1. Introduction
The human gene for epidermal growth factor 2 (HER2) [also known as ERBB2 or epidermal growth factor receptor (EGFR)2] encodes a 185 kDa transmembrane glycoprotein with tyrosine kinase activity. It is overexpressed and/or amplified in 15-20% of all breast cancers. Amplification of HER2 has been shown to be related to tumor size, lymph node metastasis, high S-phase fraction, aneuploidy, and low levels of steroid hormone receptors, factors that may increase the rate of tumor cell proliferation. Recent studies show a str
ong correlation between HER2 gene amplification and tamoxifen resistance. Anti-HER2 therapy consists of the administration of the monoclonal antibody trastuzumab (Herceptin), which is effective in the case of metastasis and/or in combination with chemotherapy [
1].
According to the guidelines of the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP), there are now established techniques for determining HER2 status in primary tumours based on immunohistochemistry (IHC) or fluorescence/chromogenic in situ hybridization analysis (FISH/CISH) [
2]. Nevertheless, there is increasing evidence of equivocal samples with a negative FISH result, which are therefore not eligible for antiHER2 therapies. Initial estimates put the number of such cases at around 8 [
3]
The aim of the study is to introduce the determination of HER2 status using molecular biology methods, both at the level of the number of copies of the HER2 gene and at the level of its gene expression, so that we can evaluate with the advantage of two levels (both DNA and cDNA), which will help us to read particularly ambiguous and doubtful cases accurately.
2. Materials and methods
We examined samples of formalin-fixed, paraffin-embedded (FFPE) primary breast cancer tumors obtained from the Pathological Anatomy Department of Bulovka College Hospital (FNB). The samples were routine surgical specimens that were fixed in formalin, processed and stored according to standard histologic protocols and the rules of the local medical ethics committee of the FNB. The selection of tumor material was based on a previous clinical examination for HER2 performed by immunohistochemistry (IHC).
2.1. DNA/RNA preparation
On the original slides of each of the samples, a representative area containing tumor cells was marked by a pathologist (Dr. Z. Špurkova). Thereafter, 3-5 microtome slices (diameter 5 µM) were obtained from the corresponding area in each paraffin block, and RNA/DNA was isolated using CE-IVD certified diagnostics: FFPE RNA Purification Kit Dx (Norgen Biotek, Thorold, Ontario, CA) or AmoyDx FFPE DNA/RNA kit (Xiamen, China). The clear advantage of the AmoyDx kit was the simultaneous isolation of both RNA and DNA without affecting RNA yields compared to the Norgen Biotek extraction. The concentration of the DNA/RNA was determined using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Fluorometric analysis was performed by Quant-iTTM dsDNA HS Assay Kit Quant -iTTM RNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) for, respective DNA and RNA.
2.2. Targets
Real-time PCR assays for the target gene HER2 and the reference gene APP (encoding amyloid precursor protein, were run on both levels, as DNA so cDNA. As expected, HER2 overexpression was present in all tumors with HER2 gene amplification but was uncommon in breast cancers without gene amplification [
4]. The APP gene has been selected as a „housekeeping“/“backbone“ marker. Design of primers for gene expression study was performed in the ExonSurfer software allowing to avoid amplification of genomic template (
Table n. 1) [
5].
2.3. Reverse transription and real-time PCR of cDNA (gene expression level)
cDNA was synthesized using ProtoScript II First Strand cDNA Synthesis Kit (New England BioLab., Ipswich, MA, USA) according to the manufacturer’s instruction. The Random Primer Mix yields shorter cDNAs on average and can be used for the detection of multiple short RT-PCR products, appropriately designed for fragmented nucleic acids.
The 20-µl reaction mixture contained 2x Luna® Universal qPCR Master Mix (New England BioLab., Ipswich, MA, USA), 0,05 µM forward primer, 0,05 µM reverse primer (all from Generi-Biotech s.r.o., H. Kralove, CZ) and 2,5 ul DNA or cDNA. See the New England Biolabs for the exact protocol [
6]. In general, the volume of cDNA product should not exceed 1/10 of the PCR reaction volume. The PCR program started with one cycle at 95°C for 2 mins. The amplification was run for 40 cycles with short denaturation at 95°C for 15sec, and annealing with extension at 60°C for 45 sec. All reactions were performed in a Connect CFX real-time PCR system (Bio-Rad, Hercules, CA, USA).
