Submitted:
18 December 2024
Posted:
19 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bee pollen collected and beebread sampling
2.2. Processing of samples
2.3. Isolation and identification of yeasts
2.4. Potential probiotic features of isolated yeasts
3. Results
3.1. Yeast, bacterial and mold occurrence
3.2. Yeast isolates identification
3.2. Probiotic features, antimicrobial activity and safety tests
4. Discussion
5. Conclusions
Author Contributions
References
- Gilliam, M. MICROBIOLOGY OF POLLEN AND BEE BREAD : THE GENUS BACILLUS. Apidologie 1979, 10, 269–274. [Google Scholar] [CrossRef]
- Campos, M.G.R.; Bogdanov, S.; De Almeida-Muradian, L.B.; Szczesna, T.; Mancebo, Y.; Frigerio, C.; Ferreira, F. Pollen Composition and Standardisation of Analytical Methods. J. of Api. Res 2008, 47, 154–161. [Google Scholar] [CrossRef]
- Denisow, B.; Denisow-Pietrzyk, M. Biological and Therapeutic Properties of Bee Pollen: A Review. J Sci Food Agric 2016, 96, 4303–4309. [Google Scholar] [CrossRef] [PubMed]
- Mărgăoan, R.; Stranț, M.; Varadi, A.; Topal, E.; Yücel, B.; Cornea-Cipcigan, M.; Campos, M.G.; Vodnar, D.C. Bee Collected Pollen and Bee Bread: Bioactive Constituents and Health Benefits. Antioxidants 2019, 8, 568. [Google Scholar] [CrossRef]
- Zakaria, Z.; Othman, Z.A.; Suleiman, J.; Nna, V.; Mohamed, M. Pollen and Bee Bread and Liver Health. In; 2022; pp. 283–314 ISBN 978-0-323-85400-9.
- Végh, R.; Csóka, M.; Sörös, C.; Sipos, L. Food Safety Hazards of Bee Pollen – A Review. Trends in Food Science & Technology 2021, 114, 490–509. [Google Scholar] [CrossRef]
- Fuenmayor B, C.; Zuluaga D, C.; Díaz M, C.; Quicazán De C, M.; Cosio, M.; Mannino, S. Evaluation of the Physicochemical and Functional Properties of Colombian Bee Pollen. Rev MVZ Córdoba 2014, 19, 4003–4014. [Google Scholar] [CrossRef]
- Kieliszek, M.; Piwowarek, K.; Kot, A.M.; Błażejak, S.; Chlebowska-Śmigiel, A.; Wolska, I. Pollen and Bee Bread as New Health-Oriented Products: A Review. Trends in Food Science & Technology 2018, 71, 170–180. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored Pollen of Honey Bees: Many Lines of Evidence Are Consistent with Pollen Preservation, Not Nutrient Conversion. Molecular Ecology 2014, 23, 5904–5917. [Google Scholar] [CrossRef] [PubMed]
- Detry, R.; Simon-Delso, N.; Bruneau, E.; Daniel, H.-M. Specialisation of Yeast Genera in Different Phases of Bee Bread Maturation. Microorganisms 2020, 8, 1789. [Google Scholar] [CrossRef]
- Gilliam, M. Identification and Roles of Non-Pathogenic Microflora Associated with Honey Bees. FEMS Microbiology Letters 1997, 155, 1–10. [Google Scholar] [CrossRef]
- Stringini, M.; Comitini, F.; Taccari, M.; Ciani, M. Yeast Diversity in Crop-Growing Environments in Cameroon. International Journal of Food Microbiology 2008, 127, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B. Identification of Yeasts by RFLP Analysis of the 5.85 rRNA Gene and the Two Ribosomal Internal Transcribed Spacers. International Journal of Systematic Bacteriology.
