition of rVACV Nluc/GFP multiplication was assessed based on GFP expression levels (green signal) using Celigo machine. Screenshots, single plate of the three (plates) biological replicates is shown, were used to capture the plate results (left side panel). CCS were collected, diluted into 1:10 (total of 150µL) in DMEM containing 10% FBS and 50 µL used to infect fresh A549 cells (seeded at 4 x 104/well in 96 clear well format). After 60 minutes adsorption, virus inoculum was removed and fresh DMEM containing 10% FBS (100 µL/96 well) added to cells. At 48 hpi, cells were fixed with 4% PFA and imaged with Celigo machine (right panel), single plate of the three (plates) biological replicates is shown. B. The heat maps of synergy matrices, dose response, three-dimensional synergy plots and scores were obtained, using SynergyFinder+ software, for the three biological replicates. Barplots of the synergistic dose-response were obtained using RStudio software (version2024.04.2 Build 764). The % inhibition option was set for GFP signals.
Figure 3.
A. Synergistic anti-VACV activity of MMF or IMP-1088 in combination with TPOXX: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and the indicated drug combinations and concentrations added to the cells. At 48 hpi, inhibition of rVACV Nluc/GFP multiplication was assessed based on GFP expression levels (green signal) using Celigo machine. Screenshots, single plate of the three (plates) biological replicates is shown, were used to capture the plate results (left side panel). CCS were collected, diluted into 1:10 (total of 150µL) in DMEM containing 10% FBS and 50 µL used to infect fresh A549 cells (seeded at 4 x 104/well in 96 clear well format). After 60 minutes adsorption, virus inoculum was removed and fresh DMEM containing 10% FBS (100 