Submitted:
02 December 2024
Posted:
03 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
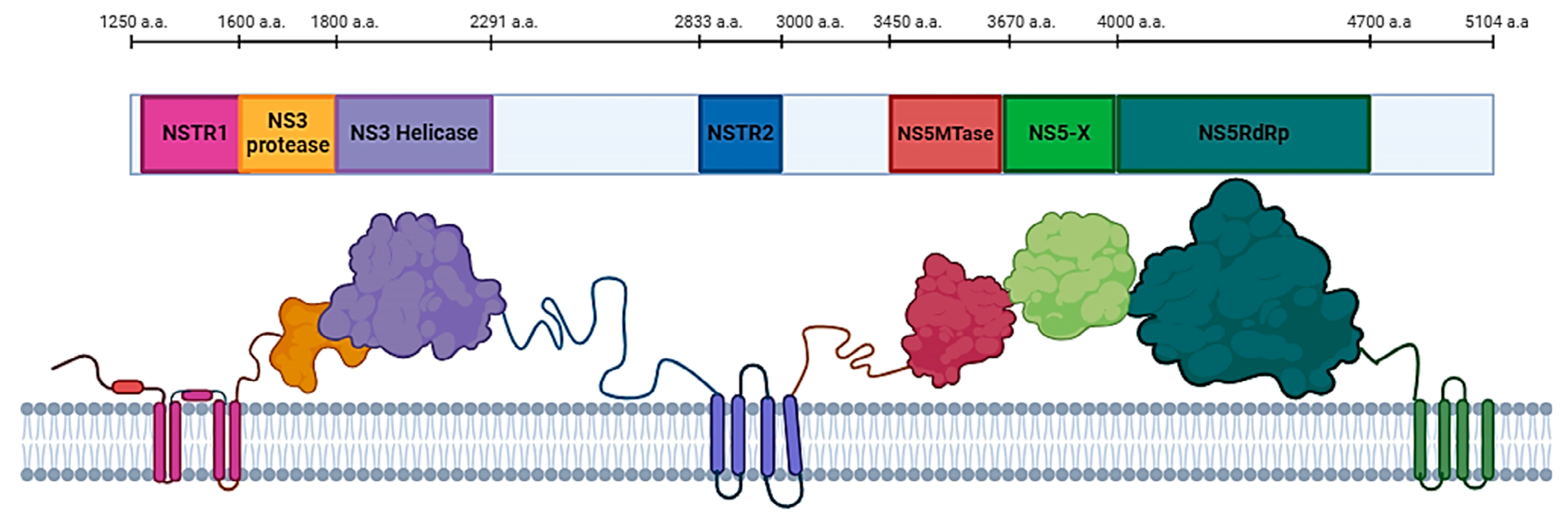
2.1. Determination of Localization of Haseki Tick Virus Nonstructural Proteins
2.1.1. Search for Putative HSTV Nonstructural Transmembrane Proteins
2.1.2. Search for putative HSTV nonstructural cytoplasmic proteins
