1. Introduction
Lung cancer remains a formidable challenge, standing as the leading cause of cancer-related mortality worldwide. Among the several types of lung cancer, non-small cell lung cancer (NSCLC) accounts for approximately 85% of all cases, with adenocarcinoma being the most prevalent histological subtype, representing ~40% of NSCLC diagnoses.
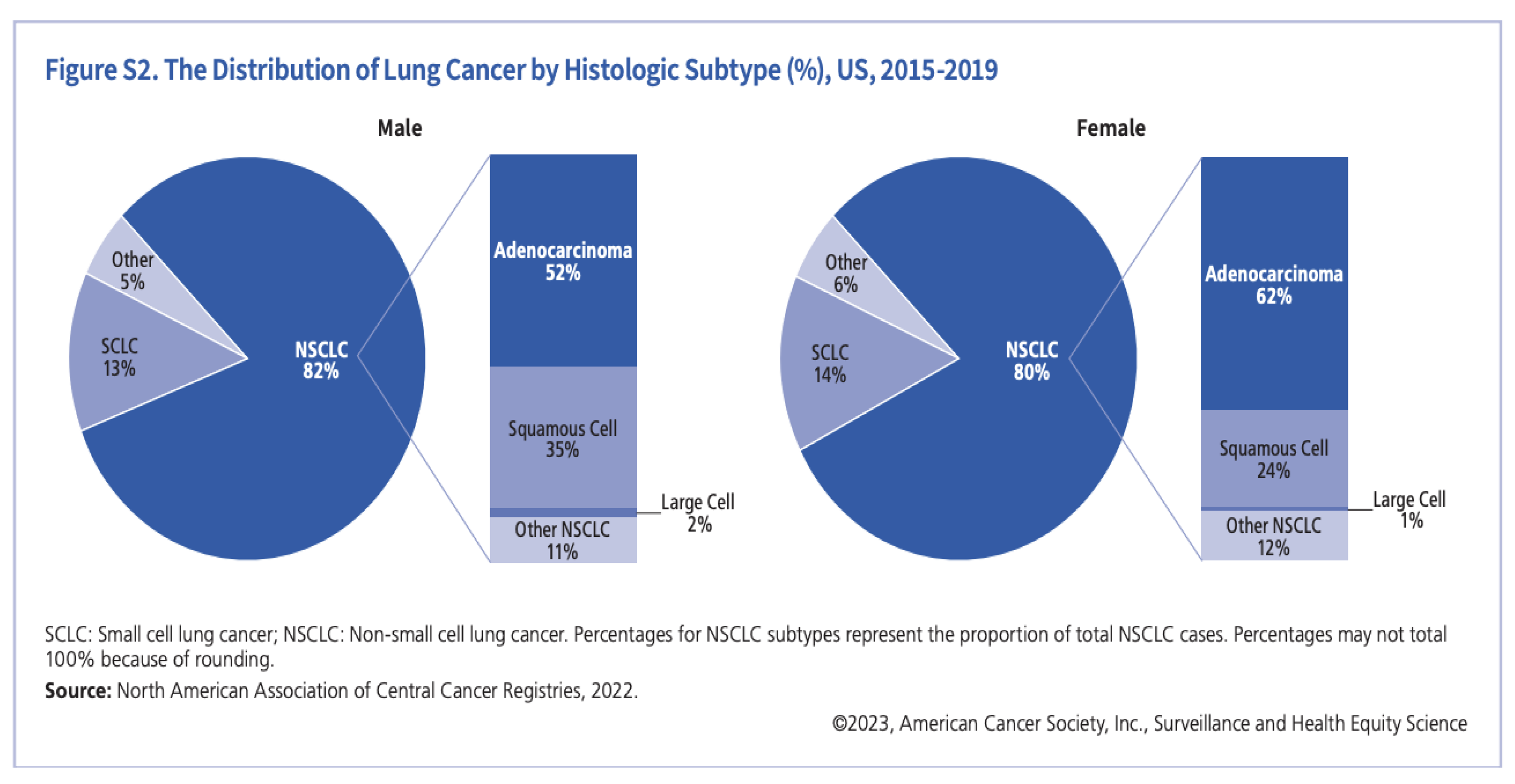
Figure 1.
