1. Introduction
The prevalence of bacterial resistance has become one of the most serious public health crises because of the limited availability of effective antibiotics to treat them. The rise and spread of antibiotic-resistant infections in the hospitals and communities have challenged the effectiveness of the current antibiotic stewardship strategists to face this obstacle [
1,
2,
3].
The Center for Disease Control has reported that more than 2.8 million antibiotic-resistant infections occur in the U.S. each year, with more than 35,000 people mortality [
4]. Multidrug-resistant bacteria are responsible for roughly half of the 37,000 deaths a year in the 27 member states of the European Union that are caused by infections associated with hospital care [
5]. The study only focuses on infections related to hospital care and does not count community-acquired infections. Thus, these data show that the discovery of novel treatment alternatives to commercially available antibiotics is urgently required to tackle the challenges of bacterial resistance.
Methicillin-Resistant
Staphylococcus aureus (MRSA) can cause skin infection [
6], bacteremia, pneumonia, endocarditis, and osteomyelitis [
7].
K. pneumoniae can cause illnesses, such as pneumonia, bloodstream infections, wound or surgical site infections, and meningitis [
8]. Recently,
Klebsiella pneumoniae (
K. pneumoniae, KPC) has developed antimicrobial resistance against carbapenems [
8], known as broad-spectrum antibiotics as the last line of defense against infections.
Multidrug-resistant
Pseudomonas aeruginosa (
P. aeruginosa, PSA) caused approximately 32,600 infections among hospitalized patients and approximately 2,700 deaths in the United States in 2017 [
9].
P. aeruginosa can cause infections in the blood, lungs (pneumonia), or other parts of the body after surgery [
9].
Escherichia coli (
E. coli) can cause diarrhea, urinary tract infections, respiratory illness, and other illnesses [
10]. There have been a lot of
E. Coli outbreaks that have brought a lot of attention in the media, warning people about eating raw and uncooked foods to stop the bacteria from spreading.
Enterococcus, especially
Enterococcus faecalis (
E. faecalis), can survive harsh environmental conditions, including a very dry, high-temperature environment and exposure to some antiseptics [
11], and can cause endocarditis and urinary tract infections [
12,
13].
Streptococcus pneumoniae (
S. pneumoniae) has been studied extensively, and many vaccines have been discovered covering 23 different serotypes. However, it still remains a major cause of morbidity and mortality in children and older adults worldwide as a principal bacterial cause of otitis media, pneumonia, meningitis and an important cause of community-acquired bacteremia [
14].
Antimicrobial peptides (AMPs) are part of the defense system of the infected organism. AMPs are considered promising candidates to fight multi-drug resistant bacterial pathogens due to their excellent broad-spectrum antibacterial activity and non-specific bacterial membrane rupture mechanism, which does not allow bacterial or fungal pathogens to develop resistance [
15,
16,
17,
18]. The endogenous AMPs of plants and animals are typically cationic (i.e. contain excess lysine and arginine residues) amphipathic molecules [
19]. Two major classes of peptides in human skin are reported β-defensins and cathelicidins, which have antimicrobial activity against bacterial, fungal, and viral pathogens. These peptides, produced by keratinocytes in the skin, disrupt the membrane of the target microbe or penetrate the microbial membrane [
20]. This unspecific mode of action is suggested to be responsible for the broad-spectrum activity of many antimicrobial peptides [
21]. Saccharum officinarum is a plant that produces an AMP called sugarcane defensin 5, which is associated with antifungal property [
22].
The membrane permeability and forming pores in the plasma membrane to burst bacteria are mostly recognized as the well-accepted mechanisms to describe the action of cationic AMPs. Most often, cell-penetrating peptides (CPPs) and AMPs have been studied at the same time as their mechanism of action has some minor differences [
23,
24,
25]. AMPs maintain the formed pores in the plasma membrane open for a longer period of time compared to CPPs. Therefore, delivery of the intended reagents could be accomplished with the least toxicity to the cells in the case of CPPs.