- NS3 protein
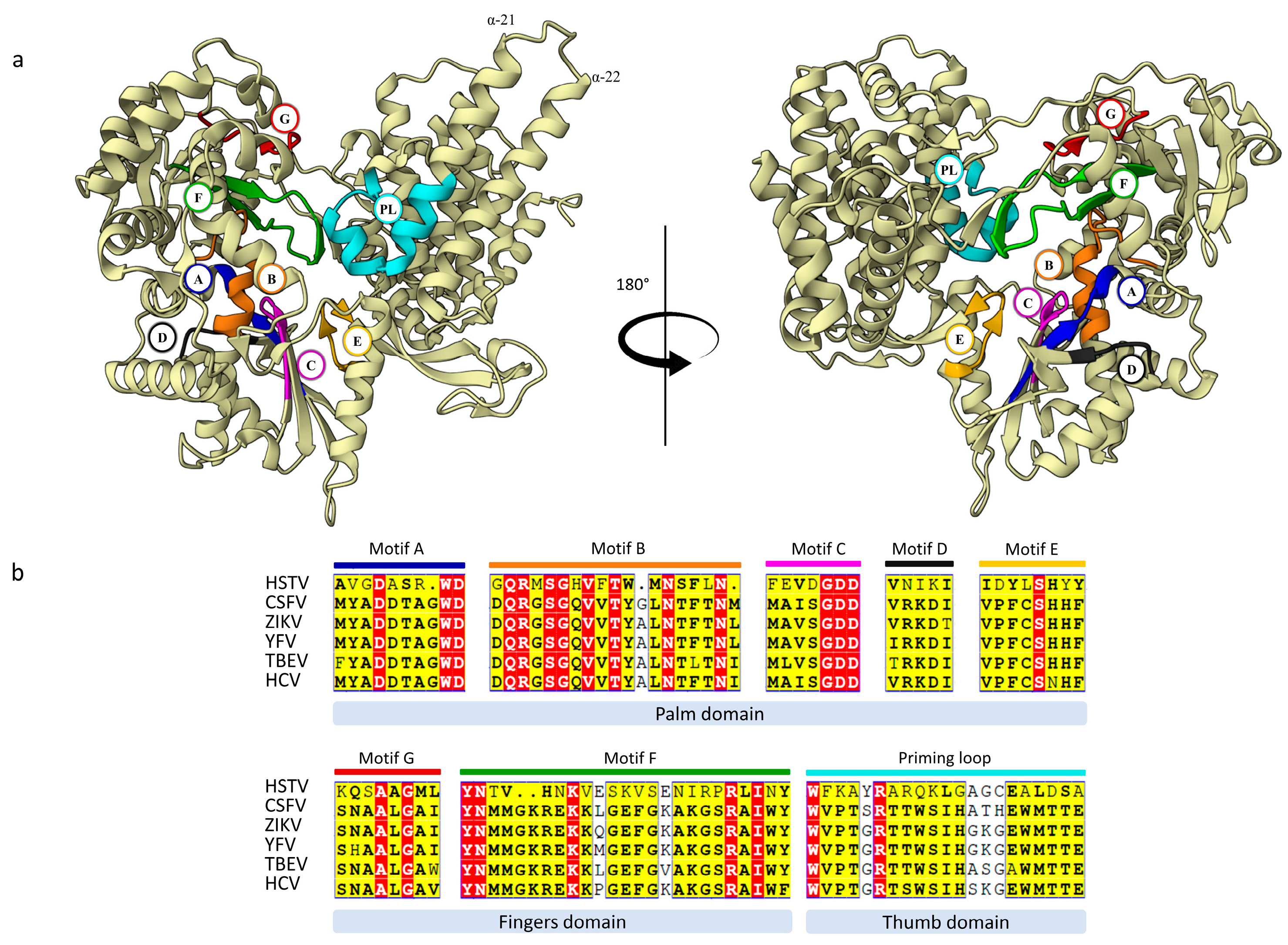
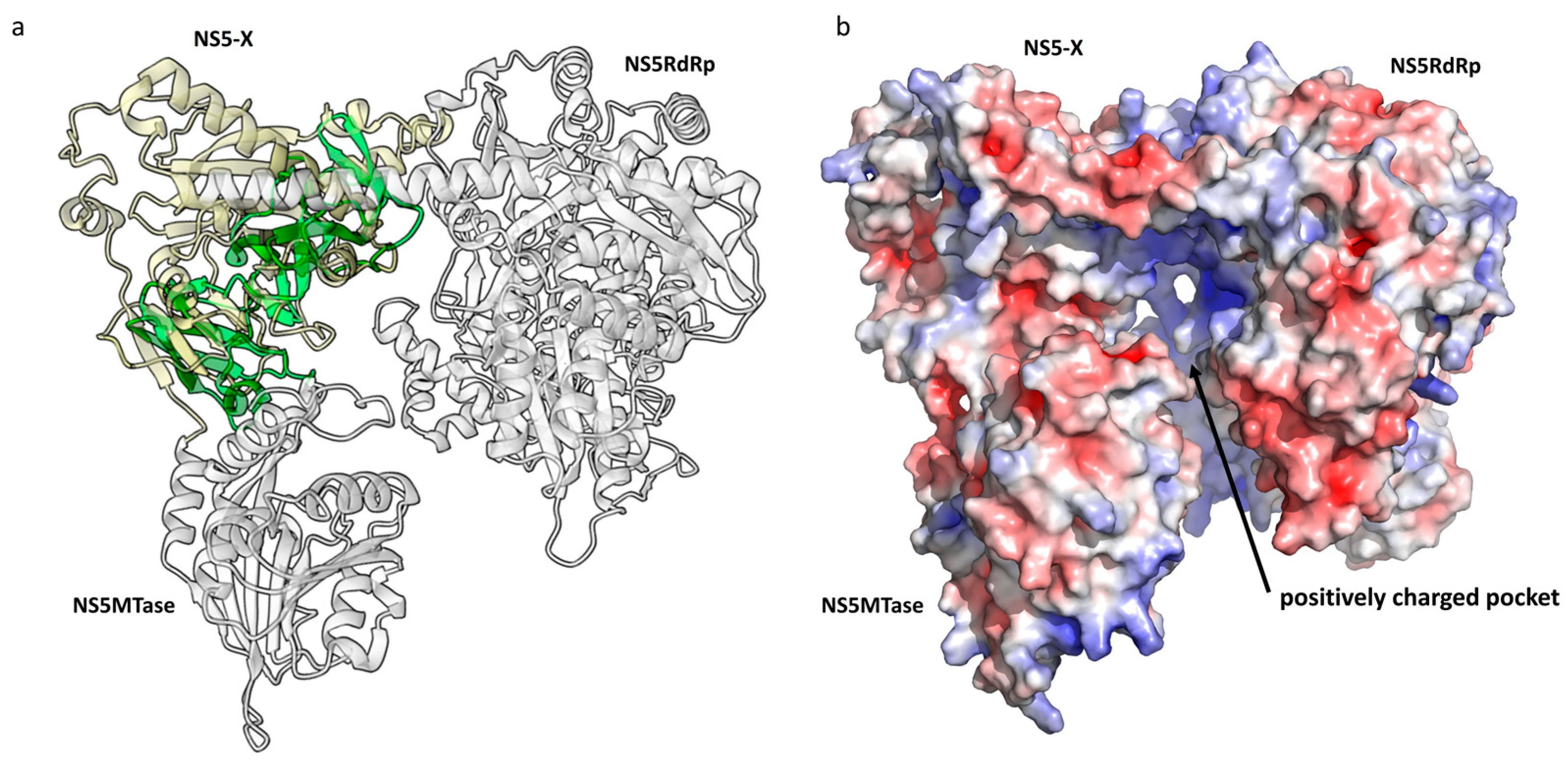
- Methyltransferase and RNA-dependent RNA-polymerase
2.2. Tertiary Structure Models of Haseki Tick Virus Nonstructural Proteins
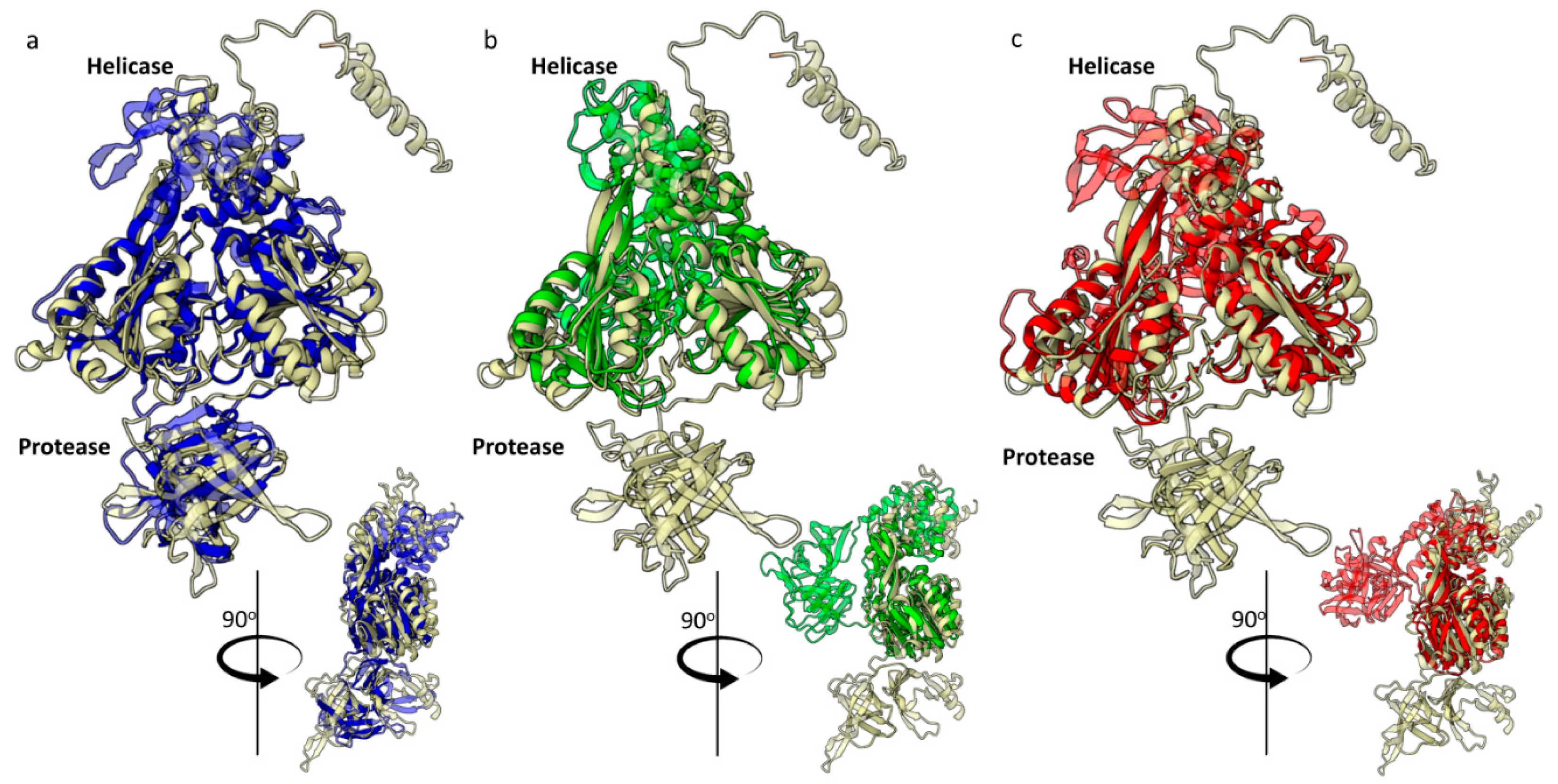
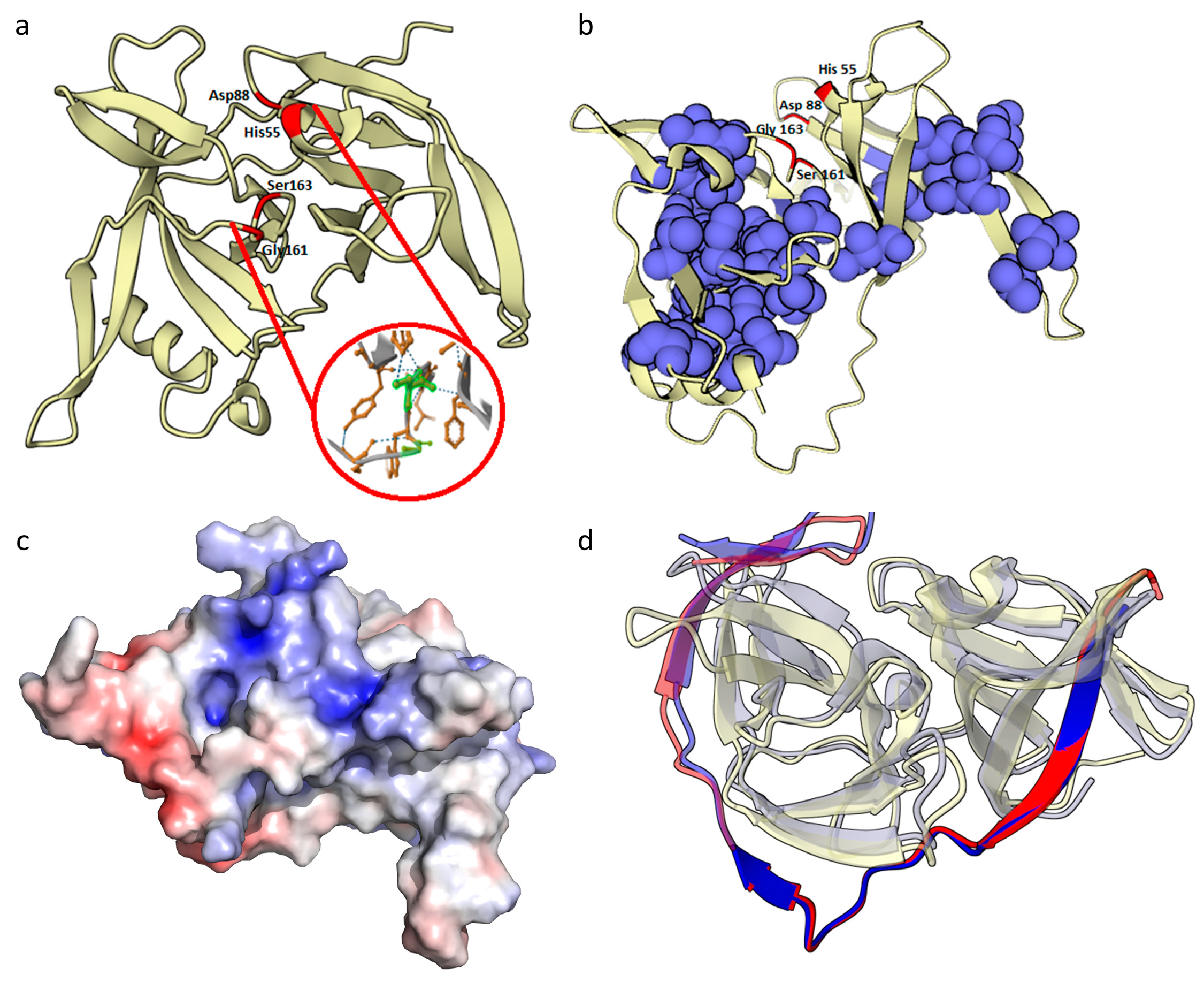
