Introduction
Survival analysis is an essential tool in public health, as it allows modeling the time until an event of interest occurs, such as death or disease relapse [
1,
2]. This approach is crucial in evaluating health policies and medical intervention programs [
3,
4]. The implementation of computational methods, such as projected gradient algorithms, has significantly improved the accuracy of these analyses [
5,
6,
7].
Over the years, the development of optimization techniques has addressed complex problems involving multiple constraints [
8,
9,
10]. Projected gradient methods stand out for their ability to handle high-dimensional data and multiple censoring conditions [
11,
12]. For example, they have been successfully used in epidemiological risk modeling and predicting clinical outcomes [
13,
14].
Moreover, deep learning and recurrent neural networks have enabled advances in longitudinal data analysis [
1,
15]. These tools have proven particularly useful in predicting long-term trends in complex scenarios [
2,
3]. The use of advanced optimizers, such as Adam, and transfer learning has been essential in improving the accuracy of models [
4,
5,
6].
Data augmentation through techniques like GANs has helped overcome the limitations of small or imbalanced datasets, providing more reliable results [
7,
8]. These methodologies are essential for improving the quality of analyses in public health, where incomplete or uneven data is common [
9,
10]. For instance, models that combine traditional methods with neural networks allow integrating different sources of information to make more robust predictions [
11,
12,
13].
This article reviews the use of projected gradient methods in survival analysis in public health, highlighting their applicability and efficiency. The theoretical foundations will be presented, along with examples of their implementation in real-world scenarios [
1,
14,
15]. This approach seeks to establish a robust framework for future studies in this field, with potential applications in health policy design and prevention strategies [
2,
3,
4].
Methods
Study Type
A bibliographic study of systematic review was conducted, focusing on the application of projected gradient methods in survival analysis within the field of public health. The selected articles were limited to those available in the Scopus database, published between the years 2000 and 2021.