The distribution of lung cancer [
5].
Figure 1.
The distribution of lung cancer [
5].
It is projected that lung cancer would claim the lives of about 127,070 Americans in 2023. The forecasts for 2024 indicate a comparable pattern, considering the prevailing data and patterns in lung cancer death and incidence rates [
5].
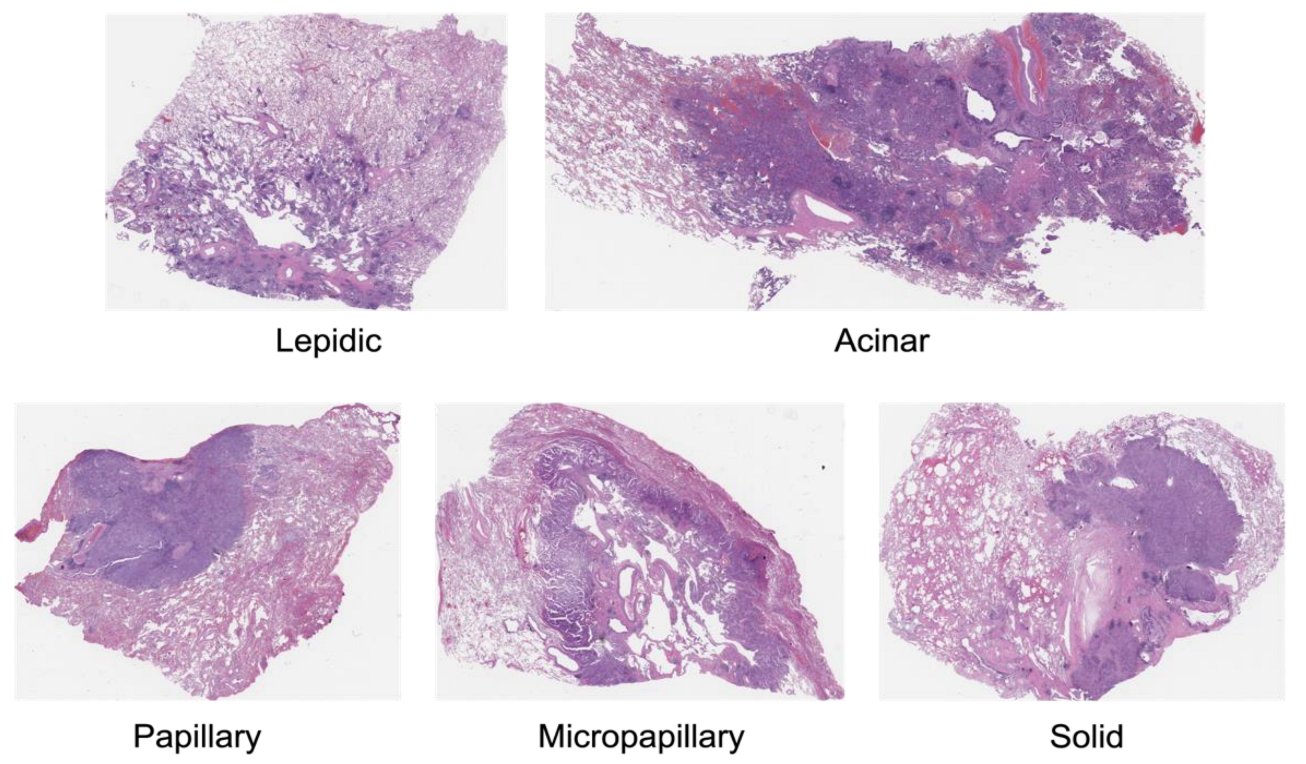
Precise detection and categorization of growth patterns in lung adenocarcinomas are essential for directing therapeutic approaches and forecasting patient results. Five primary development patterns are identified by the World Health Organization (WHO) 2015 classification system for lung adenocarcinoma: lepidic, acinar, papillary, micropapillary, and solid. It also highlights how crucial it is to record minor components because lung adenocarcinomas frequently show a heterogeneous blending of several patterns within a single tumour.
