Submitted:
19 December 2024
Posted:
20 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
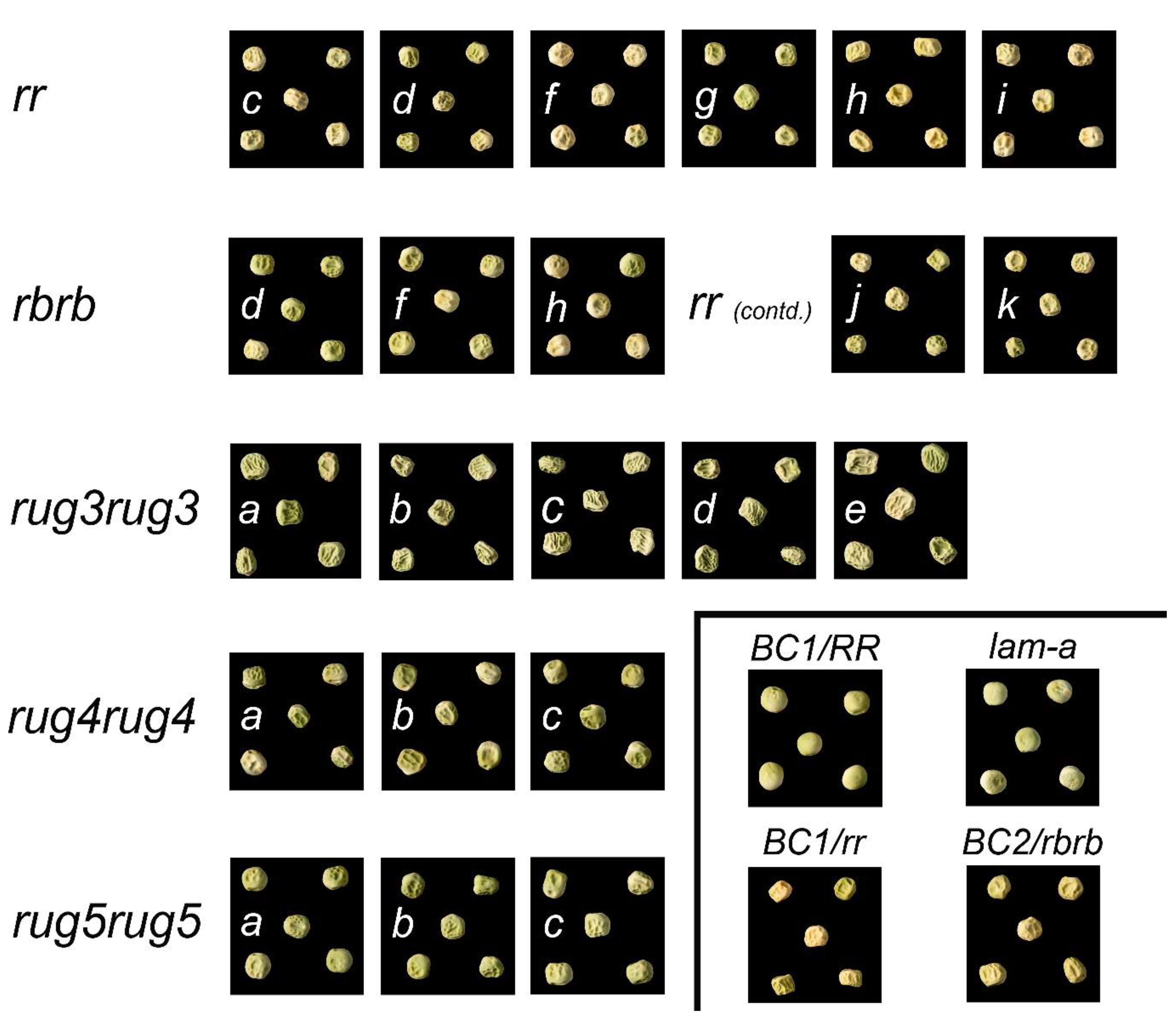
2.1. Seed Phenotypes
2.2. Unique Alleles
2.3. Protein Structure Modelling
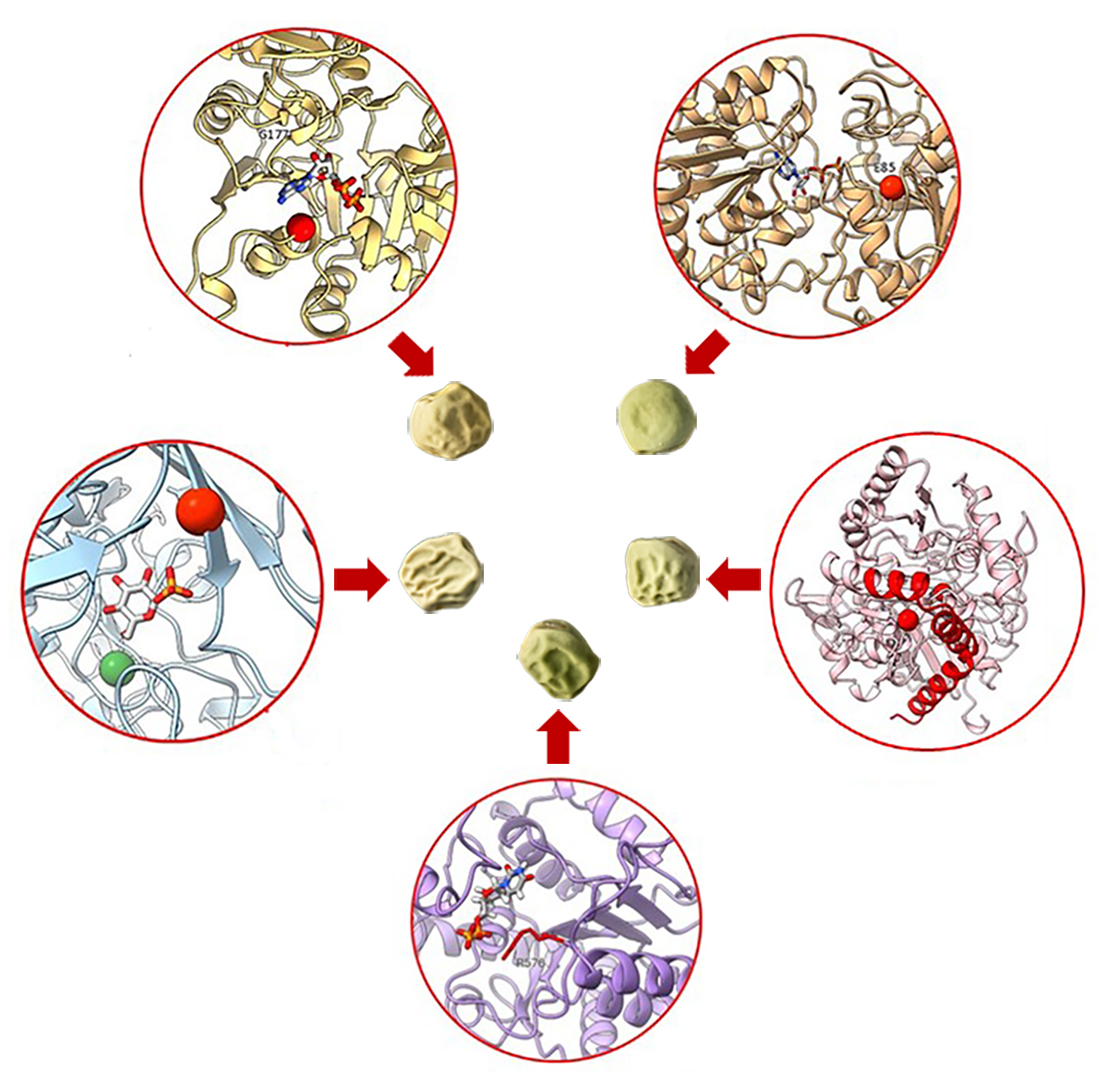
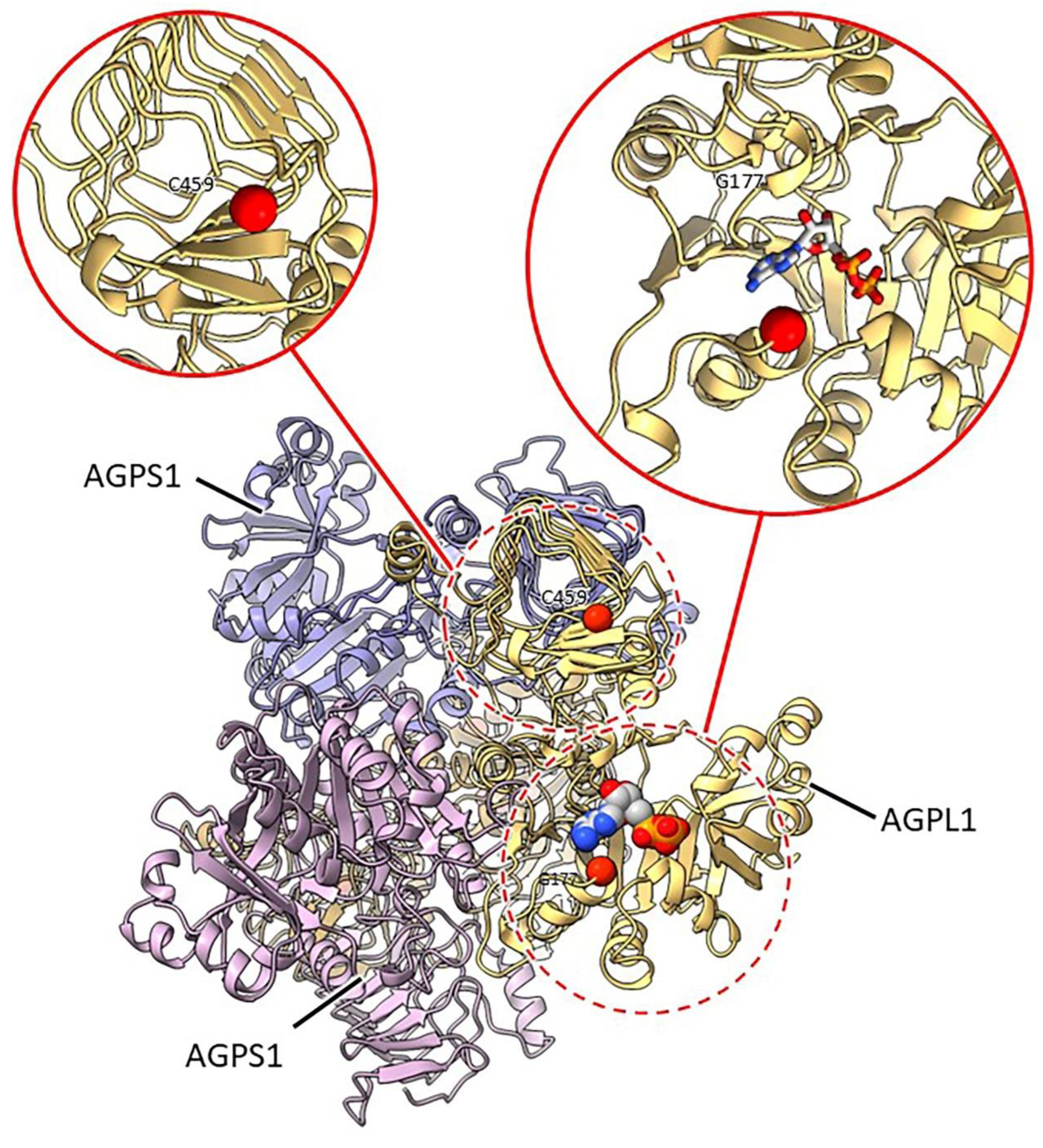