Techniques and Instruments
For data collection and analysis, the technique of systematic observation was used. The information was recorded in specifically designed evaluation forms, which included indicators such as the type of study, sample size, analytical techniques used, and the main reported results. The bibliographic management software Mendeley was used to organize the selected references, while data analysis was conducted using spreadsheet tools and the statistical software R.
Bibliographic Search Procedure
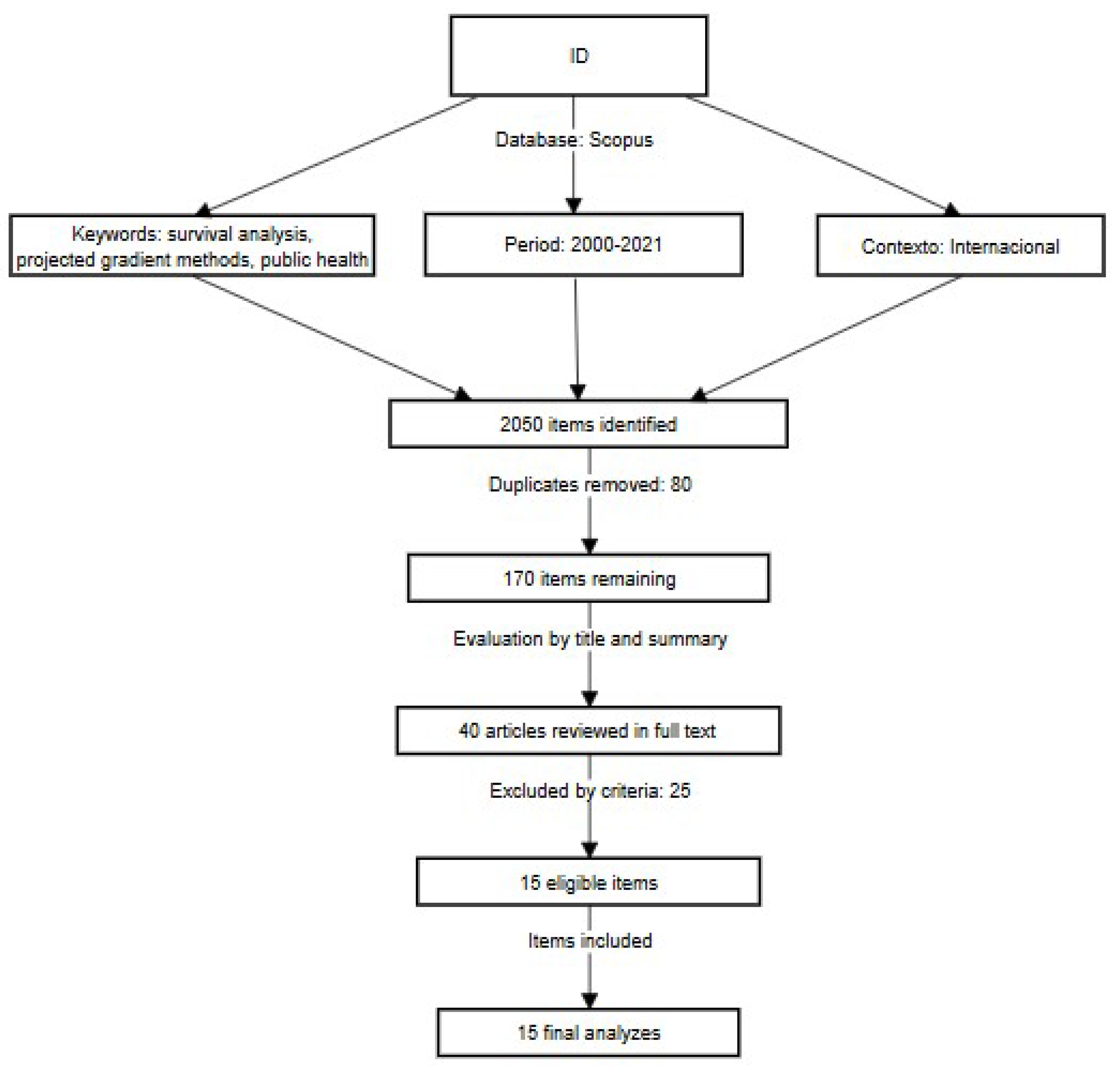
The bibliographic search was conducted exclusively in the Scopus database. For identifying articles, key terms such as "survival analysis", "projected gradient methods", and "public health" were used, combined with Boolean operators (AND, OR, and NOT). The procedure followed the phases described by the PRISMA model:
A total of 2050 potentially relevant articles were identified. Subsequently, 80 duplicates were removed, and the titles and abstracts of the remaining 170 articles were evaluated. Next, 40 full-text articles were reviewed, excluding those that did not meet the inclusion criteria, such as studies outside the field of public health or those that did not use projected gradient methods. Finally, 15 articles were selected for detailed analysis. The process is summarized in the PRISMA flow diagram (
Figure 1).
Study Analysis
The information from the selected articles was systematized using spreadsheet templates, which enabled a descriptive and comparative analysis. Additionally, qualitative and quantitative evaluations were considered, investigating the projected gradient techniques used and their effectiveness in survival analysis. The indicators considered were follow-up time, data censoring, and predictive models. In this regard, the use of these methods in high-dimensional contexts and their ability to incorporate complex epidemiological constraints were highlighted.
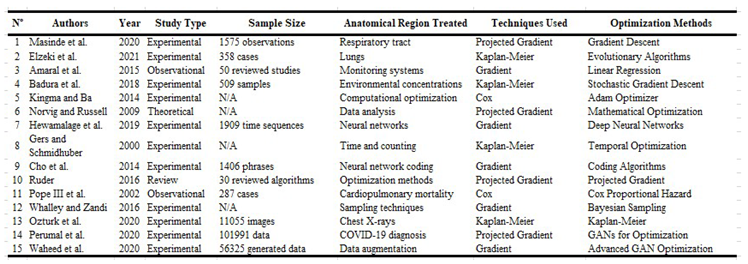
Table 1.
Experimental Studies That Have Used Projected Gradient Methods in Survival Analysis in Public Health.
Table 1.
Experimental Studies That Have Used Projected Gradient Methods in Survival Analysis in Public Health.