Antimicrobial peptides share amphiphilicity and cationic structural properties with CPPs. We have previously reported the synthesis of [E
4W
4], [KR
5], [F
4R
5], [Y
4R
4], and [R
4W
4] and tested them against bacterial pathogens. Among all the synthesized peptides, [R
4W
4] containing arginine and tryptophan residues in a sequential manner showed the most potent antibacterial activity against MRSA, exhibiting a minimal inhibitory concentration (MIC) of 2.67 μg/mL [
26,
27]. [R
4W
4] also exhibited antibacterial activity against Gram-negative bacteria and other Gram-positive [
23,
26,
27,
28,
29,
30]. [R
4W
4] demonstrated bactericidal activity against bacterial isolates with an MBC/MIC ≤ 4 and showed a synergistic effect with gentamicin against
E. coli (FICI = 0.3). The mechanism of action against MRSA involved changes in zeta potential, membrane depolarization, and binding to lipoteichoic acid (LTA), with concentration-dependent membrane perturbations [
30]. Similarly, Mohammed et al. (2022) introduced a potential strategy for treating multidrug-resistant pathogens by combining [W
4KR
5] and a variety of classical antibiotics to improve the antibacterial effectiveness [
23]. These findings suggest that cyclic peptides with sequential tryptophan and arginine residues hold promise as potential antibacterial agents.
Our laboratory also has previously reported the antibacterial activities of several peptides containing sequential R, hydrophobic residues (W, 3,3-diphenyl-L-alanine (Dip), 4,4′-biphenyl-Lalanine (Bip), 3-(2-naphthyl)-L-alanine (Nal), and a number of peptide-antibiotic conjugates [
27,
31,
32].
Bicyclic peptide [W(WR)
4K]-[W(WR)
4E] containing alternate arginine and tryptophan residues demonstrated activity against Gram-positive bacteria MRSA (ATCC BAA-1556) and
S. aureus (ATCC 29213) with MIC values of 4.8-9.6 µM [
33]. Furthermore, we reported the synthesis, cell-penetrating properties, and molecular transporter efficiency of linear and cyclic peptides containing alternative positively charged R residues and hydrophobic residues (W and Dip) [
34]. A number of peptides in this class were able to act as a CPP to deliver cargo compounds intracellularly in a physical mixture.
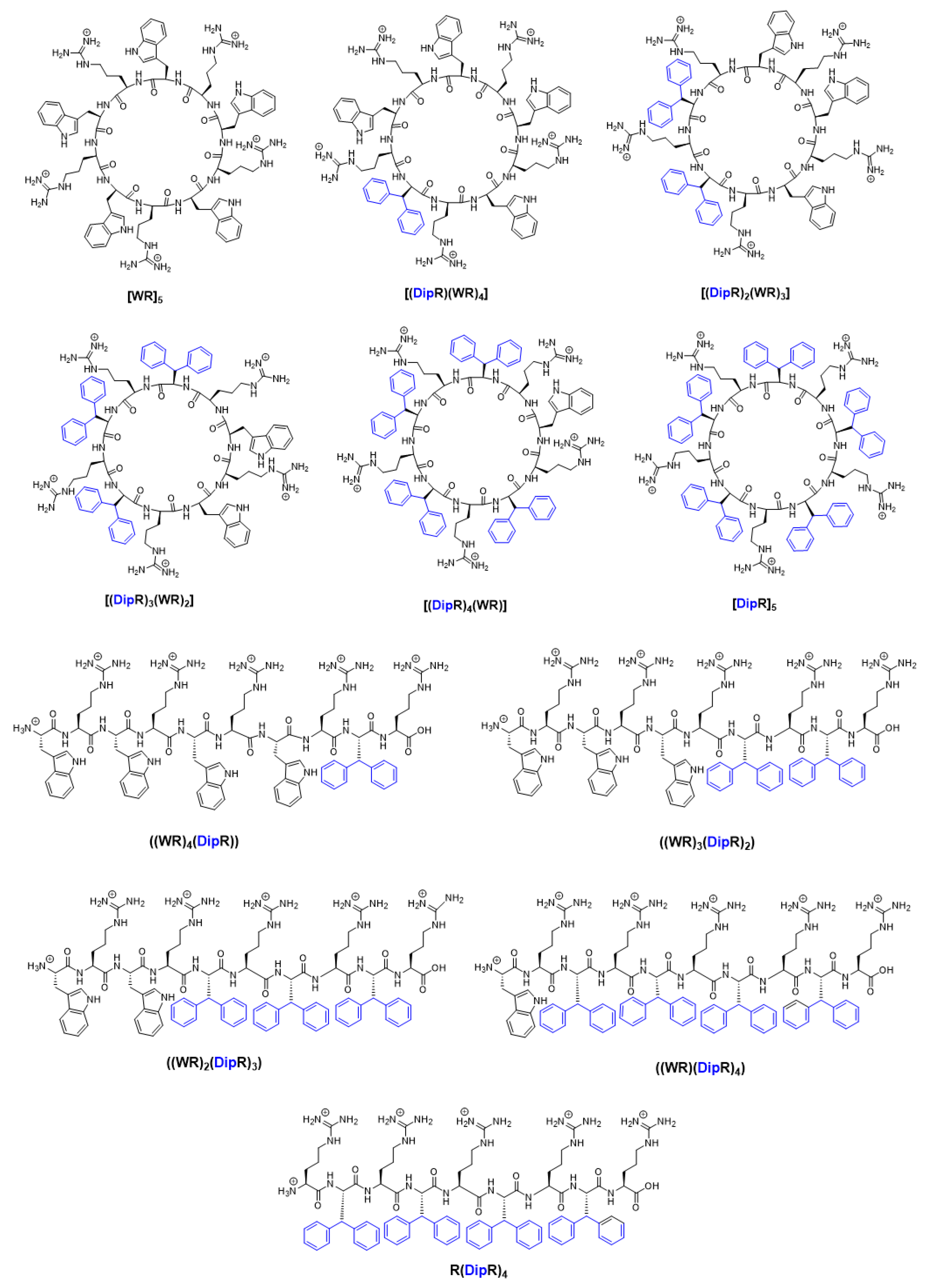