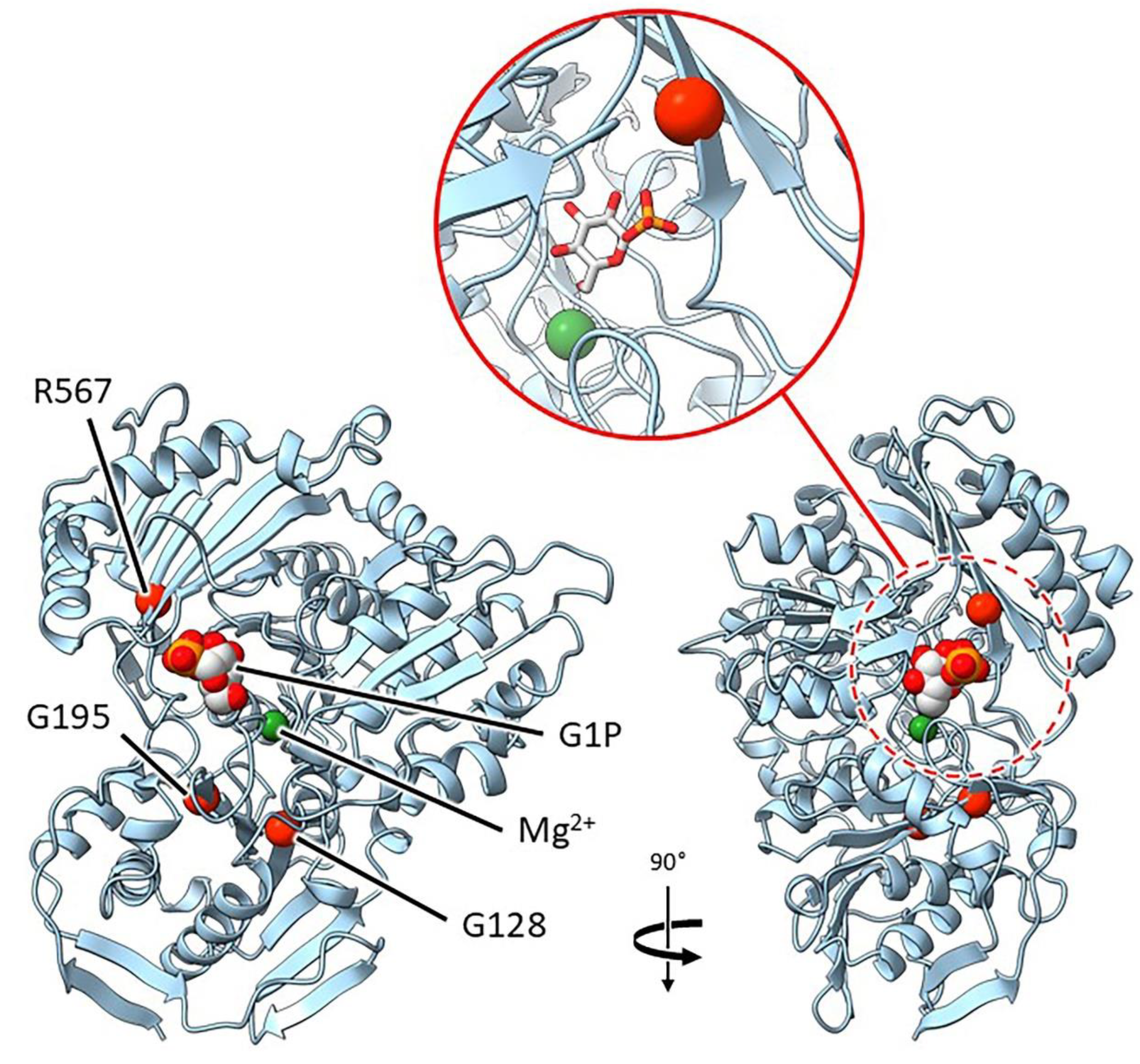
2.3.1. Mutants at the rb Locus (ADP-Glucose Pyrophosphorylase1 Large Subunit; Figure 3 and 4)
2.3.2. Mutants at the rug3 Locus (Plastidial Phosphoglucomutase; Figure 5)
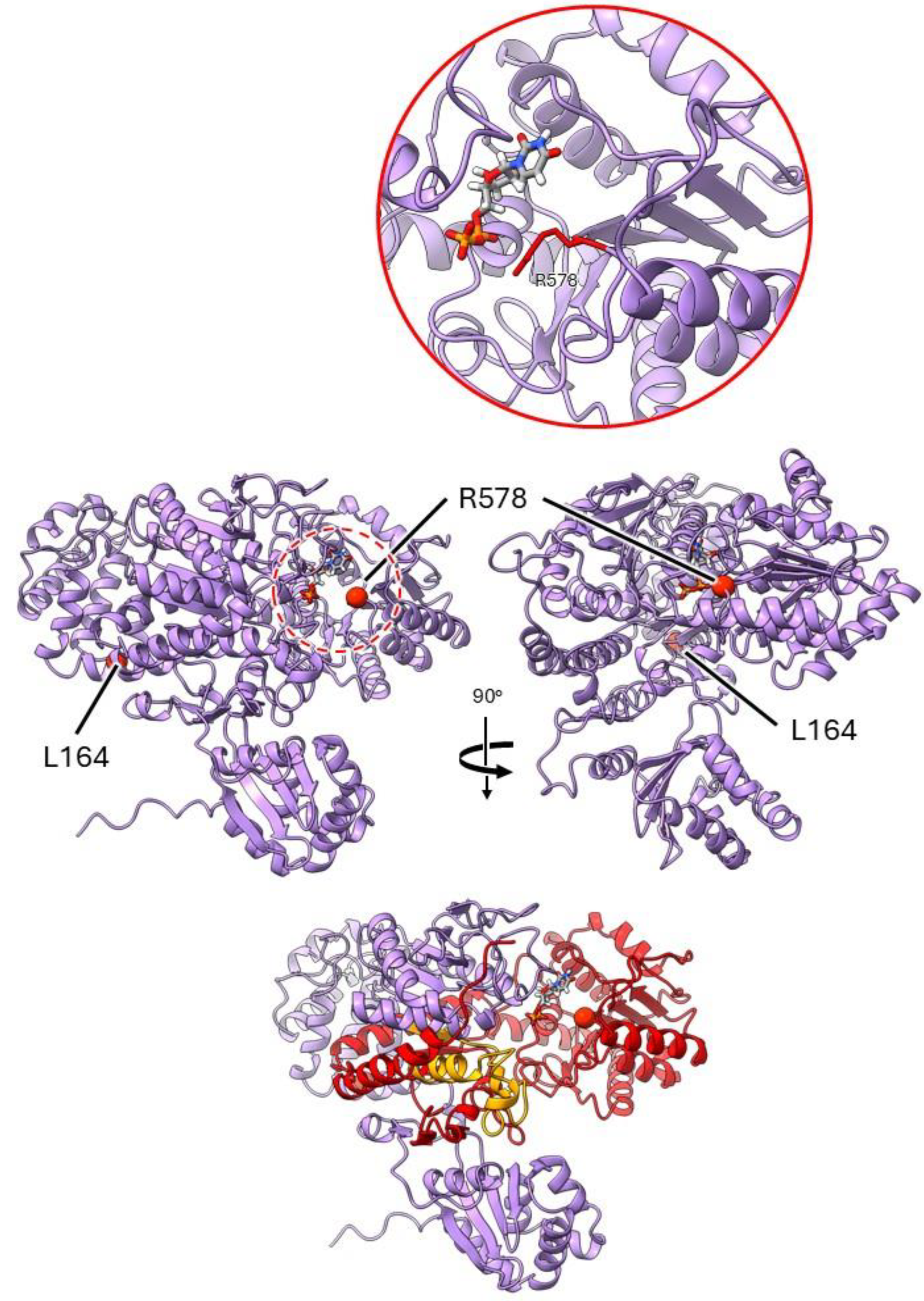
2.3.3. Mutants at the rug4 Locus (Sucrose Synthase 1; Figure 6)