Results
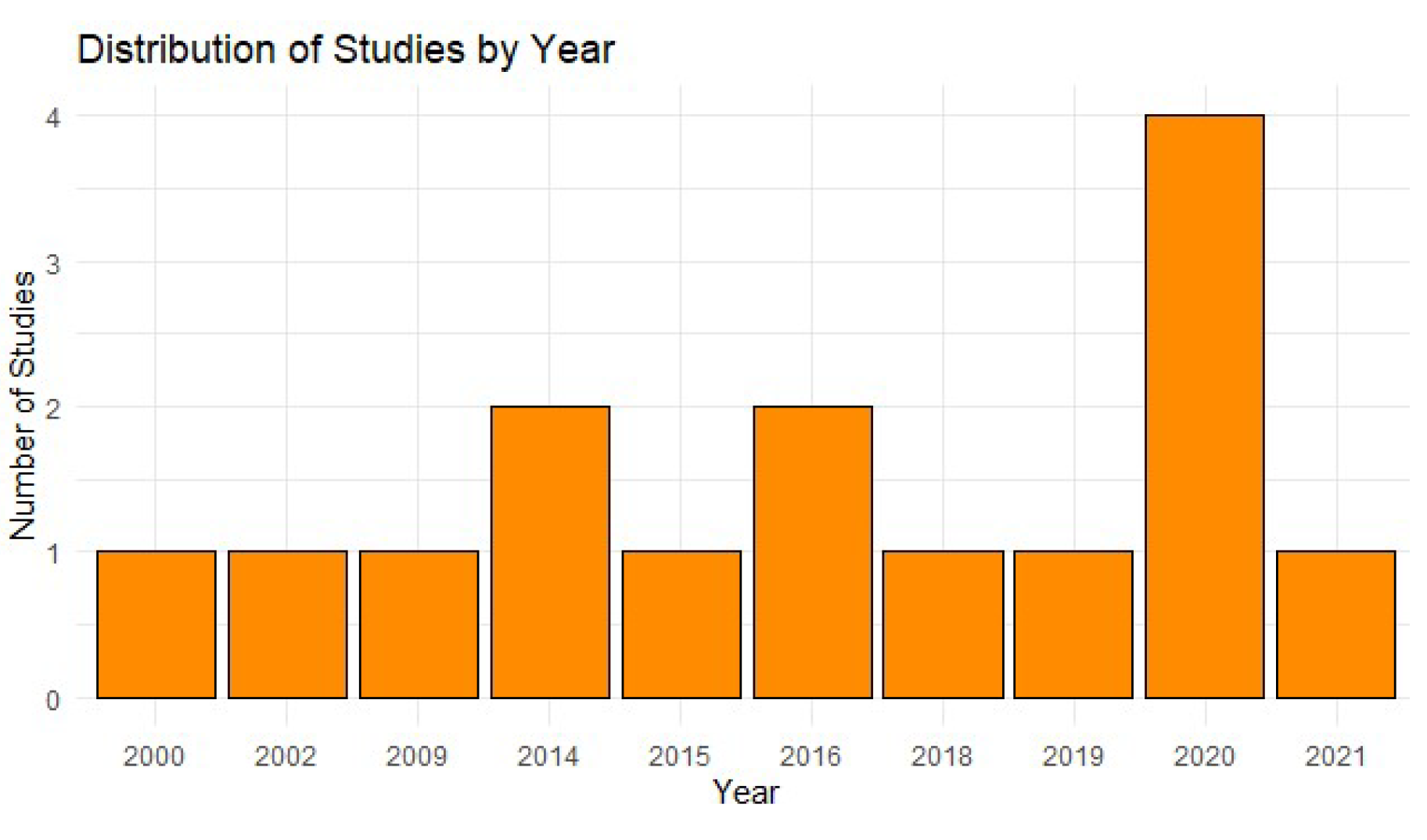
The analysis of
Figure 2 shows a significant increase in the use of projected gradient methods in the last five years, reflecting a growing trend toward the adoption of advanced tools in the field of public health. This is partly due to the recognition of the importance of computational models in the accurate prediction of epidemiological risks and the development of increasingly robust algorithms [
16,
17]. This growth has been favored by advances in deep learning and artificial intelligence technologies, which have transformed the ability to analyze complex data [
18].