In continuation of our efforts to design novel AMPs with improved properties, we report here the evaluation of the same linear and cyclic peptides containing alternative positively charged R residues and hydrophobic residues (W and Dip) (
Figure 1) against pathogenic bacteria and fungi (
Table 1 and
Table 2). Herein, we report the antibacterial activities of the same linear peptides ((DipR)
4(WR)), ((DipR)
3(WR)
2), ((DipR)
2(WR)
3), ((DipR)(WR)
4), (DipR)
4R, and cyclic peptides [(DipR)
4(WR)], [(DipR)
3(WR)
2], [(DipR)
2(WR)
3], [(DipR)(WR)
4], and [DipR]
5 to determine the effect of replacing of W with Dip in antimicrobial properties.
We hypothesized that these peptides can be used as a broad-spectrum antibiotic and antifungal agents by interacting with bacterial and fungal membrane and/or pore formation acting as AMPs. The head-to-tail cyclization and incorporation of unnatural amino acids could provide improved stability against degradation in the biological environment.
The peptides were evaluated against MRSA (ATCC BAA-1556) and some of the ESKAPE pathogens that, include Enterococcus faecium (E. faecium) (ATCC 700221), Staphylococcus aureus (S. aureus) (ATCC 29213), K. pneumoniae (ATCC BAA-1705), P. aeruginosa (ATCC 27883), and some other pathogenic bacteria, such as E. coli (ATCC 25922), E. faecalis (ATCC 29212), S. pneumoniae (ATCC 51938), and Bacillus subtilis (B. subtilis) (ATCC-6633). The peptides were also evaluated against pathogenic fungi, Candida parapsilosis (C. parapsilosis) (ATCC 22019), Aspergillus fumigatus (A. fumigatus) (Af-293), and Candida albicans (C. albicans) (ATCC 60193), to determine the broad-spectrum antifungal activities of the compounds.
2. Results and Discussion
2.1. Antibacterial Activity
The antibacterial activity of all ten synthesized linear and cyclic peptides was evaluated against MRSA (ATCC BAA-1556), S. aureus (ATCC 29213), E. faecium (ATCC 700221), S. pneumoniae (ATCC 51938), E. faecalis (ATCC 29212), B. subtilis (ATCC-6633), K. pneumoniae (ATCC BAA-1705), P. aeruginosa (ATCC 27883), and E. coli (ATCC 25922). Meropenem and daptomycin were used as positive controls for this study. The MIC was determined by micro-broth dilution protocol, where the minimal concentrations were determined to be at concentrations in wells with no visible bacterial growth.