2.4. IHC assessment
A total of 10 breast cancer tissue samples were analysed immunohistochemically using the c-erbB-2 oncoprotein antibody (DAKO, Glostrup, Denmark) with internal (on slide) positive and negative controls. Staining was performed in a Ventana immunohistochemistry staining system.
Immunostaining was evaluated by a pathologist and was performed according to ASCO guidelines [
7]: negative for 0 (no or week incomplete staining <10% tumour cells) and 1+ (faint or barely perceptible incomplete membrane staining > 10% tumour cells); equivocal for 2+ (weak to moderate complete membrane staining >10% tumour cells) and positive for 3+ (>10% tumour cells with circumferential complete, intense membrane staining). The samples were then sent to the reference laboratory of the Institute of Pathology VFN Prague (Czech Republic), where the determination was performed using the certified Ventana HER/Neu kit, clone 4B5. Some samples with a negative and indeterminate result (positivity 0+,1+,2+) were subsequently examined by FISH, whereby no amplification of the HER2 gene could be detected (
Table 2). FISH was not performed on definitely positive tumors (3+) (samples
no. 3, 7).
3. Results
3.1. Assessment of HER2 status
We developed a protocol to determine the copy number of the HER2 gene by quantitative RT-PCR/qPCR using SYBR green dye I, with which we examined 10 blind DNA samples from tumour tissue of breast cancer patients. The content of the target DNA in tumor samples were quantified by using standard curves according to the Control Genomic Human DNA (ref.no. 4312660 Themo Fisher Scientific, Waltham, MA, USA) and the same PCR protocol as for gene expression profiling (written in the section
2.3.). In order to correctly determine the ratios in FFPE samples, we were forced to replace the 168bp amplicon of the TBP reference gene (
TATA-binding protein gene) with a shorter one (APP, 124bp, see
Table 1). HER2 copy number in each sample, was normalized on the basis of its content of the „backbone“ reference gene that is located at
21q21, which has not been found to exhibit alterations in breast cancer patients [
1]. and amplification efficiency was found to be similar to HER2 gene [
8]. Standard curves for both the targets; HER2 and the APP reference gene for each run were constructed using threefold serial dilution ranging from 1 ng/µl to 50 pg/µl of the Control DNA (cat.no. 4312660, Thermo Fisher Scientific, Waltham, MA, USA) and CFX Manager software (Bio-Rad, Hercules, CA, USA). Supplementary file S2 contains such runs with constructed calibration curves, then HER2 copy number was calculated as a ratio
HER2 (ng/μl) to
APP (ng/μl). To test the reproducibility, we analyzed ten samples in duplicates. Amplification of HER2 has been already defined as 5 copies of this gene above average ploidy of the tumor sample [