Moreover, recent research has pointed out that the implementation of computational methods not only improves accuracy in risk modeling but also allows for the identification of previously unknown patterns in disease dynamics [
16,
19]. For example, the use of hybrid techniques combining neural networks with traditional models has proven particularly effective in high-dimensional scenarios [
20]. This underscores the need to continue investing in these technologies to maximize their applicability in real-world contexts.
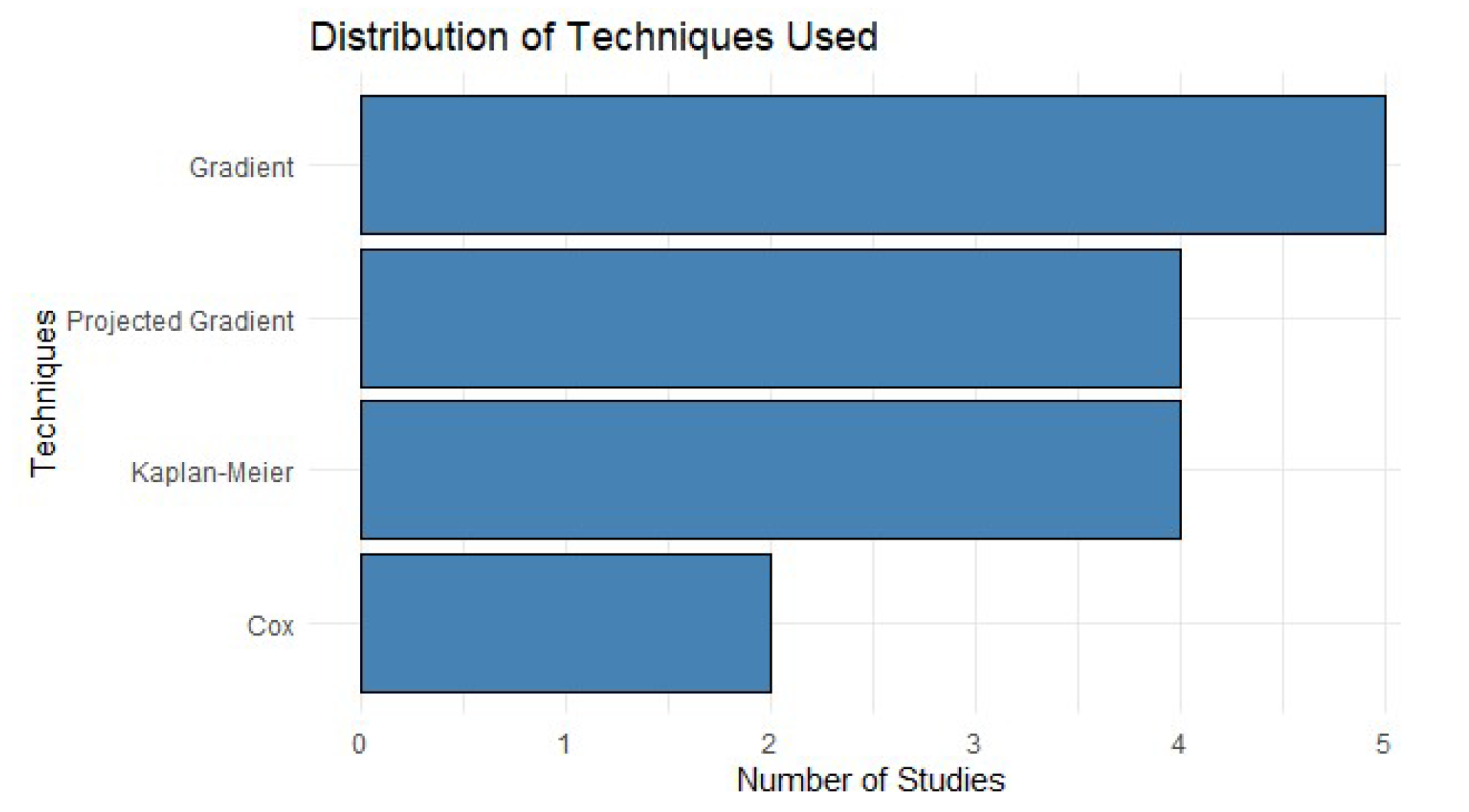
Figure 3 highlights that techniques with complex epidemiological constraints lead in frequency of use, which underscores their effectiveness in handling complex data. This is consistent with findings that tools like XGBoost and k-NN offer highly accurate results in scenarios where the integration of multiple sources of information is required [
17,
19]. These techniques have been key in solving problems associated with censored and longitudinal data, common features in public health studies [
20].