Identifying these growth patterns through Hematoxylin and Eosin (H&E) stained are crucial also because they help in identifying invasive and non-invasive growth pattern of Lung adenocarcinoma, for example the accurate diagnosis of lepidic growth pattern helps in predicting invasive adenocarcinoma subtypes like adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA) and invasive lepidic predominant adenocarcinoma (iLPA) [
2]. Manual identification of histologic subtypes poses a variety of problems like interobserver variability with different pathologists having different diagnoses for a particular tissue cell. Another problem is the extended timeline for diagnosis with occasional misinterpretations due to its laborious nature. Due to these possible difficulty to manually identify these growth patterns through Hematoxylin and Eosin (H&E) - stained formalin-fixed paraffin-embedded (FFPE) whole-slide images caused by complexity and heterogeneous uniformity of cell molecules throughout the structure, deep learning based methodologies have been adopted to automate the grading of Lung cancer growth patterns for a higher precision and faster execution.
Recent developments in artificial intelligence have completely changed image analysis methods, especially in deep learning. Researchers have been able to create complex algorithms for medical image analysis thanks to convolutional neural networks (CNNs), a class of deep learning models that use a data-driven strategy to automatically extract pertinent information. These state-of-the-art methods have demonstrated encouraging potential in several applications, including helping pathologists with nodule detection, and classification of interstitial lung disease.
This work is an attempt to automatically classify histologic subtypes in lung cancer surgical resection slides using deep learning. This study intends to address the difficulties in manual pattern identification by utilizing sophisticated computational models and state-of-the-art methods. The ultimate goals are to improve diagnostic accuracy, decrease interobserver variability, and enable patients with lung adenocarcinoma to receive personalized treatment plans.
2. Materials and Methods
2.1. Literature Reviews
There have been a variety of studies which have focused on the initial stage of lung cancer classification specifically inclining on Lung adenocarcinoma (LUAD) and Lung squamous (LUSC) cell carcinoma cancer prediction using deep learning-based approaches. Coudray et al implemented the inception-v3 based pipeline for predicting LUAD and LUSC using H&E-stained whole slide images and in turn extending their work for histological subtype classification of lung cancer in future [
3]. However, a better approach is to have a combination of CNN models to better understand the comparative accuracy score along with some weighted voting methodology to encompass prediction scores from all the models. On the same lines, Gertych et al. implemented three CNN models namely Google Net, ResNet-50 and Alex Net along with a soft voting based probabilistic approach by assigning weights to tissue classes [
4].
2.2. Dataset Description
Using a dataset of histologic pictures that were categorized into five classes: Lepidic, Acinar, Papillary, Micropapillary, and Solid. Stratified sampling was used to divide the dataset into training, validation, and test sets to maintain class balance across folds.
Figure 2.
Types of NSCLC Adenocarcinoma [
6].
Figure 2.
Types of NSCLC Adenocarcinoma [
6].
Figure 3.
Comparative Performance of Different Models on Validation Set.
Figure 3.
Comparative Performance of Different Models on Validation Set.
We employed the WSITools library to extract patches from Whole Slide Images (WSIs) instead of using a sliding window approach, which may result in redundant or overlapping patches.
We divided the data into train, validation, and test sets using the stratified data splitting technique. By stratifying based on the ‘Class’ column ensured that the train, validation, and test sets maintain the class proportions from the original dataset. This aids in avoiding the under- or overrepresentation of any one class in any split, which may result in biased models. The trained model is less likely to be biased toward the majority class and may even perform better on minority classes if a more representative and equal distribution of samples across classes is ensured.