Among the tested peptides, [DipR]
5 showed promising MIC values of 0.39-6.25 µM (0.74-11.9 µg/mL) against Gram-positive bacteria strains: MRSA
, S. aureus,
E. faecium,
E. faecalis,
S. pneumoniae, and
B. subtilis. [DipR]
5 showed moderate MIC values of 12.5-25 µM (23.8-47.6 µg/mL) against Gram-negative strains,
K. pneumoniae,
P. aeruginosa, and
E. coli (
Table 1).
Cyclic peptide [DipR]5 was found to be active against the bacteria in the order of S. pneumoniae (MIC = 0.39 µM) ˃ E. faecalis and E. faecium (MIC = 0.78 µM) > MRSA and S. aureus (MIC = 3.1 µM) > B. subtilis (MIC = 6.3 µM) > E. coli (MIC = 12.5 µM) > K. pneumoniae, and P. aeruginosa (MIC = 25 µM).
The linear peptides, on the other hand, showed different levels of activity, with some peptides, like (DipR)
4R, demonstrating potent effects against certain bacterial strains like
MRSA and
K. pneumoniae. The corresponding linear peptide (DipR)
4R showed activity in the order of
S. pneumoniae (MIC = 0.39 µM)
˃ MRSA
, S. aureus,
E. faecium, and
Bacillus subtilis (MIC = 1.6 µM)
˃ E. faecalis (MIC = 3.1 µM) >
K. pneumoniae and
E. coli (MIC = 6.3 µM) >
P. aeruginosa (MIC = 12.5 µM). These data indicate that the linear peptide (DipR)
4R was more potent against a number of bacterial strains, such as MRSA,
K. pneumoniae,
P. aeruginosa,
E. Coli,
S. aureus, and
B. Subtilis when compared with [DipR]
5. Indeed (DipR)
4R was the most potent peptide among all the compounds against MRSA
, K. pneumoniae,
E. Coli, and
S. aureus. At the same time [DipR]
5 was the most potent peptide against
E. Faecium,
E. faecalis, and
S. pneumoniae. Of note, we have previously reported that (DipR)
4R exhibited more cytotoxicity than other peptides in HEK-293 cells after 24 h incubation, especially above 10 µM [
34]
While peptides showed less potent than [DipR]5 or (DipR)4R or comparable antibacterial activities against the bacterial strains, all the peptides were more potent against S. pneumoniae (MIC = 0.39-0.78 µM) when compared with other bacteria. Furthermore, these peptides showed modest activity against E. Faecalis (MIC = 0.78-12.5 µM) and MRSA (MIC = 1.6-6.3 µM).
Linear peptide ((DipR)2(WR)3) was found to show activity against the bacteria in the order of S. pneumoniae (MIC = 0.78 µM) ˃ B. subtilis (MIC = 1.6 µM) > MRSA, S. aureus, E. faecium (MIC = 3.1 µM) > E. faecalis and E. coli (MIC = 6.3 µM) > K. pneumoniae and P. aeruginosa (MIC = 12.5 µM).
The peptides demonstrated varying degrees of potency when compared to traditional antibiotics and antifungals, such as meropenem and daptomycin. The peptides were significantly more potent than meropenem against
K. pneumoniae, E. faecium, and except ((DipR)(WR)
4), more potent than daptomycin against
K. pneumoniae,
E. Coli, and
P. aeruginosa. Furthermore, [DipR]
5 was more potent than daptomycin against
E. faecium,
E. faecalis, and
S. pneumoniae, suggesting that these peptides could serve as potential alternatives or complementary agents in treating resistant bacterial infections. Overall, these compounds can be regarded as strong candidate compounds against
S. pneumoniae and MRSA with MIC values of 0.39-0.78 µM and 1.6-6.3 µM. respectively, for further optimization. As demonstrated for [R
4W
4], the mechanism of action of these compounds against bacteria is expected to involve changes in zeta potential, membrane depolarization, and concentration-dependent membrane perturbations [
30].