2.2.1. HSTV NS3 Protein
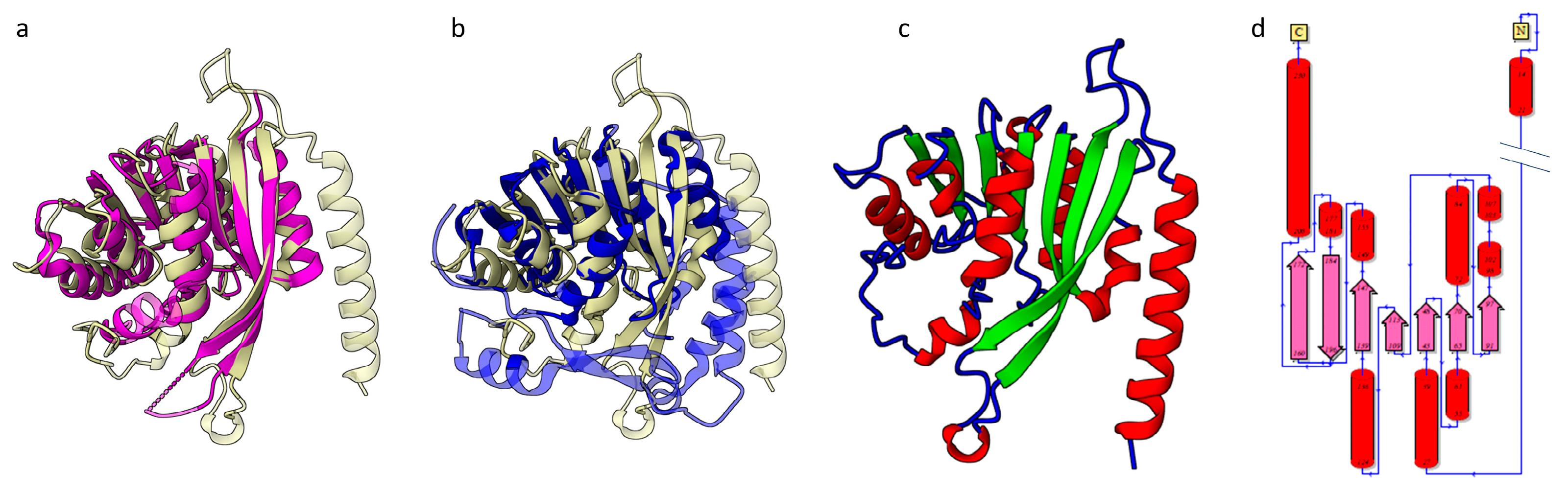
- HSTV NS3 protease
- HSTV NS3 helicase
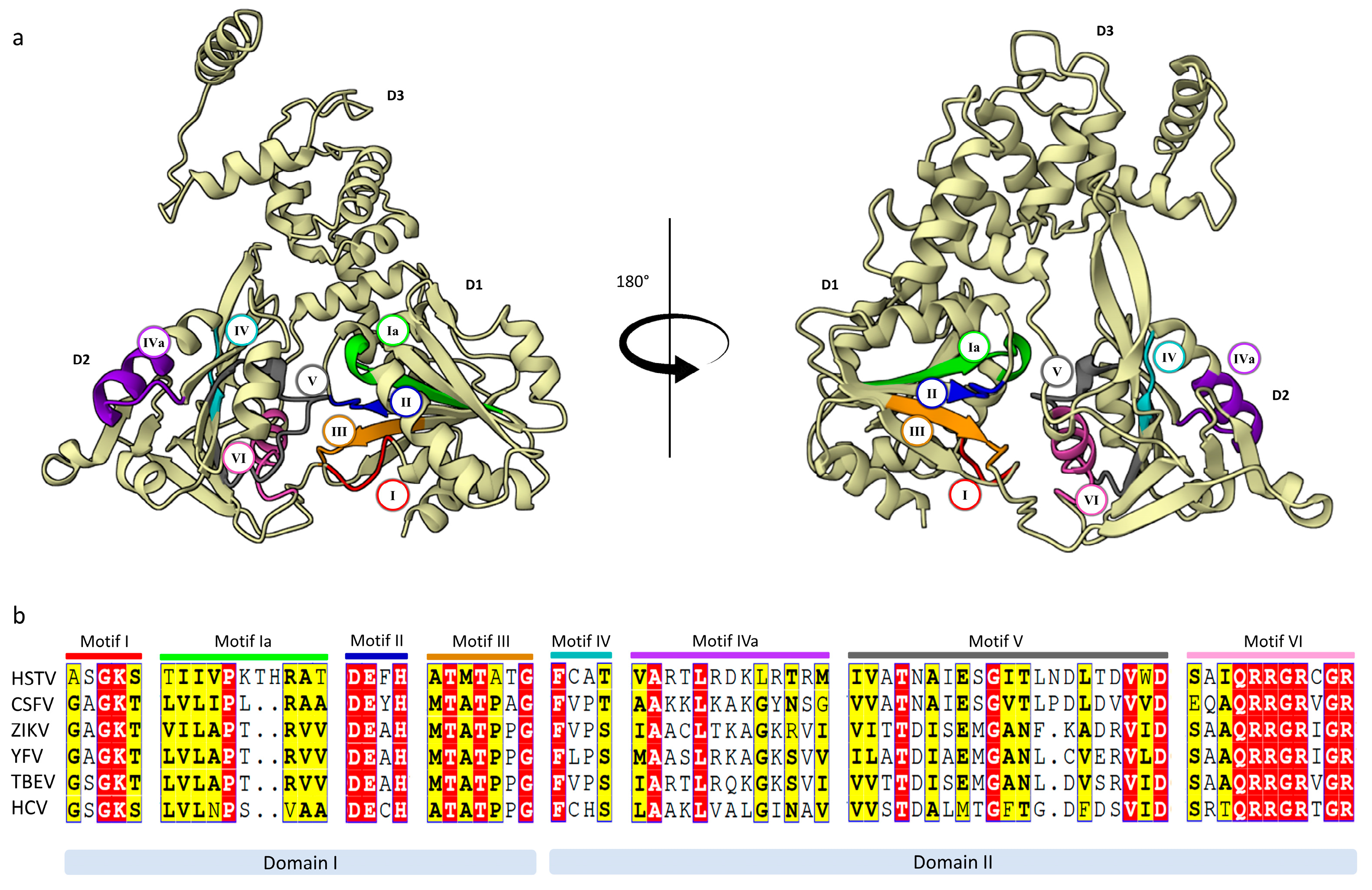
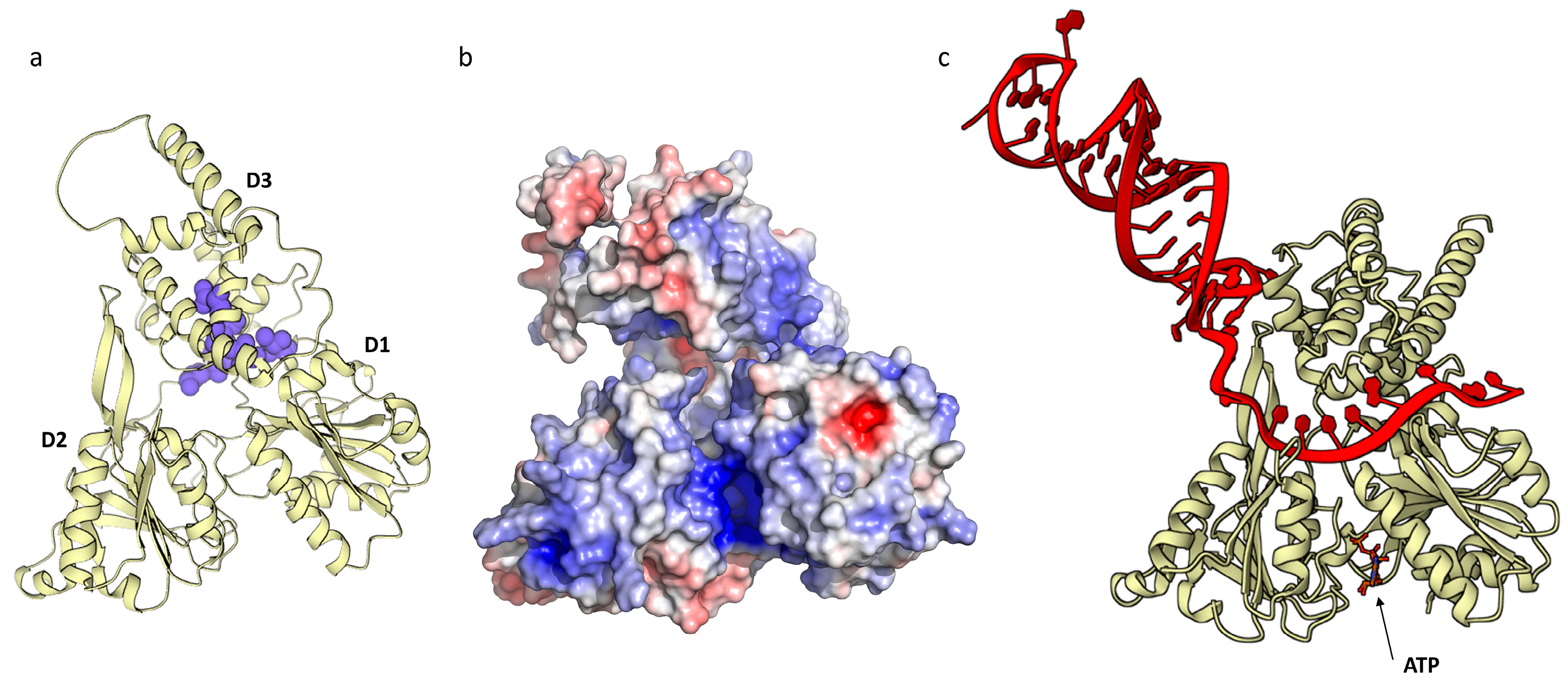
2.2.2. HSTV NS5 RNA-Dependent RNA-Polymerase
2.2.3. HSTV NS5 Methyltransferase
3. Discussion
4. Materials and Methods
4.1. HSTV Sequence
4.2. Multiple Sequence Alignment (MSA) and Analysis
4.3. Search for Transmembrane Nonstructural Proteins
4.4. Model Building Using AlphaFold 3 and Structural Alignments
4.5. Protein Structure and Function Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zerbini, F.M.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Hendrickson, R.C.; et al. Changes to Virus Taxonomy and the ICTV Statutes Ratified by the International Committee on Taxonomy of Viruses (2023). Arch Virol 2023, 168, 175. [CrossRef]
- Paul Chrystal The History of the World in 100 Pandemic; Pen and Sword History: Barnsley, United Kingdom, 2021; ISBN 9781399005432.