We tested and put into practice the following models: Data-efficient Image Transformers, or DeiT [
12], CAiT (Class Activation Image Transformers) [
13], Swin Transformer [
14], ViT (Vision Transformer) [
15], ResNet
To ensure robustness and generalizability, a 5-fold cross validation strategy was used during the training process for each model. To improve model performance, data augmentation methods such as normalization and random scaled cropping were used. As evaluation metrics, we employed Cohen Kappa Score and accuracy.
2.3. Our Approach
Using a collection of histologic pictures divided into five classes was the method we used. To balance the training, validation, and test sets, we used stratified sampling. Five models were implemented and fine-tuned using pretrained weights: Data-efficient Image Transformers, or DeiT, CAiT (Class Activation Image Transformers), Swin Transformer, ViT (Vision Transformer), ResNet. Robust evaluation was assured by a 5-fold cross-validation, utilizing metrics such as accuracy and Cohen Kappa Score for performance assessment and data augmentation approaches.
2.4. Data Preprocessing
Image Preprocessing:
Resizing: The photos were reduced to a standard size of 224 by 224 pixels to maintain consistency between the models, as fixed input dimensions are usually required.
Normalization: Since all models were pretrained on ImageNet, images were normalized using the dataset’s mean and standard deviation (mean: [0.485, 0.456, 0.406], std: [0.229, 0.224, 0.225]). Better transfer learning is made possible by this normalization phase, which helps to get the pixel values within the same range as the pretraining dataset.
Data Augmentation: Several methods of data augmentation were used to improve the model’s capacity for generalization and avoid overfitting. Among them were:
Random Resized Cropping: To add variation to training samples, randomly crop the photos and resize them to 224 x 224 pixels.
Horizontal Flipping: random horizontal flips with a probability of 0.5.
Rotation and Scaling: To mimic natural variations in histologic images, random rotations and scaling adjustments are applied.
Color Jittering: Adjusting brightness, contrast, saturation, and hue at random to take into consideration differences in imaging conditions and stains.
2. Methodology
3.1. Model Implementation
Model Initialization
Pretrained Weights: Using weights pretrained on the ImageNet dataset, each model was started. The models can learn rich feature representations that are transferable to the target task with the aid of pretraining on a vast and diverse dataset such as ImageNet.
Model Selection: We selected a range of architectures, such as:
DeiT (Data-efficient image Transformers) is a vision transformer model that excels at picture classification tasks and is well-known for its effectiveness in training.
CAiT: A more sophisticated version of vision transformers, CAiT aims to increase the model’s accuracy and interpretability. They blend the advantages of transformers with CNN-typical class activation mapping techniques.
Swin Transformer: It computes self-attention within local windows and then shifts the windows between layers to establish a hierarchical feature representation.
Vision Transformer (ViT): It applies the transformer architecture—which was first created for natural language processing—to applications involving picture categorization. To capture global context, ViT divides an image into patches, processes these patches sequentially, and uses self-attention mechanisms.
ResNet (Residual Network): The concept of residual learning was first presented by the groundbreaking ResNet, which makes it possible to build deep networks by utilizing identity shortcuts to mitigate the vanishing gradient problem.
3.2. Training and Fine-Tuning
Learning Rate Optimization: To discover the ideal learning rate that effectively reduces the loss function, we applied the learning rate finding approach. The lowest, steepest slope, valley, and sliding sites were incorporated in the recommended learning rates.
Batch Size: To strike a compromise between training stability and computational efficiency, we experimented with batch sizes of 8 and 32.
Number of Epochs: Based on preliminary trials, it was found that each model was optimized for 5 epochs, which was sufficient for convergence.