2.2. Antifungal Activity
The antifungal activity of all ten synthesized linear and cyclic peptides was evaluated against C. albicans (ATCC 60193), C. parapsilosis (ATCC 22019), and A. fumigatus (AF-293). Fluconazole and amphotericin B were used as a positive control for this study. The MIC was determined at concentrations in wells with no visible fungal growth.
Cyclic peptide [DipR]
5 showed MIC values of 1.6-6.6 µM (3.0-12.5 µg/mL) against C. parapsilosis and A. fumigatus (
Table 2). Cyclic peptide [DipR]
5 was found to be active against the fungi in the order of A. fumigatus (MIC = 1.6 µM) ˃ C. parapsilosis (MIC = 6.6 µM) > C. albicans (MIC = 13.1 µM). Cyclic peptide [(DipR)
4(WR)] also showed activity in the order of A. fumigatus (MIC = 1.7 µM) ˃ C. albicans and C. parapsilosis (MIC = 6.7 µM).
Linear peptide ((DipR)4(WR)) was found to show activity against the fungi in the order of A. fumigatus (MIC = 1.6 µM) ˃ C. albicans and C. parapsilosis (MIC = 6.6 µM). All the peptides exhibited potency against A. fumigatus (MIC = 1.6-3.5 µM) when compared with other fungi. Furthermore, these peptides showed modest activity against C. parapsilosis (MIC = 6.6-14.1 µM) and C. albicans (MIC = 3.4-28.2 µM). Fluconazole exhibited no activity at the highest concentration tested, with a MIC greater than 209 µM.
While the antifungal activity of these peptides was generally more potent against A. fumigatus, they exhibited varying degrees of activity against the Candida species. Their activity against A. fumigatus, C. albicans, and C. parapsilosis suggests potential applications as a lead candidate for further investigation as antifungal agents, with particular strength against A. fumigatus.
We have previously reported noticeable cytotoxicity above 25 µM concentration for HEK-293 cells, but no significant toxicity was recorded at 10 µM concentration for the peptides [
34]. This provides a relatively acceptable therapeutic window for further development. The results indicate that the peptides could be used as broad-spectrum antifungal and antibacterial agents against pathogenic fungi and bacteria with minimal cytotoxicity. However, further evaluations of the compounds against other normal cell lines and animal studies are still needed.
3. Materials and Methods
C. albicans (ATCC 60193), C. parapsilosis (ATCC 22019), MRSA (AATCC BAA-1556), K. pneumoniae (ATCC BAA-1705), P. aeruginosa (ATCC 27883), E. coli (ATCC 25922), S. aureus (ATCC 29213), E. faecium (ATCC 700221), E. faecalis (ATCC 29212), S. pneumoniae (ATCC 51938), and B. subtilis (ATCC-6633) were obtained from ATCC (Manassas, VA), and propagated as per the recommendation of American Type Culture Collection (ATCC; USA). A. fumigatus Af-293 was obtained from Dr. Eric Pearlman from the University of California, Irvine. RPMI 1640 (Lot # RNBG5842, Sigma Aldrich, Sigma Aldrich, St. Louis, MO, USA) with L-glutamine supplemented with 2% D-Glucose (Lot # 160725, Fisher Scientific, Kansas City, MO, USA) and adjusted pH with HEBES buffer (Ref 15630-080, Gibco ThermoFisher Scientific, Grand Island, New York, USA) were used as bacteria media. Yeast Peptone Dextrose (YPD) agar (1% w/v yeast extract (Pcode 102436548, Sigma Aldrich, St. Louis, MO, USA), 2% w/v peptone (Pcode 102475393, Sigma Aldrich, St. Louis, MO, USA), 2% w/v D-Glucose, 1.5% Agar (Pcode 102425215, Sigma Aldrich, St. Louis, MO, USA)) were used as fungal media. Synthesis of all the peptides was conducted according to the previously reported procedure by us [
34].