- Nomburg, J.; Doherty, E.E.; Price, N.; Bellieny-Rabelo, D.; Zhu, Y.K.; Doudna, J.A. Birth of Protein Folds and Functions in the Virome. Nature 2024, 633, 710–717. [CrossRef]
- Ergunay, K.; Bourke, B.P.; Reinbold-Wasson, D.D.; Nikolich, M.P.; Nelson, S.P.; Caicedo-Quiroga, L.; Vaydayko, N.; Kirkitadze, G.; Chunashvili, T.; Long, L.S.; et al. The Expanding Range of Emerging Tick-Borne Viruses in Eastern Europe and the Black Sea Region. Sci Rep 2023, 13, 19824. [CrossRef]
- Kartashov, M.Y.; Gladysheva, A. V.; Shvalov, A.N.; Tupota, N.L.; Chernikova, A.A.; Ternovoi, V.A.; Loktev, V.B. Novel Flavi-like Virus in Ixodid Ticks and Patients in Russia. Ticks Tick Borne Dis 2023, 14, 102101. [CrossRef]
- Shi, M.; Lin, X.-D.; Vasilakis, N.; Tian, J.-H.; Li, C.-X.; Chen, L.-J.; Eastwood, G.; Diao, X.-N.; Chen, M.-H.; Chen, X.; et al. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J Virol 2016, 90, 659–669. [CrossRef]
- Temmam, S.; Chrétien, D.; Bigot, T.; Dufour, E.; Petres, S.; Desquesnes, M.; Devillers, E.; Dumarest, M.; Yousfi, L.; Jittapalapong, S.; et al. Monitoring Silent Spillovers Before Emergence: A Pilot Study at the Tick/Human Interface in Thailand. Front Microbiol 2019, 10. [CrossRef]
- Zakham, F.; Albalawi, A.E.; Alanazi, A.D.; Truong Nguyen, P.; Alouffi, A.S.; Alaoui, A.; Sironen, T.; Smura, T.; Vapalahti, O. Viral RNA Metagenomics of Hyalomma Ticks Collected from Dromedary Camels in Makkah Province, Saudi Arabia. Viruses 2021, 13, 1396. [CrossRef]
- Bratuleanu, B.E.; Temmam, S.; Chrétien, D.; Regnault, B.; Pérot, P.; Bouchier, C.; Bigot, T.; Savuța, G.; Eloit, M. The Virome of Rhipicephalus , Dermacentor and Haemaphysalis Ticks from Eastern Romania Includes Novel Viruses with Potential Relevance for Public Health. Transbound Emerg Dis 2022, 69, 1387–1403. [CrossRef]
- Sameroff, S.; Tokarz, R.; Vucelja, M.; Jain, K.; Oleynik, A.; Boljfetić, M.; Bjedov, L.; Yates, R.A.; Margaletić, J.; Oura, C.A.L.; et al. Virome of Ixodes Ricinus, Dermacentor Reticulatus, and Haemaphysalis Concinna Ticks from Croatia. Viruses 2022, 14, 929. [CrossRef]
- Zhang, J.; Zheng, Y.-C.; Chu, Y.-L.; Cui, X.-M.; Wei, R.; Bian, C.; Liu, H.-B.; Yao, N.-N.; Jiang, R.-R.; Huo, Q.-B.; et al. Skin Infectome of Patients with a Tick Bite History. Front Cell Infect Microbiol 2023, 13. [CrossRef]
- Duan, Y.; Zeng, M.; Jiang, B.; Zhang, W.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; et al. Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication. Viruses 2019, 11, 929. [CrossRef]
- Du Pont, K.E.; McCullagh, M.; Geiss, B.J. Conserved Motifs in the Flavivirus <scp>NS3 RNA</Scp> Helicase Enzyme. WIREs RNA 2022, 13. [CrossRef]
- Li, R.; Niu, Z.; Liu, Y.; Bai, X.; Wang, D.; Chen, C. Crystal Structure and Cap Binding Analysis of the Methyltransferase of Langat Virus. Antiviral Res 2022, 208, 105459. [CrossRef]
- Chen, S.; Harris, M. NS5A Domain I Antagonises PKR to Facilitate the Assembly of Infectious Hepatitis C Virus Particles. PLoS Pathog 2023, 19, e1010812. [CrossRef]
- Shiryaev, S.A.; Cieplak, P.; Cheltsov, A.; Liddington, R.C.; Terskikh, A. V. Dual Function of Zika Virus NS2B-NS3 Protease. PLoS Pathog 2023, 19, e1011795. [CrossRef]
- Benarroch, D.; Selisko, B.; Locatelli, G.A.; Maga, G.; Romette, J.-L.; Canard, B. The RNA Helicase, Nucleotide 5′-Triphosphatase, and RNA 5′-Triphosphatase Activities of Dengue Virus Protein NS3 Are Mg2+-Dependent and Require a Functional Walker B Motif in the Helicase Catalytic Core. Virology 2004, 328, 208–218. [CrossRef]
- Luo, D.; Xu, T.; Watson, R.P.; Scherer-Becker, D.; Sampath, A.; Jahnke, W.; Yeong, S.S.; Wang, C.H.; Lim, S.P.; Strongin, A.; et al. Insights into RNA Unwinding and ATP Hydrolysis by the Flavivirus NS3 Protein. EMBO J 2008, 27, 3209–3219. [CrossRef]