µL/96 well) added to cells. At 48 hpi, cells were fixed with 4% PFA and imaged with Celigo machine (right panel), single plate of the three (plates) biological replicates is shown. B. The heat maps of synergy matrices, dose response, three-dimensional synergy plots and scores were obtained, using SynergyFinder+ software, for the three biological replicates. Barplots of the synergistic dose-response were obtained using RStudio software (version2024.04.2 Build 764). The % inhibition option was set for GFP signals.
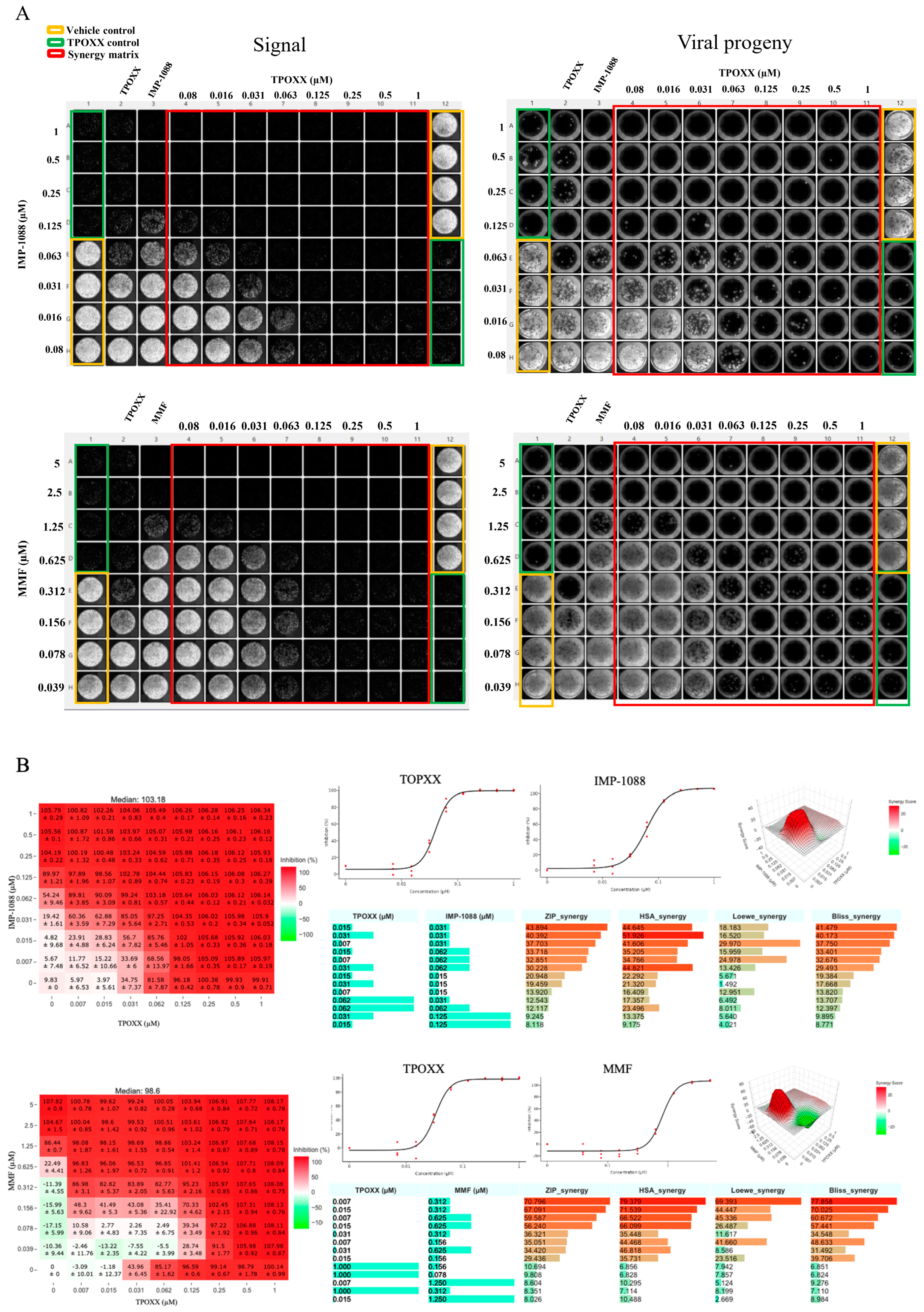
Figure 4.
Effect of combination therapy of MMF or IMP-1088 with TPOXX on VACV multi-step growth kinetics: A549 cells were seeded at 1.25 x 105/well in optical 24 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and media containing the indicated drugs and concentrations (0.05 µM TPOXX, 1.5 µM MMF and 1.5 µM IMP-1088 as single drug or combinations of TPOXX and MMF or IMP-1088) added to cells. At 48 hpi CCS were collected and cells fixed with 4% PFA. Images showing GFP expression were obtained using Keyence XZ-710 (A). Virus titers in CCS were determined by FFA using A549 cells in a 96 well plate format (B).
Figure 4.
Effect of combination therapy of MMF or IMP-1088 with TPOXX on VACV multi-step growth kinetics: A549 cells were seeded at 1.25 x 105/well in optical 24 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and media containing the indicated drugs and concentrations (0.05 µM TPOXX, 1.5 µM MMF and 1.5 µM IMP-1088 as single drug or combinations of TPOXX and MMF or IMP-1088) added to cells. At 48 hpi CCS were collected and cells fixed with 4% PFA. Images showing GFP expression were obtained using Keyence XZ-710 (A). Virus titers in CCS were determined by FFA using A549 cells in a 96 well plate format (B).
Figure 5.