3.1. Antibacterial Assays
All bacterial experiments were carried out in a laminar flow hood (Labconco, Kansas City, MO, USA). The bacterial strains were cultured following the guidelines set by the Clinical Laboratory Standards Institute (CLSI). Antibacterial assays were performed using a standard microtiter dilution method as described in previous studies [
23]. The minimum inhibitory concentration (MIC) refers to the lowest peptide concentration that inhibits bacterial growth. Bacteria were cultured overnight in 6 mL of Luria Broth (LB). A stock solution of the peptide was prepared in water and DMSO, and a test compound solution with a concentration of 256 µg/mL was made in LB. Meropenem, with a solubility of 8 mg/mL, was prepared as a stock solution at 4 mg/500 µL. An aliquot of the overnight bacteria culture was diluted in normal saline (NS, 1 mL) until achieving a 0.5 McFarland turbidity (1.5 × 10
8 bacterial cell CFU/mL). McFarland compared solution (60 µL) was added to Mueller Hinton Broth (MH) 8,940 µL to generate a 1/150 dilution. A 200 µL test compound solution was added to the first well of a 96-well plate, and the remaining wells (2-12) were filled with 100 µL of MH media using a multi-channel pipette from a sterile reservoir.
A 100 µL sample from well 1 was pipetted into well 2, and the solution was mixed thoroughly by pipetting up and down. Next, 100 µL was transferred from well 2 to well 3, followed by thorough mixing. This 2-fold serial dilution process continued through well 11, with no additions made to well 12. The final 100 µL from well 11 was discarded. After completing the serial dilution, each well (1-12) contained 100 µL of solution. The bacterial culture in MH media was vortexed and transferred to a sterile reservoir, from which 100 µL was added to each well using a 12-channel pipette. (Note: The dilutions were further diluted upon adding the bacterial solution; for example, the concentration in well 1 started at 512 µg/mL but became 256 µg/mL after the addition of the bacterial culture.) The plates were incubated overnight at 37 ºC for 18-24 hours. All experiments were performed in triplicate, with controls consisting of bacteria in MH media and positive controls of daptomycin or meropenem.
3.2. Antifungal Assays
A. fumigatus (Af-293) was cultured on Sabouraud dextrose agar (SDA) (Lot #3431660, Oxoid) for 48-72 h at 35 °C to obtain fresh and mature colonies which reached the proper sporulation then used to prepare the inoculum. The colonies were covered with Phosphate Buffered Saline (PBS) containing 0.025% Tween-20, scraped from the plate, and carefully collected into a sterile tube. The suspension was centrifuged at 3000 RPM for 5 min. after the separation between conidia and hyphae. The supernatant was discarded, and the pellet was resuspended in 5 ml of PBS.
Peptide concentrations of 50, 25, 12.5, 6.25, and 3.1 μg/mL were tested in triplicate. Each well of a 96-well plate was inoculated with the prepared inoculum suspension, which was diluted with RPMI 1640 medium containing L-glutamine, supplemented with 2% D-glucose, and the pH adjusted using HEPES buffer. The plates were incubated for 50 min. at 35 °C to achieve a final cell concentration of 106 cells/mL. The control wells contained the same inoculum without any compound. The plates were then incubated for 24 and 48 h at 35 °C. Fungal growth was visualized, and the minimum inhibitory concentration (MIC) was determined as the lowest peptide concentration, where no growth was observed.
In this study, C. albicans and C. parapsilosis were cultured on Yeast Peptone Dextrose (YPD) agar containing 1% w/v yeast extract, 2% w/v peptone, 2% w/v D-glucose, and 1.5% agar. A single colony from the agar was selected and cultured in Tryptone soy broth supplemented with 0.1% D-glucose, then incubated for 24 h at 37 °C. The cells were washed twice with PBS by centrifugation at 2000 RPM for 5 minutes. The pellet was resuspended in 5 mL of PBS, diluted in RPMI 1640 with L-glutamine, supplemented with 2% D-glucose, and the pH adjusted using HEPES buffer. The suspension was then incubated for 50 minutes at 35 °C to achieve a final cell concentration of 106 cells/mL.
Peptide concentrations of 50, 25, 12.5, 6.25, and 3.1 μg/mL were tested in triplicate. Each well of a 96-well plate was inoculated with the prepared inoculum suspension, which was diluted with RPMI media to a final volume of 200 μL per well. The control well received the same inoculum without any compound. The plates were incubated for 24 h at 37 °C. Fungal growth was visually observed in the control wells, and the minimum inhibitory concentration (MIC) was determined as the lowest concentration at which no growth was detected in the wells. Fluconazole was used as standard antifungal drugs.