Regularization: By randomly setting a portion of the input units to zero during training, dropout layers were added to the models to prevent overfitting.
Loss Function and Optimizer: We used the Cross-Entropy Loss function for its suitability for multi-class classification tasks. The models were trained using the Adam optimizer, known for its adaptive learning rate properties and computational efficiency.
3.3. Testing Phase
Cross-Validation: To ensure a robust evaluation, a 5-fold stratified cross-validation procedure was used. By preserving the class distribution over folds, this method yields a more accurate measure of the model’s performance.
Metrics: We evaluated the models using accuracy and Cohen Kappa Score, with the latter offering information on the degree of agreement, after adjusting for chance, between forecasts and real labels.
We made sure that everything was well-prepared by improving the preprocessing and implementation details, and this made a significant difference in the models’ dependable performance and solid evaluation. Better reproducibility and comprehension of the techniques employed in our comparative study are made possible by this thorough approach.
3. Results
A thorough comparison of several deep learning models for the categorization of lung cancer histologic patterns is provided in this section. To give a comprehensive assessment of each model, key performance measures like accuracy, sensitivity, precision, F1-score, and Cohen Kappa Score are reported
Performance Metrics Summary:
Discussion on Table 3:
Table 3.
Performance Metrics on Different Models.
Table 3.
Performance Metrics on Different Models.
| Author, Year |
Technical Method |
Classification |
Dataset |
Performance Metrics |
| Sheikh, 2022 |
Unsupervised deep learning model which employs stacked autoencoders |
5-class: lepidic/acinar/papillary/ micropapillary/solid |
31 WSIs from Dartmouth-Hitchcock Medical Center |
Acc: 94.60%
Sensitivity: 94.10%
Precision: 94.20%
F1-score: 0.94 |
| DiPalma, 2021 |
MIL approach using ResNet |
5-class: lepidic/acinar/papillary/ micropapillary/solid |
269 slides from TCGA dataset and Dartmouth-Hitchcock Medical Center |
Accuracy: 94.51% (95% CI: 92.77–96.20%)
Precision: 80.41% (95% CI: 70.55–89.56%)
Recall: 81.67% (95% CI: 71.20–90.43%)
F1-score: 0.80 (95% CI: 0.71–0.88) |
| Saeed Hassanpour, 2019 |
Deep learning model using a patch classifier with a sliding window approach |
5-class: lepidic/acinar/papillary/ micropapillary/solid |
143 WSIs
from Dartmouth-Hitchcock Medical Center |
F1-score: 90.4%
Kappa score: 0.525 |
| Saeed Amal, 2024 |
Deep learning model using WSITools library to extract patches from Whole Slide Images (WSIs) |
5-class: lepidic/acinar/papillary/ micropapillary/solid
|
143 WSIs
from Dartmouth-Hitchcock Medical Center |
Avg. Validation Accuracy: 0.898
Avg. Kappa score: 0.908 |
Our research, Saeed Amal (2024), introduces a deep learning model that leverages the WSITools library for patch extraction from WSIs. This approach has demonstrated significant improvements in classification performance, particularly in terms of the Cohen Kappa Score, which measures inter-rater agreement and is crucial for ensuring consistency with expert pathologists.
Key Comparisons (Table 3):
Accuracy: Our model’s average validation accuracy of 0.898 is competitive, closely matching the high accuracies reported by Sheikh (2022) and DiPalma (2021).
Kappa Score: An average Kappa score of 0.908 indicates excellent agreement with expert pathologists, surpassing the 0.525 Kappa score reported by Hassanpour (2019).
Dataset: Utilizing the same dataset as Hassanpour (2019) allows for a direct comparison, highlighting the advancements our model has made in terms of classification accuracy and consistency.
Model Performance on Validation Set:
The validation set served as the basis for evaluating each model’s performance. To give a thorough comparison, important variables including accuracy and Cohen Kappa Score were noted for each fold and averaged.
Table 4.