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Research 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Agarbati, A.; Moretti, L.; Canonico, L.; Ciani, M.; Comitini, F. Agro-Ecosystem of Honeybees as Source for Native Probiotic Yeasts. World J Microbiol Biotechnol 2024, 40, 147. [Google Scholar] [CrossRef]
- Perricone, M.; Bevilacqua, A.; Altieri, C.; Sinigaglia, M.; Corbo, M. Challenges for the Production of Probiotic Fruit Juices. Beverages 2015, 1, 95–103. [Google Scholar] [CrossRef]
- Speranza, B. Biofilm Formation by Potentially Probiotic Saccharomyces Cerevisiae Strains. Food Microbiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Ogunremi, O.R.; Agrawal, R.; Sanni, A. Production and Characterization of Volatile Compounds and Phytase from Potentially Probiotic Yeasts Isolated from Traditional Fermented Cereal Foods in Nigeria. Journal of Genetic Engineering and Biotechnology 2020, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Lei, X.G. An Improved Method for a Rapid Determination of Phytase Activity in Animal Feed1. Journal of Animal Science 2005, 83, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Agarbati, A.; Canonico, L.; Marini, E.; Zannini, E.; Ciani, M.; Comitini, F. Potential Probiotic Yeasts Sourced from Natural Environmental and Spontaneous Processed Foods. Foods 2020, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Pereira, W.A.; Mendonça, C.M.N.; Urquiza, A.V.; Marteinsson, V.Þ.; LeBlanc, J.G.; Cotter, P.D.; Villalobos, E.F.; Romero, J.; Oliveira, R.P.S. Use of Probiotic Bacteria and Bacteriocins as an Alternative to Antibiotics in Aquaculture. Microorganisms 2022, 10, 1705. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic Mechanisms of Action. Ann Nutr Metab 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Czerucka, D.; Piche, T.; Rampal, P. Review Article: Yeast as Probiotics – Saccharomyces Boulardii. Aliment Pharmacol Ther 2007, 26, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Kiepś, J.; Dembczyński, R. Current Trends in the Production of Probiotic Formulations. Foods 2022, 11, 2330. [Google Scholar] [CrossRef]
- Staniszewski, A.; Kordowska-Wiater, M. Probiotic and Potentially Probiotic Yeasts—Characteristics and Food Application. 2021.
- Baky, M.H.; Abouelela, M.B.; Wang, K.; Farag, M.A. Bee Pollen and Bread as a Super-Food: A Comparative Review of Their Metabolome Composition and Quality Assessment in the Context of Best Recovery Conditions. Molecules 2023, 28, 715. [Google Scholar] [CrossRef] [PubMed]
- Agarbati, A.; Gattucci, S.; Canonico, L.; Ciani, M.; Comitini, F. Yeast Communities Related to Honeybees: Occurrence and Distribution in Flowers, Gut Mycobiota, and Bee Products. Appl Microbiol Biotechnol 2024, 108, 175. [Google Scholar] [CrossRef]
- Giger-Reverdin, S.; Bezault, N.; Sauvant, D.; Bertin, G. Effects of a Probiotic Yeast in Lactating Ruminants: Interaction with Dietary Nitrogen Level. Animal Feed Science and Technology 1996, 63, 149–162. [Google Scholar] [CrossRef]
- Busscher, H.J. Effect of Probiotic Bacteria on Prevalence of Yeasts in Oropharyngeal Bio®lms on Silicone Rubber Voice Prostheses in Vitro.