9]. However, as we found small differences between APP and HER2 amplification efficiency, the threshold for a positive result was appropriately shifted (
Supplementary Figure S1a).
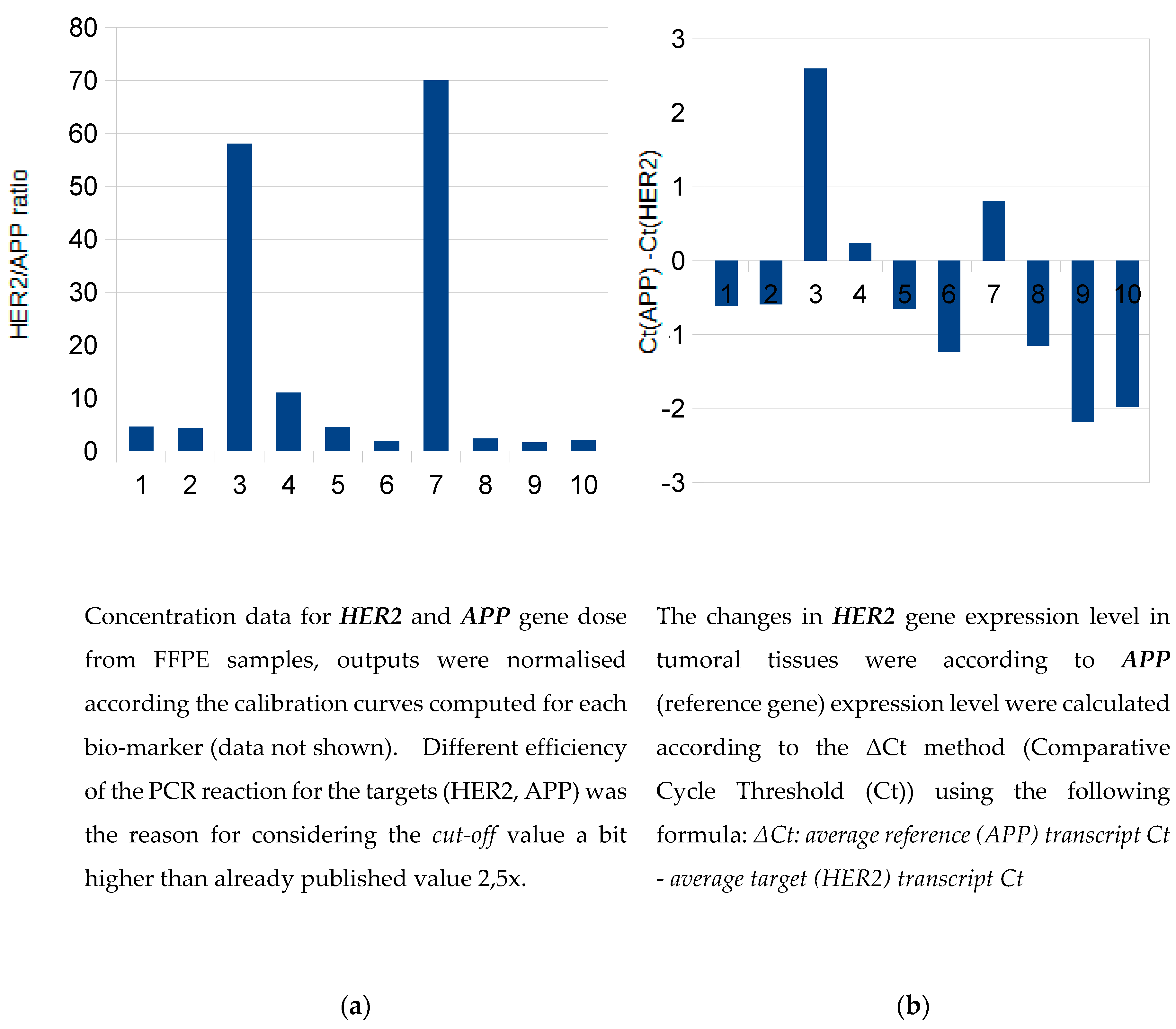
The samples 3 and 7 were strongly positive (in agreement with IHC assessment 3+) and the
sample no. 4 appeared slightly above the
cut-off value (
Figure 1a). Dual concept for evalutation revealed concordant results also on gene expression level.
(Figure 1b).
Relative quantification of HER2 expression was performed using the ΔCt method. For HER2-positive samples, the increase in the difference between the Ct values of HER2 and APP transcripts in tumour tissue essentially corresponded to the overexpression of the HER2 gene (
Figure 1b). The real-time PCR gene expression data for another positive sample (
no. 7) are shown in
Supplementary Figure S3.
Different studies demonstrate that quantitative real-time PCR approaches are valuable tools for the assessment of HER2 gene overexpression and can provide a reliable alternative to FISH and IHC determinations [
10,
11,
12]. Generally, PCR-based assays can detect changes in HER2 gene number as well as gene expression. Q-PCR assays based on DNA/cDNA analyses are describing as sensitive enough, faster, easy to perform, specific and cost-effective to measure HER2 alterations and able to analyse multiple samples simultaneously [
13]. The high sensitivity of real-time qPCR means that even minute amounts of DNA or RNA can be detected in FFPE tissue [
4], opening up the possibility of performing retrospective clinical and molecular studies on the large sample archives stored in pathology institutes [
14]. The design of short amplicon primers and the use of random primers in reverse transcription appear to be crucial both for long-term archived FFPE blocks and for the precise calculation of ratios between the focused biomarkers. In addition, linkage to reference genes enables the HER2-status to be monitored in time course and therefore it can reflect
on therapy efficiency. Determined gene expression ratios are also useful in HER2-Low breast cancer patients [
15].
Strongly positive samples from IHC decising (samples no. 3, 7) were also positive using molecular method determination, consistently at both DNA and RNA levels. Regarding to the requirements on content of tumor tissue, usually it is about 20% [
16] it seems in agreement with another studies [
3] that molecular approaches are rather sensitive and less demanding on percentage of tumour cells, with fractions of amplified cells as small as 5%. Our system offers a decisive advantage: the possibility to perform a double evaluation, which facilitates the interpretation of the results, enhances result interpretation flexibility and quality control (
Figure 2).