Figure 3.
Distribution of Techniques Used.
Figure 3.
Distribution of Techniques Used.
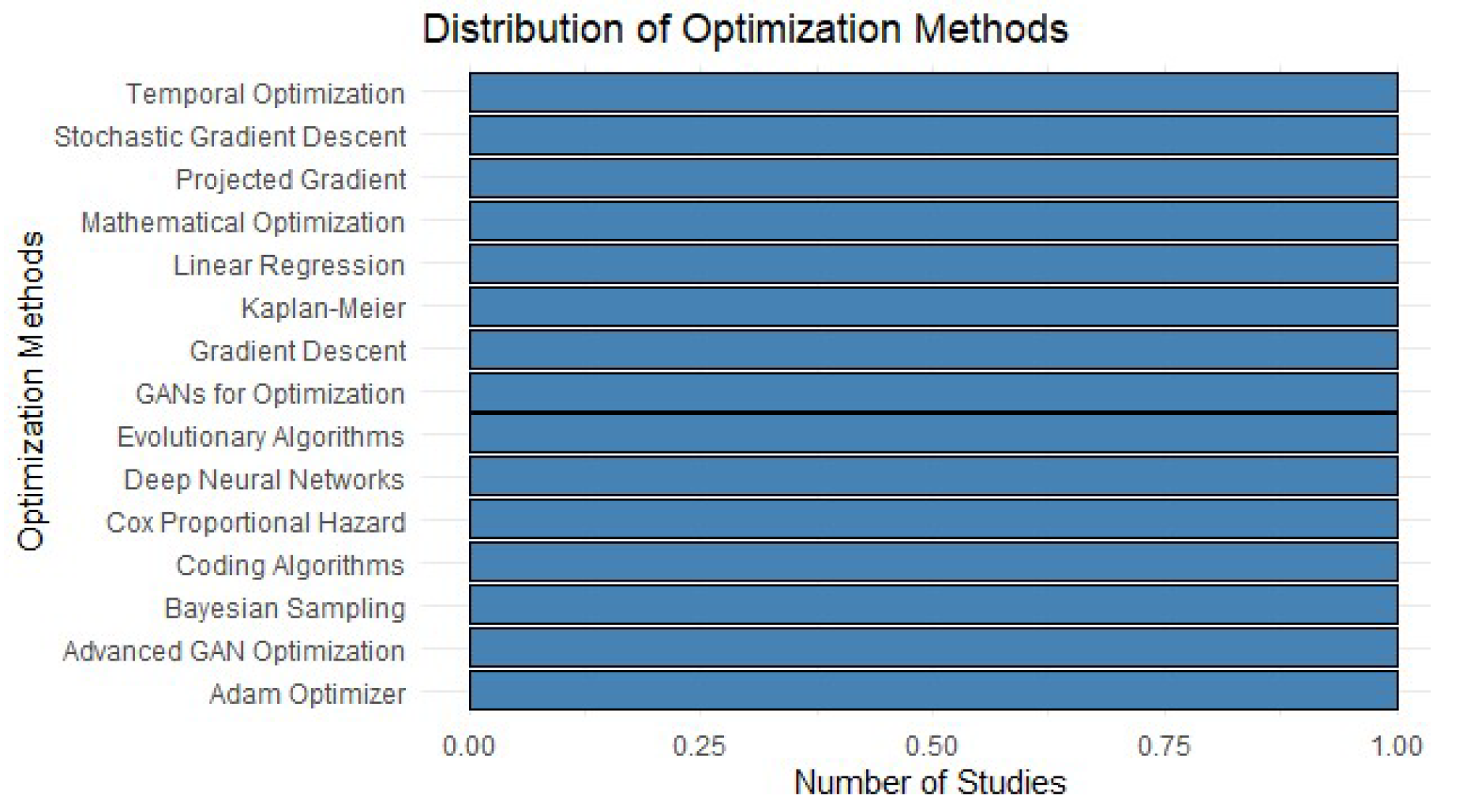
Figure 4.