- Gusils, C.; Chaia, A.P.; González, S.; Oliver, G. Lactobacilli Isolated from Chicken Intestines: Potential Use as Probiotics. Journal of Food Protection 1999, 62, 252–256. [Google Scholar] [CrossRef]
- Jakobsen, M.; Narvhus, J. Yeasts and Their Possible Beneficial and Negative Effects on the Quality of Dairy Products. International Dairy Journal 1996, 6, 755–768. [Google Scholar] [CrossRef]
- Pozo, M.I.; Van Kemenade, G.; Van Oystaeyen, A.; Aledón-Catalá, T.; Benavente, A.; Van Den Ende, W.; Wäckers, F.; Jacquemyn, H. The Impact of Yeast Presence in Nectar on Bumble Bee Behavior and Fitness. Ecological Monographs 2020, 90, e01393. [Google Scholar] [CrossRef]
- Rosa, C.; Lachance, M.; Silva, J.; Teixeira, A.; Marini, M.; Antonini, Y.; Martins, R. Yeast Communities Associated with Stingless Bees. FEMS Yeast Research 2003, 4, 271–275. [Google Scholar] [CrossRef]
- Caruffo, M.; Navarrete, N.; Salgado, O.; Díaz, A.; López, P.; García, K.; Feijóo, C.G.; Navarrete, P. Potential Probiotic Yeasts Isolated from the Fish Gut Protect Zebrafish (Danio Rerio) from a Vibrio Anguillarum Challenge. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Raggi, P.; Lopez, P.; Diaz, A.; Carrasco, D.; Silva, A.; Velez, A.; Opazo, R.; Magne, F.; Navarrete, P.A. D Ebaryomyces Hansenii and R Hodotorula Mucilaginosa Comprised the Yeast Core Gut Microbiota of Wild and Reared Carnivorous Salmonids, Croaker and Yellowtail. Environmental Microbiology 2014, 16, 2791–2803. [Google Scholar] [CrossRef]
- Saber, A.; Alipour, B.; Faghfoori, Z.; Yari Khosroushahi, A. Cellular and Molecular Effects of Yeast Probiotics on Cancer. Critical Reviews in Microbiology 2017, 43, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bai, X.; Zhang, Y.; Gao, Q.; Bu, X.; Xu, Y.; Guo, N. Evaluation of the Potential Probiotic Yeast Characteristics with Anti-MRSA Abilities. Probiotics & Antimicro. Prot. 2022, 14, 727–740. [Google Scholar] [CrossRef]
 ), molds (
), molds (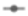 ) and yeasts as osmophilic (
) and yeasts as osmophilic (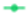 ) or non-osmophilic (