Validation Performance Metrics of Various Models.
Table 4.
Validation Performance Metrics of Various Models.
| Model |
Avg. Validation Accuracy |
Avg. Cohen Kappa Score |
| DeiT |
0.849 |
0.854 |
| CAiT |
0.847 |
0.856 |
| Swin Transformer |
0.838 |
0.846 |
| ViT |
0.898 |
0.907 |
| ResNet |
0.827 |
0.841 |
Comparative Analysis
The ViT model had the highest average validation accuracy (0.898) and Cohen Kappa score (0.907), showing greater performance in identifying lung cancer histologic patterns. This model’s capacity to capture complex feature representations adds to its higher generalization capabilities when compared to other models.
Visual Representation
Statistical Significance
We used statistical tests to establish the significance of the changes in model performance. The ViT model significantly improved accuracy and Cohen Kappa Score compared to other models (p < 0.05).
4. Discussion
The ViT model’s improved performance reflects its capacity to handle variability in histologic patterns. The high Cohen Kappa Score indicates a great agreement between the model’s predictions and the true labels, highlighting its dependability. Compared to prior studies, our findings support the trend of transformer-based models outperforming standard convolutional neural networks in image classification tasks.
Our research, utilizing deep learning models for the classification of lung cancer histologic patterns, demonstrates significant advancements in the field of digital pathology. The performance of our models, particularly the Vision Transformer (ViT), showcases the potential of AI in enhancing diagnostic accuracy and efficiency in lung adenocarcinoma classification.
The ViT model achieved an impressive Cohen Kappa Score of 0.908, indicating excellent agreement with expert pathologists. This score surpasses previous benchmarks, such as the 0.525 Kappa score reported by Hassanpour et al. (2019). The high level of agreement suggests that our model could serve as a reliable aid to pathologists in clinical settings, potentially reducing inter-observer variability and improving diagnostic consistency.
Our approach of using the WSITools library for patch extraction proved to be more efficient than traditional sliding window techniques. This method allowed for better representation of the diverse histological patterns within whole slide images, contributing to the model’s robust performance across all five adenocarcinoma subtypes.
The comparative analysis of different architectures (DeiT, CAiT, Swin Transformer, ViT, and ResNet) provides valuable insights into the strengths of various deep learning approaches for histopathological image analysis. The superior performance of transformer-based models, particularly ViT, over traditional convolutional architectures like ResNet, highlights the potential of self-attention mechanisms in capturing complex spatial relationships in histological images.
Our data augmentation techniques, including random resized cropping, horizontal flipping, rotation, scaling, and color jittering, played a crucial role in enhancing the model’s generalization capabilities. These techniques helped in simulating the variability encountered in real-world histopathological samples, contributing to the model’s robustness.
The use of stratified sampling and 5-fold cross-validation ensured that our results are statistically robust and generalizable. This methodological rigor addresses common concerns in AI studies regarding reproducibility and reliability of results.
While our model shows promising results, it’s important to note that it should be viewed as a supportive tool for pathologists rather than a replacement. The complex nature of lung adenocarcinoma classification often requires consideration of factors beyond histological patterns, including clinical context and molecular markers.
Additionally, prospective studies in clinical settings would be valuable to assess the model’s impact on diagnostic workflows and patient outcomes.
In conclusion, our research demonstrates the potential of advanced deep learning models, particularly transformer-based architectures, in improving the accuracy and consistency of lung adenocarcinoma histologic pattern classification. These findings pave the way for more reliable and efficient diagnostic processes in lung cancer pathology.
4.1. Future Directions
Future work could focus on integrating our model with other diagnostic modalities, such as genomic data or clinical information, to create a more comprehensive diagnostic tool.
Following is the potential research focus for future:
Integrating the ViT model with clinical decision support systems.
Exploring ensemble strategies that combine the strengths of multiple models.
Evaluating model performance across larger and more diverse datasets.