Synergistic antiviral activity of MMF and IMP-1088 combination treatment: A549 cells were seeded at 4 x 104/well in 96 optical well format, next day, cells were infected rVACV Nluc/GFP at MOI of 0.01 and adsorbed for 60 minutes, a 2x of 1:2 serially dilution of the indicated drugs combined into 1x, then replaced adsorption inocula and incubated at 37oC. At 48 hpi, inhibition of rVACV Nluc/GFP viral replication was assessed with fluorescence imaging of GFP (green signal) using Celigo machine and screenshot the plate results (left side panel) and cell viability, DAPI signal, (right side panel) (A). The signal depiction obtained using BioTek Cytation 5 for both GFP and DAPI signal (B). The inhibition of rVACV Nluc/GFP viral replication was assessed by quantifying fluorescent GFP and cell viability determined by DAPI staining using BioTek Cytation 5. The heat maps of synergy matrices (C), dose response (E), three-dimensional synergy plot and scores (F), and barplot of the synergetic dose-response (G) were obtained using SynergyFinder+ software or its R code using RStudio software (version2024.04.2 Build 764), the inhibition setting was set for GFP signals. The Z’ score were calculated using the following equation: Z’=1-(3(standard deviation of vehicle control-standard deviation of negative control))/(average of vehicle control-average of negative control). These experiments were done in three biological replicates with Z` averaged 0.75 (three biological replicates with 24 technical replicates for vehicle control and negative control).
Figure 5.
Synergistic antiviral activity of MMF and IMP-1088 combination treatment: A549 cells were seeded at 4 x 104/well in 96 optical well format, next day, cells were infected rVACV Nluc/GFP at MOI of 0.01 and adsorbed for 60 minutes, a 2x of 1:2 serially dilution of the indicated drugs combined into 1x, then replaced adsorption inocula and incubated at 37oC. At 48 hpi, inhibition of rVACV Nluc/GFP viral replication was assessed with fluorescence imaging of GFP (green signal) using Celigo machine and screenshot the plate results (left side panel) and cell viability, DAPI signal, (right side panel) (A). The signal depiction obtained using BioTek Cytation 5 for both GFP and DAPI signal (B). The