- Lu, G.; Gong, P. A Structural View of the RNA-Dependent RNA Polymerases from the Flavivirus Genus. Virus Res 2017, 234, 34–43. [CrossRef]
- Wu, J.; Liu, W.; Gong, P. A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family. Int J Mol Sci 2015, 16, 12943–12957. [CrossRef]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D.; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Structural Basis for RNA Replication by the Hepatitis C Virus Polymerase. Science (1979) 2015, 347, 771–775. [CrossRef]
- Shu, B.; Gong, P. Structural Basis of Viral RNA-Dependent RNA Polymerase Catalysis and Translocation. Proceedings of the National Academy of Sciences 2016, 113. [CrossRef]
- Krejčová, K.; Krafcikova, P.; Klima, M.; Chalupska, D.; Chalupsky, K.; Zilecka, E.; Boura, E. Structural and Functional Insights in Flavivirus NS5 Proteins Gained by the Structure of Ntaya Virus Polymerase and Methyltransferase. Structure 2024, 32, 1099-1109.e3. [CrossRef]
- Zayed, A.A.; Lücking, D.; Mohssen, M.; Cronin, D.; Bolduc, B.; Gregory, A.C.; Hargreaves, K.R.; Piehowski, P.D.; White III, R.A.; Huang, E.L.; et al. Efam: An e Xpanded, Metaproteome-Supported HMM Profile Database of Viral Protein Fam Ilies. Bioinformatics 2021, 37, 4202–4208. [CrossRef]
- Durairaj, J.; Waterhouse, A.M.; Mets, T.; Brodiazhenko, T.; Abdullah, M.; Studer, G.; Tauriello, G.; Akdel, M.; Andreeva, A.; Bateman, A.; et al. Uncovering New Families and Folds in the Natural Protein Universe. Nature 2023, 622, 646–653. [CrossRef]
- Barrio-Hernandez, I.; Yeo, J.; Jänes, J.; Mirdita, M.; Gilchrist, C.L.M.; Wein, T.; Varadi, M.; Velankar, S.; Beltrao, P.; Steinegger, M. Clustering Predicted Structures at the Scale of the Known Protein Universe. Nature 2023, 622, 637–645. [CrossRef]
- Murray, C.L.; Jones, C.T.; Rice, C.M. Architects of Assembly: Roles of Flaviviridae Non-Structural Proteins in Virion Morphogenesis. Nat Rev Microbiol 2008, 6, 699–708. [CrossRef]
- Dietz, C.; Maasoumy, B. Direct-Acting Antiviral Agents for Hepatitis C Virus Infection—From Drug Discovery to Successful Implementation in Clinical Practice. Viruses 2022, 14, 1325. [CrossRef]
- Goh, J.Z.H.; De Hayr, L.; Khromykh, A.A.; Slonchak, A. The Flavivirus Non-Structural Protein 5 (NS5): Structure, Functions, and Targeting for Development of Vaccines and Therapeutics. Vaccines (Basel) 2024, 12, 865. [CrossRef]
- Xu, J.; Zhang, Y. How Significant Is a Protein Structure Similarity with TM-Score = 0.5? Bioinformatics 2010, 26, 889–895. [CrossRef]
- Sinha, S.; Singh, K.; Ravi Kumar, Y.S.; Roy, R.; Phadnis, S.; Meena, V.; Bhattacharyya, S.; Verma, B. Dengue Virus Pathogenesis and Host Molecular Machineries. J Biomed Sci 2024, 31, 43. [CrossRef]
- Osawa, T.; Aoki, M.; Ehara, H.; Sekine, S. Structures of Dengue Virus RNA Replicase Complexes. Mol Cell 2023, 83, 2781-2791.e4. [CrossRef]
- Dubrau, D.; Tortorici, M.A.; Rey, F.A.; Tautz, N. A Positive-Strand RNA Virus Uses Alternative Protein-Protein Interactions within a Viral Protease/Cofactor Complex to Switch between RNA Replication and Virion Morphogenesis. PLoS Pathog 2017, 13, e1006134. [CrossRef]
- Teramoto, T.; Choi, K.H.; Padmanabhan, R. Flavivirus Proteases: The Viral Achilles Heel to Prevent Future Pandemics. Antiviral Res 2023, 210, 105516. [CrossRef]
- Saraste, M.; Sibbald, P.R.; Wittinghofer, A. The P-Loop — a Common Motif in ATP- and GTP-Binding Proteins. Trends Biochem Sci 1990, 15, 430–434. [CrossRef]
- Banroques, J.; Doère, M.; Dreyfus, M.; Linder, P.; Tanner, N.K. Motif III in Superfamily 2 “Helicases” Helps Convert the Binding Energy of ATP into a High-Affinity RNA Binding Site in the Yeast DEAD-Box Protein Ded1. J Mol Biol 2010, 396, 949–966. [CrossRef]