) or non-osmophilic ( ). Bubble marked with * indicate the group to which the probiotic yeasts belong.
). Bubble marked with * indicate the group to which the probiotic yeasts belong.
 ), molds (
), molds (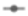 ) and yeasts as osmophilic (
) and yeasts as osmophilic (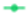 ) or non-osmophilic (
) or non-osmophilic ( ). Bubble marked with * indicate the group to which the probiotic yeasts belong.
). Bubble marked with * indicate the group to which the probiotic yeasts belong.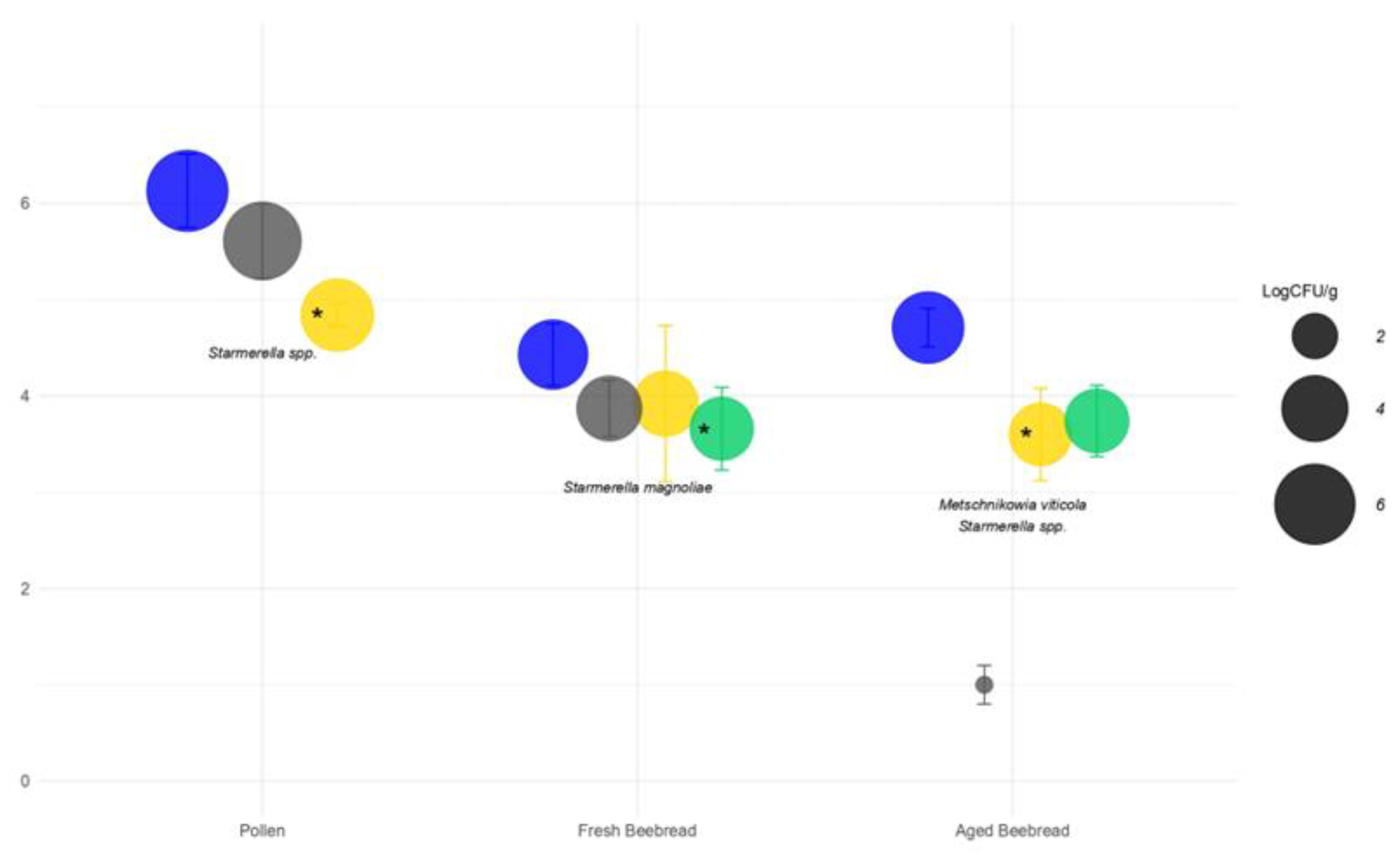
| Source | Isolate’s code | Yeast species identification | GeneBank accession number | Grouping | |
|---|---|---|---|---|---|
| Bee pollen | 1BP | Cryptococcus aureus | PQ571343 | Group 1 | |
| Bee pollen | 3BP | Starmerella spp. | PQ571344 | Group 2 | |
| Bee pollen | 9BP | Bullera alba | PQ571345 | Group 3 | |
| Bee pollen | 15BP | Starmerella spp. | PQ571346 | Group 4 | |
| Bee pollen | 17BP | Microstroma album | PQ571347 | Group 5 | |
| Fresh beebread | 52BB | Starmerella magnoliae | PQ571351 | Group 6 | |
| Fresh beebread | 54BB | Zygosaccharomyces pseudorouxii | PQ571352 | Group 7 | |
| Fresh beebread | 55BB | Metschnikowia rancensis | PQ571353 | Group 8 | |
| Fresh beebread | 65BB | Zygosaccharomyces siamensis | PQ571354 | Group 9 | |
| Fresh beebread | 67BB | Starmerella magnoliae | PQ571355 | Group 10 | |
| Aged beebread | 18BB | Starmerella spp. | PQ571348 | Group 11 | |
| Aged beebread | 20BB | Aureobasidium pullulans | PQ571349 | Group 12 | |
| Aged beebread | 88BB | Metschnikowia viticola | PQ571356 | Group 13 | |
| Aged beebread | 91BB | Kodamaea ohmeri | PQ571357 | Group 14 | |
| Aged beebread | 93BB | Starmerella spp. | PQ571358 | Group 15 | |
| Aged beebread | 94BB | Moniliella spp. | PQ571359 | Group 16 | |
| Aged beebread | 21H | Aureobasidium pullulans | PQ571350 | Group 17 | |