Three samples that were classified as positive by real-time PCR were also positive in the IHC detection. This indicates that real-time PCR does not give false-positive results, which is consistent with a previously published study [
17]. We hope that the
LDT -assay developed by our laboratory shows better analytical performance than FISH (also detectable in the current study) and would be useful also in patients with HER2-low breast cancer (sample no. 4), so that more patients could benefit from anti-HER2 treatment. The consideration here is that the combination of the recommended IHC and FISH/CISH methods still leads to results that fall into the equivocal range and could exclude true positive samples, and that qPCR methods could be useful as a complement.
5. Conclusions
While IHC requires knowledge for reading to achieve reproducibility, long-term skilled techniques and use of expensive antibodies and reagents that threaten to pro-expire; on the contrary, molecular-biological approaches would consider where large numbers of patients are not expected. We anticipate their use especially in small laboratories without the necessary equipment (a fluorescent microscope), everywhere the risk of overexposure to chemistry increases and the huge manual workload may compromise the quality of results.
The increasing number of publications and new diagnostics (e.g. MammaTyper®, OncotypeDX® test, Xpert® Breast Cancer STRAT4) on the successful implementation of HER2 determination by molecular biology methods speaks more and more for the idea that not the inherent limitations of the method but its good standardization is important for its complementary use as a third alternative.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
“Conceptualization, Z.K.; methodology, E.R.; software E.R.; validation, E.R., Z.S. and K.V.; formal analysis E.R..; investigation, E.R., K.V. and Z.S.; resources, Z.S.; data curation, E.R.; writing—original draft preparation, E.R.; writing—review and editing, E.R.; visualization, E.R.; supervision, Z.K.; project administration, E.R.; funding acquisition, Z.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Third Faculty of Medicine, Charles University, Prague, Czech Republic.
Institutional Review Board Statement
The samples were routine surgical specimens that had been fixed in formalin, processed, and stored according to standard histological protocols and rules of the local medical ethics committee at University Hospital Bulovka. The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of the University Hospital Bulovka (protocol code 10742/EK-Z and date of approval 7.02.2023).
Informed Consent Statement
The human DNA control was purchased directly as a commercial product from pooled donors; hence, informed consent was not required. Likewise, a cDNA pool for testing amplification efficiency was prepared by mixing cDNA from 10 volunteers, who signed informed consent (on the rules of the local medical committee at University Hospital Bulovka). Research studies were performed in accordance with Regulation 2016/679 of the European Parliament and the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, following Directive 95/46/EC (General Data Protection Regulation).
Data Availability Statement
The original contributions presented in the study are included in the article, futher inquiries can be directed to the corresponding author/s.
Acknowledgments
We thank to the Carolina Biosystems s.r.o. for their material support in the field of nucleic acid isolation from FFPE samples.
Conflicts of Interest
Ema Ruszova, Katerina Vlcanova, Zuzana Spurkova and Ziad Khaznadar declare no conflicts of interest.
References
- Gunnarsson, C.; Jansson; Holmlund; Olsson, H. Methods for Evaluating HER2 Status in Breast Cancer: Comparison of IHC, FISH, and Real-Time PCR Analysis of Formalin-Fixed Paraffin-Embedded Tissue. PLMI 2013, 31. [CrossRef]
- Sauter, G.; Lee, J.; Bartlett, J.M.S.; Slamon, D.J.; Press, M.F. Guidelines for Human Epidermal Growth Factor Receptor 2 Testing: Biologic and Methodologic Considerations. JCO 2009, 27, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Zoppoli, G.; Garuti, A.; Cirmena, G.; Di Cantogno, L.V.; Botta, C.; Gallo, M.; Ferraioli, D.; Carminati, E.; Baccini, P.; Curto, M.; et al. Her2 Assessment Using Quantitative Reverse Transcriptase Polymerase Chain Reaction Reliably Identifies Her2 Overexpression without Amplification in Breast Cancer Cases. J Transl Med 2017, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Bièche, I.; Onody, P.; Laurendeau, I.; Olivi, M.; Vidaud, D.; Lidereau, R.; Vidaud, M. Real-Time Reverse Transcription-PCR Assay for Future Management of ERBB2-Based Clinical Applications. Clin Chem 1999, 45, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Monfort-Lanzas, P.; Rusu, E.C.; Parrakova, L.; Karg, C.A.; Kernbichler, D.-E.; Rieder, D.; Lackner, P.; Hackl, H.; Gostner, J.M. ExonSurfer: A Web-Tool to Design Primers at Exon–Exon Junctions. BMC Genomics 2024, 25, 594. [Google Scholar] [CrossRef] [PubMed]
- Luna® Universal qPCR Master Mix Protocol.