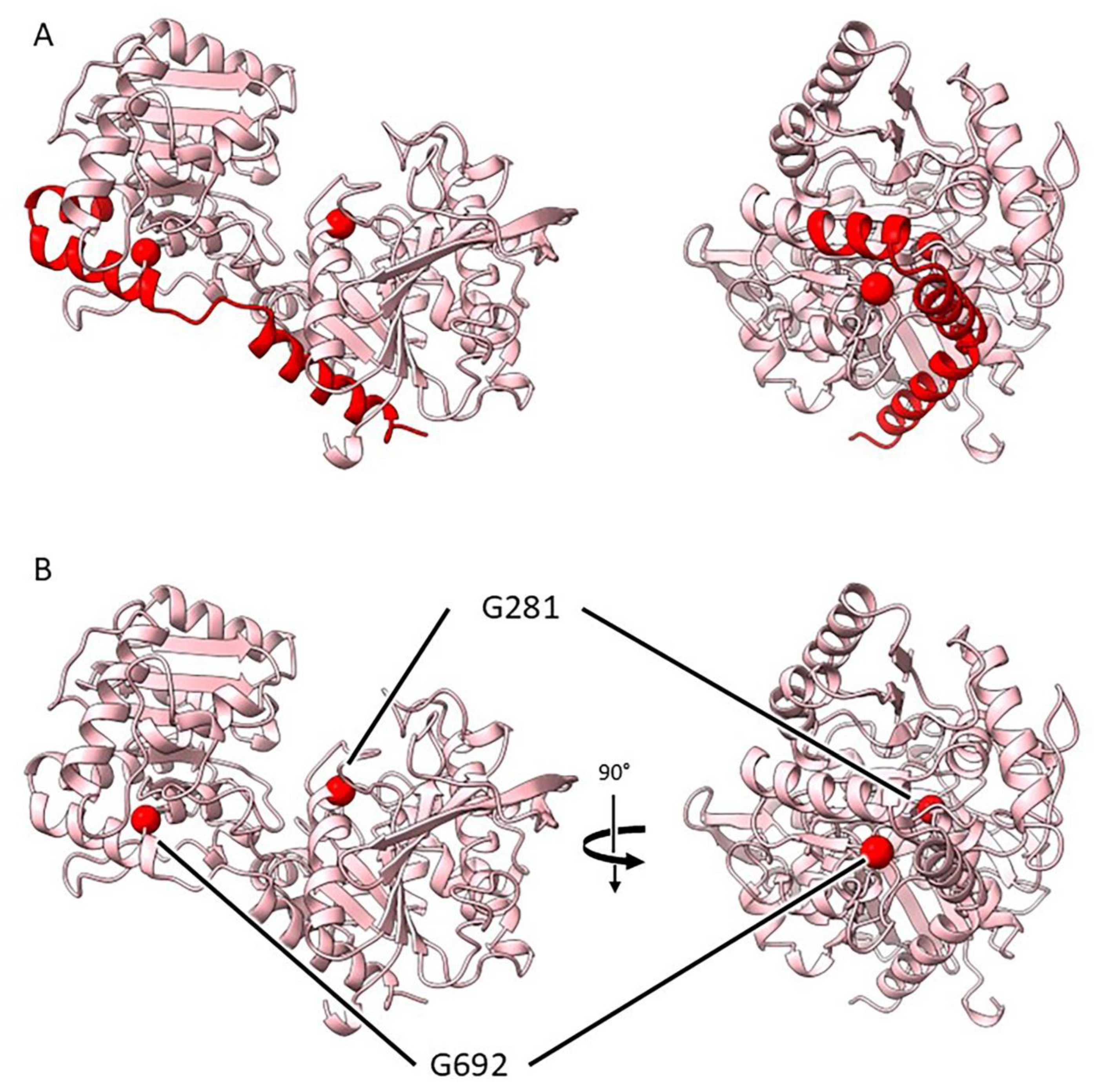
2.3.4. Mutants at the rug5 Locus (Starch Synthase 2; Figure 7)
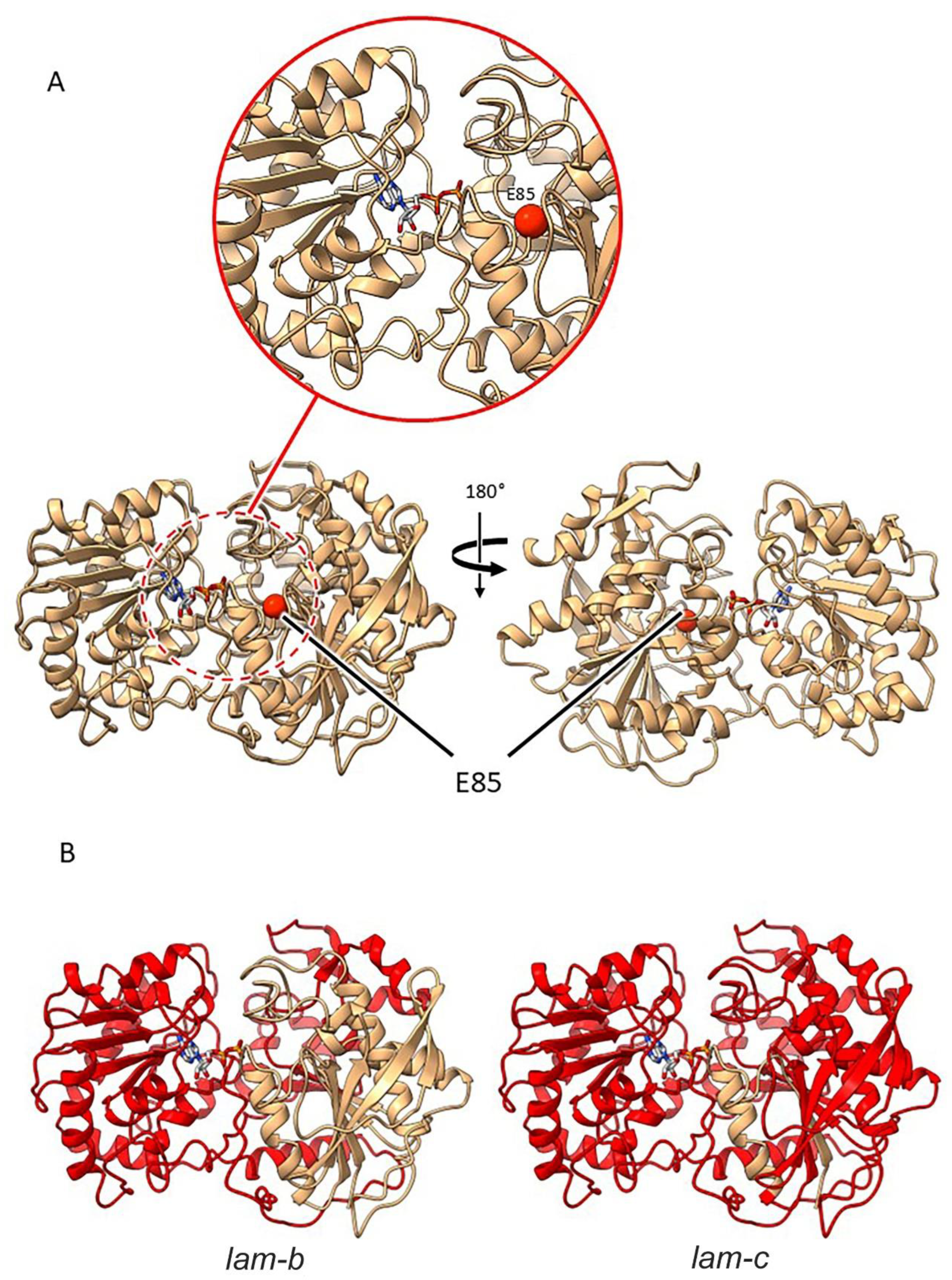
2.3.4. Mutants at the Lam Locus (Granule-Bound Starch Synthase 1; Figure 8)
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
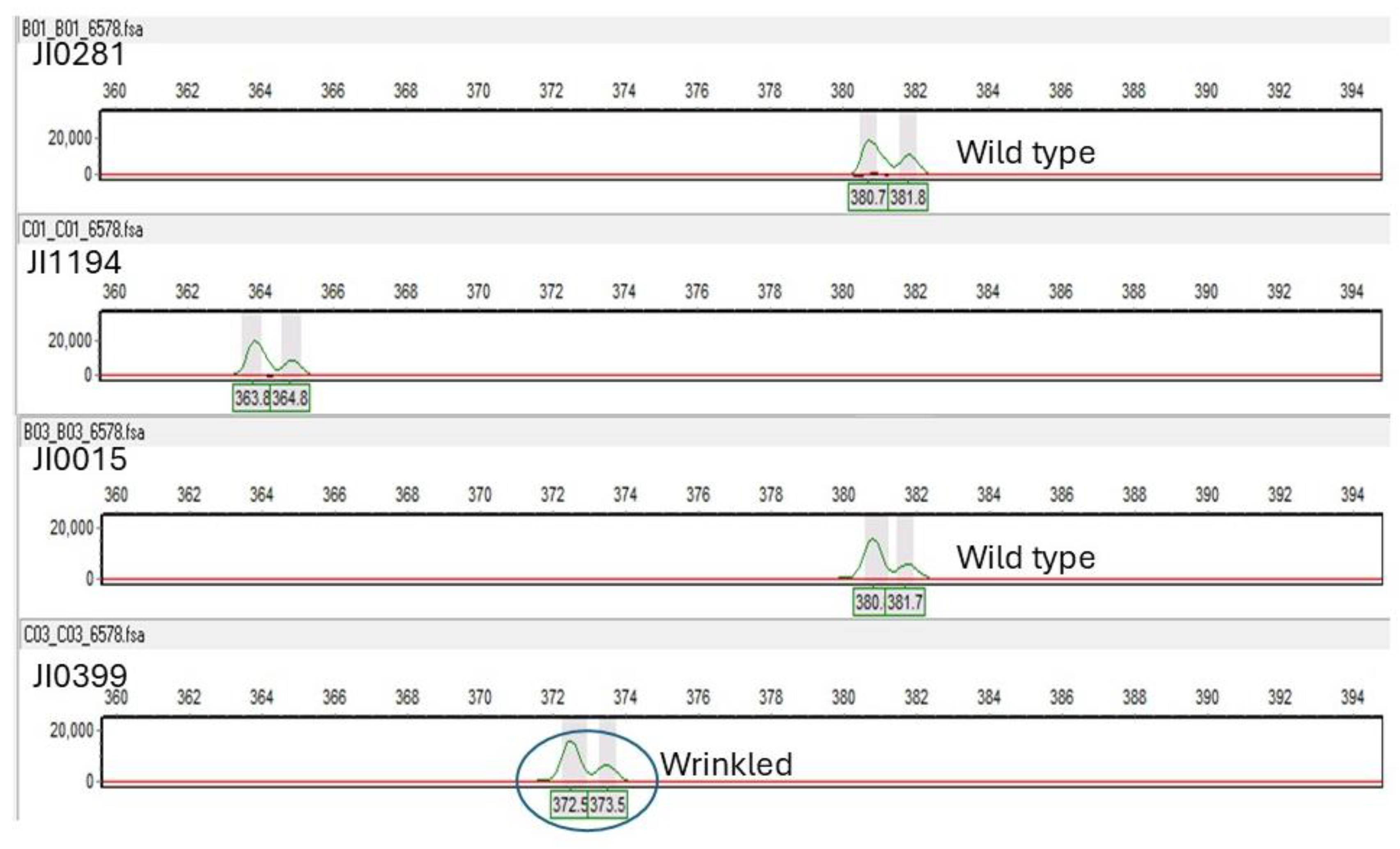
5.2. Detection of Mutations
5.3. Protein Structure Modelling
Supplementary Materials
Author Contributions
Acknowledgements
Conflicts of Interest
References
- Plant-based protein market by source (soy, wheat, pea, canola, oats, rice, potato, beans and seeds, fermented protein), type (concentrates, isolates, textured, by application (food and feed) – global forecast to 2029. MarketsandMarkets. https://www.marketsandmarkets.com/Market-Reports/plant-based-protein-market-14715651.html#:~:text=The%20global%20plant%2Dbased%20protein,2027%20in%20terms%20of%20value (accessed 16 Dec 2024).