| Probiotic features | 3BP | 15BP | 18BB | 20BB | 21H | 52BB | 54BB | 55BB | 65BB | 67BB | 88BB | 91BB | 93BB | Codex |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pollen | Pollen | Aged beebread | Aged beebread | Aged beebread | Fresh beebread | Fresh beebread | Fresh beebread | Fresh beebread | Fresh beebread | Aged bebread | Fresh beebread | Fresh beebread | Commercial strain | |
| 37 °C pH 2.0 (Log CFU/ml) |
5.74 ± 0.04 | 5.18 ± 0.15 | 5.73 ± 0.10 | 0 | 0 | 5.88 ± 0.03 | 0 | 5.24 ± 0.00 | 0 | 5.28 ± 0.03 | 4.68 ± 0.38 | 5.37 ± 0.17 | 6.44 ± 0.09 | 5.24 ± 0.26 |
| Pepsin (Log CFU/ml) |
6.10 ± 0.07 | 5.48 ± 0.15 | 5.67 ± 0.03 | 0 | 0 | 5.89 ± 0.03 | 0 | 5.47 ± 0.01 | 0 | 5.49 ± 0.10 | 4.53 ± 0.26 | 5.32 ± 0.31 | 5.93 ± 0.19 | 5.47 ± 0.25 |
| Bile salts (Log CFU/ml) |
5.74 ± 0.04 | 5.82 ± 0.08 | 6.00± 0.14 | 0 | 0 | 5.67 ± 0.04 | 0 | 5.56 ± 0.23 | 0 | 5.36 ± 0.33 | 4.28 ± 0.03 | 5.52 ± 0.13 | 4.20 ± 0.15 | 4.20 ± 0.15 |
| Hydrophobicity (%) | 59.86 ± 4.03 | 50.00 ± 4.17 | 6.21 ± 1.12 | 12.40±0.80 | 50.71 ± 1.94 | 61.34 ± 4.89 | 5.09±1.33 | 11.71±2.48 | 62.30 ± 0.18 | |||||
| Auto-aggregation (%) | 20.70 ± 0.03 | 74.56 ± 0.01 | 54.43 ± 0.02 | 35.54 ± 0.05 | 72.98 ± 0.02 | 37.51 ± 0.04 | 88.18 ± 0.04 | 81 ± 0.01 | 55.37 ± 0.03 | 91.99 ± 0.02 | ||||
| Caco-2 adhesion (%) | 36.43 ± 0.17 | 61.46 ± 0.02 | 89.92± 0.58 | 47.42 ± 0.00 | 77.33 ± 0.06 | 63.76 ± 0.15 | 84.99 ± 0.06 | 26.50 ± 0.01 | 31.70 ± 0.03 | 90.28 ± 0.01 | ||||
| Biofilm formation (%) | 0.62 ± 0.06 | 12.69 ± 0.01 | 0.26 ± 0.02 | 0.02 ± 0.03 | 0.03 ± 0.03 | |||||||||
| Phytase activity | - | - | - | - | + | |||||||||
| Antimicrobial activity | ||||||||||||||
| E. coli | ++ | + | + | - | + | ++ | + | |||||||
| L. monocytogenes | + | + | - | - | + | + | + | |||||||
| S. enterica | - | + | + | - | + | + | + | |||||||
| S. aureus | - | - | +/- | - | +/- | - | - | |||||||
| C. albicans | + | + | - | - | + | + | + | |||||||
| Safety tests | ||||||||||||||
| Haemolytic activity | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Gelatinase activity | - | - | - | - | - | - | - | - | - | - | - | - | ||
| DNase activity | - | - | - | - | - | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