inhibition of rVACV Nluc/GFP viral replication was assessed by quantifying fluorescent GFP and cell viability determined by DAPI staining using BioTek Cytation 5. The heat maps of synergy matrices (C), dose response (E), three-dimensional synergy plot and scores (F), and barplot of the synergetic dose-response (G) were obtained using SynergyFinder+ software or its R code using RStudio software (version2024.04.2 Build 764), the inhibition setting was set for GFP signals. The Z’ score were calculated using the following equation: Z’=1-(3(standard deviation of vehicle control-standard deviation of negative control))/(average of vehicle control-average of negative control). These experiments were done in three biological replicates with Z` averaged 0.75 (three biological replicates with 24 technical replicates for vehicle control and negative control).
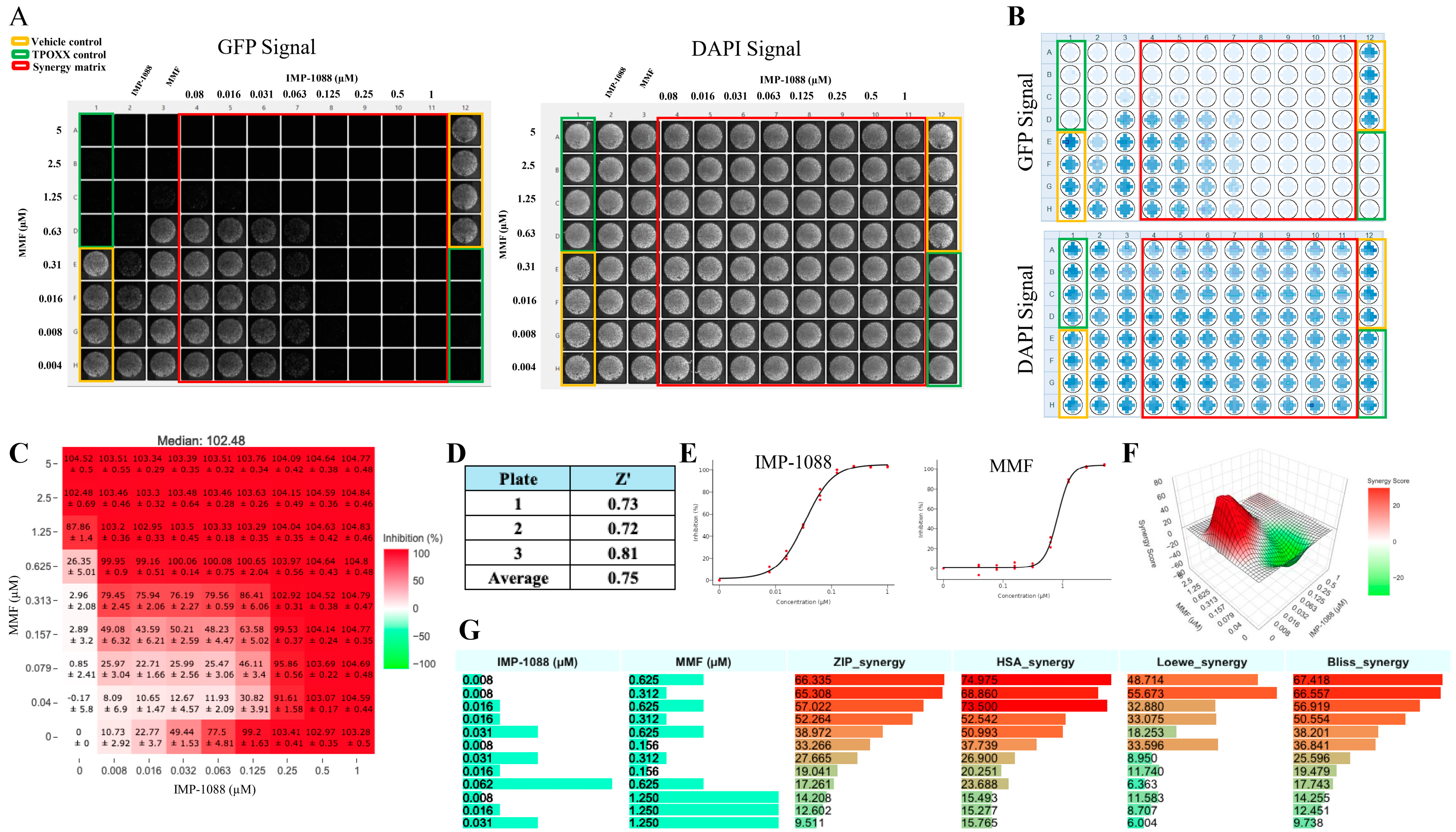
Figure 6.
Combination treatment against MPXV: A549 cells were seeded at 4 x 104/well in 96 optical well format. Next day, cells were infected with MPXV at MOI 0.01-0.02 and adsorbed for 60 minutes, a 2x of 1:2 serially dilution of the indicated drugs combined into 1x, then replaced adsorption inocula and incubated at 37oC. At 48 hpi, inhibition of MPXV replication was assessed with immunofluorescence imaging using Bioreader 7000 Fz, One of the three biological replicates plates, is shown (A). The heat maps of synergy matrices, dose response of monotherapy, three-dimensional ZIP synergy plot, Z score and barplot of the scores of four synergistic models for TPOXX and MMF (B), and IMP-1088 and MMF (C). Barplot of the synergetic dose-response were obtained using SynergyFinder+ software or its R code using RStudio software (version2024.04.2 Build 764), the inhibition setting was selected for IF signals. The Z’ score were calculated using the following equation: Z’=1-(3(standard deviation of vehicle control-standard deviation of negative control))/(average of vehicle control-average of negative control). These experiments were done in three biological replicates with Z` averaged 0.73 (for TPOXX and MMF) and 0.65 (for IMP-1088 and MMF) (three biological replicates with 24 technical replicates for vehicle control and negative control).
Figure 6.