- Aimutis, W.R. Plant-Based Proteins: The Good, Bad, and Ugly. Annu. Rev. Food Sci. Technol. 2022, 13, 1–17. [Google Scholar] [CrossRef]
- Domoney, C. Reaching for the pulse of the planet and its population. Biochemist (Lond) 2021, 43, 26–30. [Google Scholar] [CrossRef]
- McClements, D.J. ; Grossmann,L. A brief review of the science behind the design of healthy and sustainable plant-based foods. npj Sci. Food 2021, 5, 17. [Google Scholar] [CrossRef]
- Wang, T.L.; Bogracheva, T.Ya.; Hedley, C.L. Starch: as simple as A, B, C? J. Exp. Bot. 1998, 49, 481–502. [Google Scholar] [CrossRef]
- Bogracheva, T.Ya.; Cairns, P.; Noel, T.R.; Hulleman, S.; Wang, T.L.; Morris, V.J.; Ring, S.G.; Hedley, C.L. The effect of mutant genes at the r, rb, rug3, rug4, rug5 and lam loci on the granular structure and physico-chemical properties of pea seed starch. Carb. Polymers 1999, 39, 303–314. [Google Scholar] [CrossRef]
- Hedley, C.L.; Bogracheva. T.Ya.; Wang, T.L. A Genetic Approach to studying the Morphology, Structure and Function of Starch Granules using Pea as a Model. Starch/Staerke 2002, 54, 235–42. [Google Scholar] [CrossRef]
- Morales-Hernández, N. ; Mondragón-Cortéz, P; Prieto-Vázquez del Mercado, P.A. Pea starch: functionality and potential applications. In Non-Conventional Starch Sources: Properties, Functionality, and Applications, Lorenzo, J.M., Bangar, S.P., Eds.; Academic Press: London, UK, 2024; 495-521; ISBN 9780443189821. [Google Scholar]
- Fenn, D.; Wang, N.; Maximiuk, L. Physicochemical, anti-nutritional, and functional properties of air-classified protein concentrates from commercially grown Canadian yellow pea (Pisum sativum) varieties with variable protein levels. Cereal Chemistry 2022, 99, 157–168. [Google Scholar] [CrossRef]
- Farshi, P.; Mirmohammadali, S.N.; Rajpurohit, B.; Smith, J.S.; Li, Y. Pea protein and starch: Functional properties and applications in edible films J. Agri. Food Res. 2024, 15, 100927. [Google Scholar] [CrossRef]
- Petropoulou, K.; Salt, L.J.; Edwards, C.H.; Warren, F.J.; Garcia-Perez, I.; Chambers, E.S.; Alshaalan, R.; Khatib, M.; Perez-Moral, N.; Cross, K.L.; Kellingray, L.; Stanley, R.; Koev, T.; Khimyak, Y.Z.; Narbad, A.; Penney, N.; Serrano-Contreras, J.I.; Charalambides, M.N.; Miguens Blanco, J.; Castro Seoane, R.; McDonald, J.A.K.; Marchesi, J.R.; Holmes, E.; Godsland, I.F.; Morrison, D.J.; Preston, T.; Domoney, C.; Wilde, P.J.; Frost, G.S. A natural mutation in Pisum sativum L. (pea) alters starch assembly and improves glucose homeostasis in humans. Nat. Food. 2020, 1, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Olías, R.; Rayner, T.; Clemente, A.; Domoney, D, Combination of three null mutations affecting seed protein accumulation in pea (Pisum sativum L. ) impacts positively on digestibility. Food Res. Int. 2023, 169, 112825. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.K.; Desforges, N.; Selwood, C.; Smith, R.; Speirs, C.I.; Sinnaeve, G.; Gorton, P.G.; Wiseman, J.; Jumel, K.; Harding, S.E.; Hill, S.E.; Street, V.; Wang, T.L.; Hedley, C.L. Genes affecting starch biosynthesis exert pleiotropic effects on the protein content and composition of pea seeds. J. Sci. Food Agric. 2001, 81, 877–882. [Google Scholar] [CrossRef]
- Yu, B.; Xiang, D.; Mahfuz, H.; Patterson, N.; Bing, D. Understanding Starch Metabolism in Pea Seeds towards Tailoring Functionality for Value-Added Utilization. Int. J. Mol. Sci. 2021, 22, 8972. [Google Scholar] [CrossRef] [PubMed]
- Mendel, G. Versuche über pflanzen-hybriden. Verhandlungen des Naturforschenden Vereines in Brünn 1865-6,4, 3–47.