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.S.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. JCO 2013, 31, 3997–4013. [Google Scholar] [CrossRef] [PubMed]
- Lyng, M.B.; Lænkholm, A.-V.; Pallisgaard, N.; Ditzel, H.J. Identification of Genes for Normalization of Real-Time RT-PCR Data in Breast Carcinomas. BMC Cancer 2008, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.S.; Fakih, M.; Ali, S.M.; Elvin, J.A.; Schrock, A.B.; Suh, J.; Vergilio, J.-A.; Ramkissoon, S.; Severson, E.; Daniel, S.; et al. Targeting HER2 in Colorectal Cancer: The Landscape of Amplification and Short Variant Mutations in ERBB2 and ERBB3. Cancer 2018, 124, 1358–1373. [Google Scholar] [CrossRef] [PubMed]
- Noske, A.; Loibl, S.; Darb-Esfahani, S.; Roller, M.; Kronenwett, R.; Müller, B.M.; Steffen, J.; von Toerne, C.; Wirtz, R.; Baumann, I.; et al. Comparison of Different Approaches for Assessment of HER2 Expression on Protein and mRNA Level: Prediction of Chemotherapy Response in the Neoadjuvant GeparTrio Trial (NCT00544765). Breast Cancer Res Treat 2011, 126, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Borges, P.C.C.; Spencer, H.B.; Barbosa, C.; Costa, V.; Furtado, A.; Leal, M.C.; Lopes, C.; Ferreira, D.; Carvalho, A.L.; Dos-Santos-Silva, I.; et al. XPERT® Breast Cancer STRAT4 as an Alternative Method of Identifying Breast Cancer Phenotype in Cape Verde (Preliminary Results). Ecancermedicalscience 2023, 17, 1530. [Google Scholar] [CrossRef] [PubMed]
- Koudelakova, V.; Berkovcova, J.; Trojanec, R.; Vrbkova, J.; Radova, L.; Ehrmann, J.; Kolar, Z.; Melichar, B.; Hajduch, M. Evaluation of HER2 Gene Status in Breast Cancer Samples with Indeterminate Fluorescence in Situ Hybridization by Quantitative Real-Time PCR. J Mol Diagn 2015, 17, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Skálová, H.; Dundr, P.; Povýšil, C.; Velenská, Z.; Petruželka, L.; Tvrdík, D. Study of the Effect of Neoadjuvant Chemotherapy on the Status of Her2/Neu. Folia Biol (Praha) 2011, 57, 191–199. [Google Scholar] [PubMed]
- Gjerdrum, L.M.; Sorensen, B.S.; Kjeldsen, E.; Sorensen, F.B.; Nexo, E.; Hamilton-Dutoit, S. Real-Time Quantitative PCR of Microdissected Paraffin-Embedded Breast Carcinoma: An Alternative Method for HER-2/Neu Analysis. J Mol Diagn 2004, 6, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Gaudio, M.; Jacobs, F.; Benvenuti, C.; Saltalamacchia, G.; De Sanctis, R.; Santoro, A.; Zambelli, A. 53P HER2 Low by Immunohistochemistry (IHC) and Gene Expression by qRT-PCR Using OncotypeDX in ER+ Early Breast Cancer. ESMO Open 2023, 8, 101277. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Hovelson, D.H.; Suga, J.M.; Anderson, D.M.; Koh, H.A.; Dees, E.C.; McNulty, B.; Burkard, M.E.; Guarino, M.; Khatri, J.; et al. Real-World Performance of a Comprehensive Genomic Profiling Test Optimized for Small Tumor Samples. JCO Precision Oncology 2021, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Schlemmer, B.O.; Sorensen, B.S.; Overgaard, J.; Olsen, K.E.; Gjerdrum, L.M.; Nexo, E. Quantitative PCR--New Diagnostic Tool for Quantifying Specific mRNA and DNA Molecules: HER2/Neu DNA Quantification with LightCycler Real-Time PCR in Comparison with Immunohistochemistry and Fluorescence in Situ Hybridization. Scand J Clin Lab Invest 2004, 64, 511–522. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).