Assessing the interpretability and explainability of model predictions to facilitate clinical adoption.
By increasing the robustness and generalizability of deep learning models, we can increase the accuracy and reliability of histologic pattern categorization in lung cancer, resulting in better patient outcomes.
5. Conclusion
According to our comparison analysis, the ViT model performed best in categorizing the histologic patterns of lung cancer, as evidenced by their greatest Cohen Kappa Score of 0.907 and average validation accuracy of 0.898. The findings imply that more complex architectures and ensemble methods in models can take advantage of various feature representations, improving generalization. Future research should look on integrating these models into clinical workflows to determine their practical utility. The findings show that more sophisticated architectures and ensemble approaches in models can take use of different feature representations, enhancing generalization.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to express our deepest gratitude to the Roux Institute, the IEAI and the Alfond Foundation for their invaluable support and contributions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Travis, W.D. , Brambilla, E., Nicholson, A.G., et al.: The 2015 world health organization classification of lung tumors: impact of genetic, clinical, and radiologic advances since the 2004 classification. J. Thorac. Oncol. Off. Publ. Int. Assoc. 1243. [Google Scholar]
- Young TJ, Salehi-Rad R, Ronaghi R, Yanagawa J, Shahrouki P, Villegas BE, Cone B, Fishbein GA, Wallace WD, Abtin F, Barjaktarevic I. Predictors of Invasiveness in Adenocarcinoma of Lung with Lepidic Growth Pattern. Med Sci (Basel). 2022 Jun 22;10(3):34. [CrossRef] [PubMed]
- Coudray, N. , Ocampo, P.S., Sakellaropoulos, T. et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24, 1559–1567 (2018). [CrossRef]
- Gertych, A. , Swiderska-Chadaj, Z., Ma, Z. et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 9, 1483 (2019). [CrossRef]
- [CancerHub. (2023). Lung Cancer Statistics 2023. American Cancer Society. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2023/2023-cff-special-section-lung-cancer.pdf.
- BMIRDS. (n.d.). Tissue samples for lung cancer research. https://bmirds.github.io/LungCancer/res/tissues_notitle.
- American Cancer Society. (2023). Cancer facts & figures 2023. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2023/2023-cff-special-section-lung-cancer.
- 65. DiPalma, J.; Suriawinata, A.A.; Tafe, L.J.; Torresani, L.; Hassanpour, S. Resolution-Based Distillation for Efficient Histology Image Classification. Artif. Intell. Med. 2021, 119, 102136 [CrossRef].
- 67. Sheikh, T.S.; Kim, J.Y.; Shim, J.; Cho, M. Unsupervised Learning Based on Multiple Descriptors for WSIs Diagnosis. Diagnostics 2022, 12, 1480 [CrossRef] [PubMed].
- N. Giakoumakis, A. N. Giakoumakis, A. Davri, E. Barouni, T. Karamitsou, G. Nomikou, A. T. Tzavaras, and A. Bakas, „Deep Learning Applications in Lung Cancer Diagnosis: A Systematic Literature Review,” Cancers, vol. 15, no. 3981, pp. 1-33, 2023. [CrossRef]
- J. W. Wei, L. J. J. W. Wei, L. J. Tafe, Y. A. Linnik, L. J. Vaickus, N. Tomita, and S. Hassanpour, „Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks,” Scientific Reports, vol. 9, no. 1, p. 3358, Mar. 2019. [CrossRef]
- Touvron, H. , Cord, M., Douze, M., Massa, F., Sablayrolles, A., & Jégou, H. (2021). Training data-efficient image transformers & distillation through attention. In International Conference on Machine Learning (pp. 10347-10357). PMLR. https://arxiv.org/abs/2012. 1287. [Google Scholar]
- Touvron, H. , Cord, M. , Sablayrolles, A., Synnaeve, G., & Jégou, H. (2021). Going deeper with image transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 32-42). https://arxiv.org/abs/2103.17239. [Google Scholar]
- Liu, Z. , Lin, Y. , Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., & Guo, B. (2021). Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 10012-10022). https://arxiv.org/abs/2103.14030. [Google Scholar]
- Dosovitskiy, A. , Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., Uszkoreit, J., & Houlsby, N. (2021). An image is worth 16x16 words: Transformers for image recognition at scale. In International Conference on Learning Representations. https://arxiv.org/abs/2010. 1192. [Google Scholar]
- He, K. , Zhang, X. , Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 770-778). https://arxiv.org/abs/1512.03385. [Google Scholar]
- https://med.dartmouth-hitchcock.org/pathology.