- Bhattacharyya, M.K.; Smith, A.M.; Ellis, T.H.N.; Hedley, C.; Martin, C. The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 1990, 60, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kooistra, E. On the differences between smooth and three types of wrinkled peas. Euphytica 1962, 11, 357–373. [Google Scholar] [CrossRef]
- Hylton, C.; Smith, A.M. The rb mutation of peas causes structural and regulatory changes in ADP glucose pyrophosphorylase from developing embryos. Plant Physiol. 1992, 99, 1626–1634. [Google Scholar] [CrossRef]
- Harrison, C.J.; Mould, R.M.; Leech, M.J.; Johnson, S.; Turner, L.; Schreck, S.L.; Baird, K.; Jack, P.; Rawsthorne, S.; Hedley, C.L.; Wang, T.L. The rug3 locus of pea (Pisum sativum L. ) encodes plastidial phosphoglucomutase. Plant Physiol. 2000, 122, 1187–1192. [Google Scholar]
- Harrison, C.J.; Hedley, C.L.; Wang, T.L. Evidence that the rug3 locus of pea (Pisum sativum L.) encodes plastidial phosphoglucomutase confirms that the imported substrate for starch synthesis in pea amyloplasts is glucose-6-phosphate. Plant J. 1998, 13, 753–762. [Google Scholar] [CrossRef]
- Craig, J.; Barratt, P.; Tatge, H.; Déjardin, A.; Handley, L.; Gardner, C.D.; Barber, L.; Wang, T.L.; Hedley, C.L.; Martin, C.; Smith, A.M. Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J. 1999, 17, 353–362. [Google Scholar] [CrossRef]
- Barratt, D.H.; Barber, L.; Kruger, N.J.; Smith, A.M.; Wang, T.L.; Martin, C. Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol. 2001, 127, 655–664. [Google Scholar] [CrossRef]
- Craig, J.; Lloyd, J.R.; Tomlinson, K.; Barber, L.; Edwards, A.; Wang, T.L.; Martin, C.; Hedley, C.L.; Smith, A.M. Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 1998, 10, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.; Barber, L.R.; Burton, R.; Hedley, C.L.; Hylton, C.M.; Johnson, S.; Jones, D.A.; Marshall, J.; Smith, A.M.; Tatge, H.; Tomlinson, K.; Wang, T.L. The isolation and characterisation of novel low-amylose mutants of Pisum sativum L. Plant, Cell Environ. 1995, 18, 1019–1026. [Google Scholar]
- Xing, Q.; Utami, D.P.; Demattey, M.B.; Kyriakopoulou, K.; de Wit, M.; Boom, R.M.; Schutyser, M.A.I. A two-step air classification and electrostatic separation process for protein enrichment of starch-containing legumes. Inn. Food Sci. Emerging Tech. 2020, 66, 102480. [Google Scholar] [CrossRef]
- Reichert, R. Air Classification of peas (Pisum sativum) varying widely in protein content. J. Food Sci. 2006, 47, 1263–1267. [Google Scholar] [CrossRef]
- Burstin, J.; Marget, P.; Huart, M.; Moessner, A.; Mangin, B.; Duchene, C.; Desprez, B.; Munier-Jolain, N.; Duc, G. Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiol. 2007, 144, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Warsame, A.O.; Balk, J.; Domoney, C. Identification of significant genome-wide associations and QTL underlying variation in seed protein composition in pea (Pisum sativum L.). bioRxiv 2024, 07.04.602075. [Google Scholar]
- Moreau, C.; Warren, F.J.; Rayner, T.; Perez-Moral, N.; Lawson, D.M.; Wang, T.L.; Domoney, C. An allelic series of starch-branching enzyme mutants in pea (Pisum sativum L.) reveals complex relationships with seed starch phenotypes. Carb. Polymers 2022, 288, 119386. [Google Scholar] [CrossRef]
- Wang, T.L.; Hedley, C.L. Seed development in peas: Knowing your three "r's" (or four, or five). Seed Sci. Res. 1991, 1, 3–14. [Google Scholar] [CrossRef]
- Wang, T.L.; Hedley, C.L. Seed mutants in Pisum. Pisum Genet. 1993, 25, 64–70. [Google Scholar]
- Wang, T.L.; Hadavizadeh, A.; Harwood, A.; Welham,T. J.; Harwood,W.A.; Faulks, R.; Hedley, C.L. An analysis of seed development in Pisum sativum. XIII. The chemical induction of storage product mutants. Plant Breed. 1990, 105, 311–320. [Google Scholar] [CrossRef]
- Zheng, Y.; Anderson, S.; Zhang, Y.; Garavito, R.M. The Structure of Sucrose Synthase-1 from Arabidopsis thaliana and Its Functional Implications. J. Biol. Chem. 2011, 286, 36108–36118. [Google Scholar] [CrossRef]
- Edwards, A.; Marshall, J.; Denyer, K.; Sidebottom, C.; Visser, R.G.F.; Martin, C.; Smith, A.M. Evidence that a 77-kilodalton protein from the starch of pea embryos is an isoform of starch synthase that is both soluble and granule bound. Plant Physiol. 1996, 112, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Hedley, C.L.; Wang, T.L. Seed and foliar mutations in Pisum. In Developmental Mutants in Higher Plants; Thomas, H.;Grierson, D., Eds.; Cambridge University Press, Cambridge, New York, 1987, 219-244, ISBN: 0521328446.
- Rayner, T.; Moreau, C.; Ambrose, M.; Isaac, P.G.; Ellis, N.; Domoney, C. Genetic Variation Controlling Wrinkled Seed Phenotypes in Pisum: How Lucky Was Mendel? Int. J. Mol. Sci. 2017, 18, 1205. [Google Scholar] [CrossRef] [PubMed]
- Lacey, C.N.D.; Hughes, S.G.; Harrison, C.J.; Wang, T.L.; Hedley, C.L. Method for increasing sucrose content of. plants. Patent, WO1998001574A1, 15 January 1998. [Google Scholar]
- Jones, D.A.; Arthur, A.E.; Adams, H.M.; Coxon, D.T.; Wang, T.L.; Hedley, C.L. An analysis of seed development in Pisum sativum. IX. Genetic Analysis of Lipid Content. Plant Breed. 1990, 104, 144–151. [Google Scholar] [CrossRef]
- Perez, M.D.; Chambers, S.J.; Bacon, J.R.; Lambert, N.; Hedley, C.L.; Wang, T.L. Seed protein content and composition of near-isogenic and induced mutant pea lines. Seed Sci. Res. 1993, 3, 187–194. [Google Scholar] [CrossRef]
- Casey, R.; Domoney, C.; Forster, C.; Hedley, C.L.; Hitchin, E.; Wang, T.L. The effect of modifying carbohydrate metabolism on seed protein gene expression in peas. J. Plant Physiol. 1998, 152, 636–640. [Google Scholar] [CrossRef]
- Van Oosten, J-J.M.; Kaplan, C.P. Method for increasing Vitamin C content of. plants. Patent, WO-1999053041-A2, 20 October 1999.