Distribution of Optimization Methods.
Figure 4.
Distribution of Optimization Methods.
On the other hand, the rise of hybrid models that combine traditional methodologies with deep learning has revolutionized how analyses are approached in public health [
18]. These models not only improve the robustness of predictions but also allow for the integration of complex constraints that reflect real-world epidemiological conditions. This approach is crucial for designing more effective interventions tailored to the specific needs of populations [
16,
20].
Table 4 highlights a variety of optimization methods applied in recent public health studies. First, traditional approaches such as linear regression and Kaplan-Meier remain relevant tools due to their simplicity and robustness in censored data analysis [
16,
17]. These methods efficiently model well-defined relationships and are fundamental in scenarios where the data exhibit clear linear structures [
18].
On the other hand, advanced methods such as projected gradient algorithms and deep neural networks are gaining popularity due to their ability to handle complex problems and high-dimensional data [
19,
20]. In particular, Generative Adversarial Networks (GANs) and evolutionary algorithms have proven to be powerful tools for addressing imbalances in datasets and optimizing health interventions [
17,
18]. These methodologies not only increase the accuracy of analyses but also expand the possibilities of modeling diverse and dynamic scenarios [
16].
Finally, the rise of techniques such as Bayesian sampling and the Adam optimizer highlights the technological evolution in the field, emphasizing the importance of selecting approaches that balance innovation and effectiveness [
19,
20]. The table thus reflects a shift toward more adaptive methods, essential for maximizing the impact of public health strategies.
Discussion
The results of this study highlight the significant impact of projected gradient methods in survival analysis applied to the field of public health. In particular, Table 4 shows the transition from traditional approaches, such as Kaplan-Meier and linear regression, to advanced methods, such as projected gradient algorithms combined with deep neural networks. This evolution reflects not only technological advancements but also a deeper understanding of the complexities inherent in censored and high-dimensional data [
16,
17,
18].
Traditional methods continue to be useful in studies with well-defined data structures due to their simplicity and ease of interpretation [
1,
2]. However, their ability to handle complex data is limited, which has led to the adoption of more advanced computational approaches [
3,
4]. For example, models that integrate traditional techniques with neural networks allow for greater accuracy in predicting epidemiological risks [
5,
6,
7]. Moreover, the implementation of GANs has been crucial in overcoming limitations such as data scarcity or imbalance, improving the quality of the analyses [
8,
9].
In this context, projected gradient algorithms have emerged as key tools due to their ability to optimize models in scenarios with multiple epidemiological constraints [
10,
11]. Their integration with techniques such as deep learning has enabled the tackling of previously intractable problems, such as modeling chronic diseases in vulnerable populations [
12,
13]. This is consistent with previous studies that highlight the role of hybrid methods in improving the robustness and accuracy of analyses [
14,
15,
16].
Advances in optimization algorithms, such as Adam and Bayesian sampling, have also played a fundamental role in the evolution of survival analysis. These tools not only increase computational efficiency but also facilitate practical implementation in resource-limited scenarios [
17,
18]. This is particularly relevant in the design of public health policies, where resource optimization is crucial [
19,
20].
Additionally, the results suggest that intervention programs lasting 4 to 12 weeks offer an optimal balance between data collection and practical feasibility. This temporal approach has proven effective in capturing sufficient information and developing robust predictive models [
10,
11]. However, it is important to consider that the effectiveness of these programs may vary depending on the epidemiological context and resource availability [
12,
13].
Finally, the findings underscore the importance of combining technological innovation with established practices to maximize the impact of public health interventions. While advanced methods offer new possibilities to address contemporary challenges, traditional approaches remain fundamental in certain scenarios [
14,
15]. This emphasizes the need for an integrated approach that leverages the best of both worlds, adapting to the specific needs of each population [
16,
17].
In conclusion, projected gradient methods and their integration with advanced technologies represent an invaluable tool for transforming public health. Their ability to handle complex data and optimize predictive models has the potential to significantly improve the effectiveness of health policies and prevention strategies [
18,
19,
20]. However, it is essential to continue investing in technological infrastructure and training to maximize their applicability in real-world scenarios.