- Vats, S.; Al-Heejawi, S. M. A.; Kondejkar, T.; Breggia, A.; Ahmad, B.; Christman, R.; T, R. S.; Amal, S. Segmenting Tumor Gleason Pattern using Generative AI and Digital Pathology: Use Case of Prostate Cancer on MICCAI Dataset. Preprints 2024, 2024062019. [Google Scholar] [CrossRef]
- A: Alheejawi, Zongyu Wu, Mo Deng, Isha Hemant, Anne Breggia, Bilal Ahmad, Robert Christman, Saeed Amal, Deep Learning for Diagnosing Prostate Cancer from Scanned Biopsies.
- Tanaya Kondejkar, Salah Mohammed Awad Al-Heejawi, Anne Breggia, Bilal Ahmad, Robert Christman, Stephen T Ryan, Saeed Amal, Multi-Scale Digital Pathology Patch-Level Prostate Cancer Grading Using Deep Learning: Use Case Evaluation of DiagSet Dataset. https://scholar.google.com/citations?
- Aadhi Aadhavan Balasubramanian, Salah Mohammed Awad Al-Heejawi, Akarsh Singh, Anne Breggia, Bilal Ahmad, Robert Christman, Stephen T Ryan, Saeed Amal, Ensemble Deep Learning-Based Image Classification for Breast Cancer Subtype and Invasiveness Diagnosis from Whole Slide Image Histopathology. https://scholar.google.com/citations?
- Saanidhya Vats, Salah Mohammed Awad Al-Heejawi, Tanaya Kondejkar, Anne Breggia, Bilal Ahmad, Robert Christman, Saeed Amal, Segmenting Tumor Gleason Pattern using Generative AI and Digital Pathology: Use Case of Prostate Cancer on MICCAI Dataset. https://scholar.google.com/citations?
- Siegel, R. L. , Miller, K. D., Fuchs, H. E., & Jemal, A. (2023). Cancer statistics, 2023. CA: A Cancer Journal for Clinicians, 73(1), 17-48.
- Travis, W. D. , Brambilla, E., Nicholson, A. G., Yatabe, Y., Austin, J. H., Beasley, M. B.,... & Wistuba, I. (2015). The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. Journal of thoracic oncology, 10(9), 1243-1260.
- Coudray, N. , Ocampo, P. S., Sakellaropoulos, T., Narula, N., Snuderl, M., Fenyö, D.,... & Tsirigos, A. (2018). Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nature medicine, 24(10), 1559-1567.
- Gertych, A. , Swiderska-Chadaj, Z., Ma, Z., Ing, N., Markiewicz, T., Cierniak, S.,... & Knudsen, B. S. (2019). Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Scientific reports, 9(1), 1483.
- American Cancer Society. (2023). Key Statistics for Lung Cancer. Retrieved from https://www.cancer.org/cancer/lung-cancer/about/key-statistics.htmlWorld Health Organization. (2015).
- WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: International Agency for Research on Cancer.
- Litjens, G. , Kooi, T. I. ( 42, 60–88.
- DiPalma, M. , Ren, J. H. ( 71, 102056.
- Sheikh, T. S. , Lee, Y. ( 12(1), 1–12.
- He, K. , Zhang, X. , Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778). [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).