Combination treatment against MPXV: A549 cells were seeded at 4 x 104/well in 96 optical well format. Next day, cells were infected with MPXV at MOI 0.01-0.02 and adsorbed for 60 minutes, a 2x of 1:2 serially dilution of the indicated drugs combined into 1x, then replaced adsorption inocula and incubated at 37oC. At 48 hpi, inhibition of MPXV replication was assessed with immunofluorescence imaging using Bioreader 7000 Fz, One of the three biological replicates plates, is shown (A). The heat maps of synergy matrices, dose response of monotherapy, three-dimensional ZIP synergy plot, Z score and barplot of the scores of four synergistic models for TPOXX and MMF (B), and IMP-1088 and MMF (C). Barplot of the synergetic dose-response were obtained using SynergyFinder+ software or its R code using RStudio software (version2024.04.2 Build 764), the inhibition setting was selected for IF signals. The Z’ score were calculated using the following equation: Z’=1-(3(standard deviation of vehicle control-standard deviation of negative control))/(average of vehicle control-average of negative control). These experiments were done in three biological replicates with Z` averaged 0.73 (for TPOXX and MMF) and 0.65 (for IMP-1088 and MMF) (three biological replicates with 24 technical replicates for vehicle control and negative control).
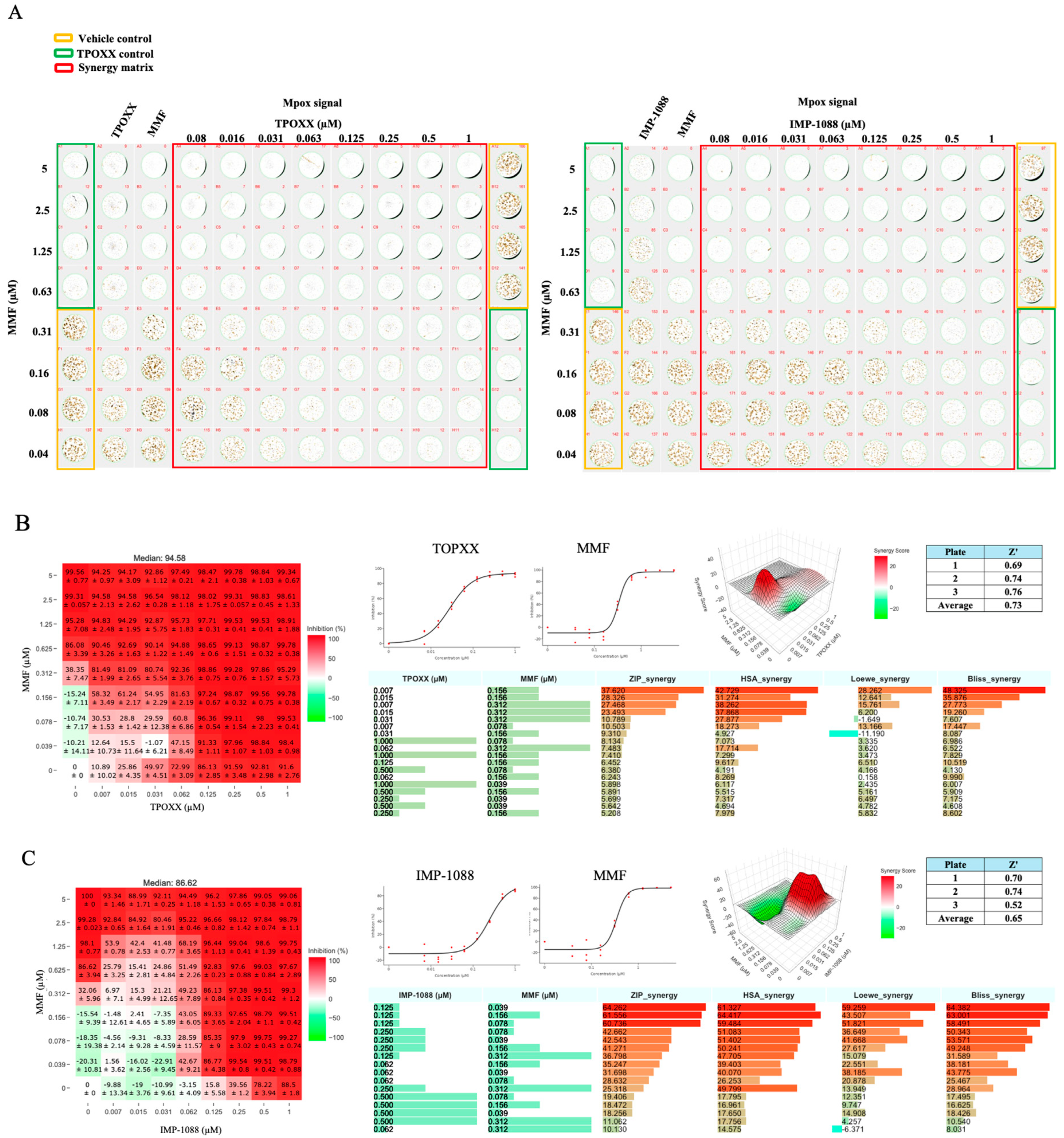
Figure 7.