- Sampath, A.; Xu, T.; Chao, A.; Luo, D.; Lescar, J.; Vasudevan, S.G. Structure-Based Mutational Analysis of the NS3 Helicase from Dengue Virus. J Virol 2006, 80, 6686–6690. [CrossRef]
- Yang, J.; Jing, X.; Yi, W.; Li, X.-D.; Yao, C.; Zhang, B.; Zheng, Z.; Wang, H.; Gong, P. Crystal Structure of a Tick-Borne Flavivirus RNA-Dependent RNA Polymerase Suggests a Host Adaptation Hotspot in RNA Viruses. Nucleic Acids Res 2021, 49, 1567–1580. [CrossRef]
- Bruenn, J.A. A Structural and Primary Sequence Comparison of the Viral RNA-Dependent RNA Polymerases. Nucleic Acids Res 2003, 31, 1821–1829. [CrossRef]
- Selisko, B.; Papageorgiou, N.; Ferron, F.; Canard, B. Structural and Functional Basis of the Fidelity of Nucleotide Selection by Flavivirus RNA-Dependent RNA Polymerases. Viruses 2018, 10, 59. [CrossRef]
- Mushegian, A. Methyltransferases of Riboviria. Biomolecules 2022, 12, 1247. [CrossRef]
- Jia, H.; Zhong, Y.; Peng, C.; Gong, P. Crystal Structures of Flavivirus NS5 Guanylyltransferase Reveal a GMP-Arginine Adduct. J Virol 2022, 96. [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat Biotechnol 2017, 35, 1026–1028. [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res 2018, 46, W200–W204. [CrossRef]
- Dobson, L.; Reményi, I.; Tusnády, G.E. CCTOP: A Consensus Constrained TOPology Prediction Web Server. Nucleic Acids Res 2015, 43, W408–W412. [CrossRef]
- Möller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of Methods for the Prediction of Membrane Spanning Regions. Bioinformatics 2001, 17, 646–653. [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Söding, J.; Steinegger, M. Fast and Accurate Protein Structure Search with Foldseek. Nat Biotechnol 2024, 42, 243–246. [CrossRef]
- Bittrich, S.; Segura, J.; Duarte, J.M.; Burley, S.K.; Rose, Y. RCSB Protein Data Bank: Exploring Protein 3D Similarities via Comprehensive Structural Alignments. Bioinformatics 2024, 40. [CrossRef]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res 2014, 42, W320–W324. [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Science 2018, 27, 14–25. [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res 2021, 49, W431–W437. [CrossRef]
- Ferruz, N.; Schmidt, S.; Höcker, B. ProteinTools: A Toolkit to Analyze Protein Structures. Nucleic Acids Res 2021, 49, W559–W566. [CrossRef]
| PDB ID | Name of virus | TM-score | RMSD, Å | Aligned residues, a.a. |
1Amino acid sequence identity1, % |
|---|---|---|---|---|---|
| NS3pro HSTV | |||||
| 2FOM | Dengue virus | 0.79 | 2.33 | 137 | 23 |
| 2F9U | Hepatitis C virus | 0.69 | 4.64 | 118 | 17 |
| 5WX1 | Classical swine fever virus | 0.60 | 3.22 | 138 | 22 |
| NS3hel HSTV | |||||
| 1A1V | Hepatitis C virus | 0.66 | 3.46 | 328 | 20 |
| 2JLS | Dengue virus | 0.63 | 4.38 | 310 | 14 |
| 4CBL | Classical swine fever virus | 0.63 | 4.45 | 283 | 18 |
| D1-D2 NS3hel HSTV | |||||
| 1A1V | Hepatitis C virus | 0.76 | 2.45 | 268 | 21 |
| 2JLS | Dengue virus | 0.74 | 3.26 | 258 | 17 |
| 4CBL | Classical swine fever virus | 0.61 | 05.02 | 183 | 15 |
| D3 NS3hel HSTV | |||||
| 1A1V | Hepatitis C virus | 0.30 | 6.16 | 47 | 5 |
| 2JLS | Dengue virus | 0.36 | 5.7 | 57 | 5 |
| 4CBL | Classical swine fever virus | 0.39 | 5.79 | 59 | 8 |
| PDB ID | Name of virus | TM-score | RMSD, Å | Aligned residues, a.a. |
1Amino acid sequence identity, % |
|---|---|---|---|---|---|
| NS5RdRp | |||||
| 7XD8 | Dengue Virus | 0.72 | 3.26 | 498 | 13 |
| 7EKJ | Classical swine fever virus | 0.66 | 3.54 | 476 | 18 |
| 6GP9 | Hepatitis C virus | 0.63 | 3.85 | 431 | 14 |
| NS5MTase | |||||
| 1WY7 | Pyrococcus horikoshii | 0.77 | 3.02 | 164 | 22 |
| 3P97 | Dengue virus | 0.60 | 4.18 | 142 | 14 |
| 2WA1 | Modoc Virus | 0.59 | 3.69 | 142 | 12 |
| 8GY4 | Alongshan virus | 0.57 | 3.89 | 140 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