- Perry J, Welham T, Brachmann A, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang Tl, Parniske M. TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methane sulfonate alleles towards glycine replacements. Plant Physiol. 2009, 151, 1281–1291. [Google Scholar] [CrossRef]
- Wang, T.L.; Bogracheva, T.Ya.; Hedley, C.L. Manipulation of starch quality in seeds: a genetic approach. In Seed Biology: Advances and Applications. Proceedings of the Sixth International Workshop on Seeds.; Black, M., Bradford, K.J., Vázquez-Ramos, J., Eds.; ; Black, M., Bradford, K.J., Vázquez-Ramos, J., Eds.; CAB International: Wallingford, UK, 2000; pp. 439–448. ISBN 0851994040. [Google Scholar]
- Figueroa, C.M.; Asencion Diez, M.D.; Ballicora, M.A.; Iglesias, A.A. Structure, function, and evolution of plant ADP-glucose pyrophosphorylase. Plant Mol. Biol. 2022, 108, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, K.; Küster, H.; Rutten, T.; Fait, A.; Fernie, A.R.; Miersch, O.; Wasternack, C.; Emery, R.J.N.; Desel, C.; Hosein, F.; Müller, M. : Saalbach, I.; Weber, H. ADP-Glucose Pyrophosphorylase-Deficient Pea Embryos Reveal Specific Transcriptional and Metabolic Changes of Carbon-Nitrogen Metabolism and Stress Responses. Plant Physiol. 2009, 149, 395–411. [Google Scholar] [CrossRef]
- Bhowmik, P.; Yan, W.; Hodgins, C.; Polley, B.; Warkentin, T.; Nickerson, M.; Ro, D.K.; Marsolais, F.; Domoney, C.; Shariati-Ievari, S.; Aliani, M. CRISPR/Cas9-mediated lipoxygenase gene-editing in yellow pea leads to major changes in fatty acid and flavor profiles. Front. Plant Sci. 2023, 14, 1246905. [Google Scholar] [CrossRef]
- Hodgins, C.L.; Salama, E.M.; Kumar, R.; Zhao, Y.; Roth, S.A.; Cheung, I.Z.; Chen, J.; Arganosa, G.C.; Warkentin, T.D.; Bhowmik, P.; Ham, B.K.; Ro, D.K. Creating saponin-free yellow pea seeds by CRISPR/Cas9-enabled mutagenesis on β-amyrin synthase. Plant Direct 2024, 8, e563. [Google Scholar] [CrossRef]
- Forster, C.; North, H.; Afzal, N.; Domoney, C.; Hornostaj, A.; Robinson, D.S.; Casey, R. Molecular analysis of a null mutant for pea (Pisum sativum L.) seed lipoxygenase-2. Plant Mol. Biol. 1999, 39, 1209–1220. [Google Scholar] [CrossRef]
- Hedley, C.L.; Smith, C.; Ambrose, M.J.; Cook, S.; Wang, T.L. An analysis of seed development in Pisum sativum. II. The effect of the r locus on the growth and development of the seed. Ann. Bot. 1986, 58, 371–379. [Google Scholar] [CrossRef]
- Hofer, J.; Turner, L.; Moreau, C.; Ambrose, M.; Isaac, P.; Butcher, S.; Weller, J.; Dupin, A.; Dalmais, M.; Le Signor, C.; Bendahmane, A; Ellis, N. Tendril-less regulates tendril formation in pea leaves. Plant Cell 2009, 21, 420–428. [Google Scholar] [CrossRef] [PubMed]
- https://bio.tools/targetp.
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb.
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: making protein folding accessible to all. Nat. Meth. 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S. A. A.; Ballard, A. J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A. W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D. Nature 2021,596, 583–589.
- EMBL-EBI Protein Data Bank in Europe. PDBeFold, https://www.ebi.ac.uk/msd-srv/ssm/.
- AlphaFill AlphaFold models enriched with ligands and co-factors https://alphafill.eu/.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.L.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Rayner, T.; Saalbach, G.; Vickers, M.; Paajanen, P.; Martins, C.; Wouters, R.H.M.; Chinoy, C.; Mulholland, F.; Bal, M.; Isaac, P.; Novak, P.; Macas, J.; Ellis, N.; Steuernagel, B.; Domoney, C. Rebalancing the seed proteome following deletion of vicilin-related genes in pea (Pisum sativum L.). J. Exp. Bot. 2024 (in press).
|
Allele |
Isolation Number (Prefix− E=EMS; M=MNU) |
Line number† |
Mutation Position Genomic Protein |
||
|
r |
N/A | BC1/rr | Exon 22 | Insertion of ‘800+’bp ‘element’ | Predicted E 860 →, ⩽30 aberrant amino acids*1 |
| r-c | M(850)7065 | SIM53 | Exon 15 | G22948A | R 571 H |
| r-d | M(718)7339 | SIM54 | Exon 13 | G20772A | G 495 D |
| r-f | E499(1677) | SIM56 | Exon 18 | G25551A | W 684 * |
| r-g | E(117)22 | SIM57 | Exon 18 | G25633A | R 712 C |
| r-h | E(349)7277 | SIM58 | Intron 15 acceptor splice site | G24153A | 10 aberrant amino acids: 586-595* |
| r-i | E(625)113 | SIM59 | Intron 8 acceptor splice site | G13242A (84 nucleotide deletion: 957-1040) | 28 amino acid deletion (319-346)2 |
| r-j | E(122)1154 | SIM61 | Exon 16 | G24218A | E 607 K |
| r-k | M(32)7325 | SIM71 | Intron 16 donor splice site | G24299A | Variants include: loss of 13 amino acids (621-633)3; four aberrant amino acids (634-637)* |
| rb | N/A | BC2/rbrb | Exon 2 | 9 bp deletion (471-479) | Amino acid loss (3: ATP or TPA) |
| rb-d | E(162)1232 | SIM15 | Exon 13 | G2802A | C 459 Y |
| rb-f | E(1663)5142 | SIM101 | Exon 8 | G1924A | 307* |
| rb-h | E(1258)3892 | SIM103 | Exon 4 | G1153A | G 177 R |
| rug3-a | E(543)1836 | SIM1 | Exon 4 | G703A | G 128 D |
| rug3-b | E(626)124 | SIM32 | Exon 20 | C4367T | R 567 C |
| rug3-c | E(476)1589 | SIM41 | Exon 7 | G1479A | G 195 S |
| rug3-d | E(549)548 | SIM42 | Exon 4 | C711T | 131* |
| rug3-e | E(1780)5702 | SIM43 | Exon 17 | CT deletion 3757/3758 | S 477 C, 479* |
| rug4-a | E(620)121 | SIM11 | Exon 4 | C788T | L 164 F |
| rug4-b | E(77)16 | SIM91 | Exon 11 | G2668A | R 578 K |
| rug4-c | M(3)545 | SIM201A | Intron 8 donor splice site | G1966A | C 432 I, 37 aberrant amino acids, 468* |
| rug5-a | E(640)2239 | SIM51 | Exon 8 | G3702A | W 708 * |
| rug5-b | E(167)1236 | SIM52 | Exon 3 | G1927A | G 281 R |
| rug5-c | E(509)1728 | SIM81 | Exon 8 | G3653A | G 692 D |
| lam-a | E(1470)4322 | SIM501 | Exon 1 | G253A | E 85 K |
| lam-b | E(0.3)1514/ E(722)149 |
SIM502/512 | Intron 6 acceptor splice site | G1654A | A 254 R, 12 aberrant amino acids, 266* |
| lam-c | E(1599)4812 | SIM503/504 | Intron 1 acceptor splice site | G454A | G 108 V, 17 aberrant amino acids, 125* |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