Effect of triple combination therapy of TPOXX, MMF, and IMP-1088 on VACV multiplication: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and the indicated drug combinations and concentrations added to the cells. At 48 hpi, inhibition of rVACV Nluc/GFP multiplication was assessed based on GFP expression levels (green signal) using BioTek Cytation 5. A. Heatmap results and plate matrix assignment. Reference vehicle control (VC) was assigned to column 1 and 2 (rows 1-4), the dose-response with four, 1:2, serial dilutions, were assigned to columns 3 and 4 for TPOXX starting at 0.05µM, columns 5 and 6 for IMP-1088 at 0.3µM, and columns 7 and 8 for MMF at 1µM. The blocks for dual therapy were assigned to columns 1-4 and 5-8 for two replicates, where rows E-H were assigned for TPOXX and IMP-1088, rows I-L for TPOXX and MMF, and rows M-P for IMP-1088 and MMF. Columns 9-24 were segregated into 4x4 blocks, and assigned the TPOXX and MMF dual therapy, then IMP-1088 was added at the indicated doses, starting with highest dose from right to left. B. IF images of GFP (top) and DAPI (bottom) signal of the plate. C. Dose-response of monotherapy for IMP-1088, TPOXX and MMF plotted using SynergyFinder+ software. D. Synergistic models showing dose response synergistic effect as calculated by SynergyFinder+ software. E. Fluorescent and DAPI examples of mono- dual-and triple therapy of IMP-1088 (0.037µM), TPOXX (0.05µM), and MMF (0.125µM). Images were obtained with Celigo machine at 4µ pixel resolution.
Figure 7.
Effect of triple combination therapy of TPOXX, MMF, and IMP-1088 on VACV multiplication: A549 cells were seeded at 4 x 104/well in optical 96 well plates, and 16 h later infected with rVACV Nluc/GFP at MOI of 0.01. After 60 min adsorption, the virus inoculum was removed and the indicated drug combinations and concentrations added to the cells. At 48 hpi, inhibition of rVACV Nluc/GFP multiplication was assessed based on GFP expression levels (green signal) using BioTek Cytation 5. A. Heatmap results and plate matrix assignment. Reference vehicle control (VC) was assigned to column 1 and 2 (rows 1-4), the dose-response with four, 1:2, serial dilutions, were assigned to columns 3 and 4 for TPOXX starting at 0.05µM, columns 5 and 6 for IMP-1088 at 0.3µM, and columns 7 and 8 for MMF at 1µM. The blocks for dual therapy were assigned to columns 1-4 and 5-8 for two replicates, where rows E-H were assigned for TPOXX and IMP-1088, rows I-L for TPOXX and MMF, and rows M-P for IMP-1088 and MMF. Columns 9-24 were segregated into 4x4 blocks, and assigned the TPOXX and MMF dual therapy, then IMP-1088 was added at the indicated doses, starting with highest dose from right to left. B. IF images of GFP (top) and DAPI (bottom) signal of the plate. C. Dose-response of monotherapy for IMP-1088, TPOXX and MMF plotted using SynergyFinder+ software. D. Synergistic models showing dose response synergistic effect as calculated by SynergyFinder+ software. E. Fluorescent and DAPI examples of mono- dual-and triple therapy of IMP-1088 (0.037µM), TPOXX (0.05µM), and MMF (0.125µM). Images were obtained with Celigo machine at 4µ pixel resolution.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).