1. Introduction
Fibrosis is a highly progressive, irreversible, and chronic inflammatory condition characterized by excessive extracellular matrix (ECM) deposition, tissue architectural remodeling and scarring. Fibroblasts are majorly responsible for synthesizing and organizing ECM proteins, playing a central role in wound healing and repair. However, persistent injury and chronic insult dysregulates and disrupts normal physiological restorative wound healing process, tipping the balance toward pathophysiological conditions [
1]. Cellular mechanism including but not limited to fibroblast proliferation, fibroblast to myofibroblast transition (FMT), epithelial injury, epithelial to myofibroblast transition (EMT) and apoptosis-resistant fibroblast phenotype is central to fibrosis pathology associated with scleroderma (skin), idiopathic pulmonary fibrosis (IPF) (lung), cardiac fibrosis (heart), renal fibrosis (kidney), and liver fibrosis amongst other organs [
2]. Currently, clinical manifestations of the disease are managed by two FDA (Food and Drug Administration) approved small molecule inhibitors, Nintedanib and Pirfenidone which provide symptomatic relief to the patients [
3]. However, both drugs are associated with severe gastrointestinal side effects such as vomiting, diarrhea, abdominal pain, appetite loss and weight loss, adversely affecting patient compliance and overall quality of life [
4]. Furthermore, the median survival rate of IPF patients after diagnosis is approximately 3-5 years, underscoring the urgent need to identify novel and promising therapeutic targets for the treatment of fibrosis.
WISP1 is a highly conserved, secreted cysteine rich multi-modular matricellular protein. Emerging evidence suggests that WISP1 is profibrotic in nature and drives myriad of cellular processes, including cell proliferation, migration, and invasion. Large-molecule therapeutic modalities, such as WISP1-neutralizing antibody, have been shown to elicit protective effects, as we and others have previously reviewed [
5,
6]. Furthermore, a recent study has identified a novel role for WISP1 in driving the progression of liver fibrosis. WISP1 knockout animals demonstrated protection in CCl
4-liver fibrosis model, highlighting WISP1 as promising therapeutic target for drug development. Besides liver fibrosis, aberrantly increased WISP1 expression in both, preclinical animal models of pulmonary fibrosis (paraquat induced model, bleomycin model) and clinically in IPF-lung as compared to healthy control, has been positively correlated with the pathophysiology of fibrosis. In fact, first in human phase-I clinical trials are underway to assess the safety, tolerability and pharmacokinetics of anti-WISP1 antibody (MTX-463) (
NCT06401213).
Although recent mechanistic studies have enhanced our understanding of the molecular mechanisms involved in WISP1 mediated liver fibrogenesis [
7], WISP1 remains an orphan ligand since its cognate receptor has yet to be discovered, limiting further mechanistic insights. Furthermore, key underlying molecular mechanisms and associated biological pathways of WISP1 mediated fibrogenesis in diseased vs non-diseased lung and dermal fibroblast still remains poorly understood.
In this study, we aim to enhance our understanding of the role of WISP1 in lung and dermal fibrosis by transcriptional profiling of WISP1 to better characterize its molecular signature in the initiation and progression of disease. By utilizing NanoString® based technology, we identified unique gene networks and pathways across multiple organ -specific fibroblasts. This allowed us to better characterize the complex role of WISP1 in both diseased and non-diseased states. For the first time, we reveal WISP1-specific gene signature and associated downstream signaling patterns for fibrosis initiation and progression in primary human healthy versus IPF-diseased, as well as primary dermal fibroblast. Our results also reveal notable differences in WISP1-mediated gene expression between healthy and IPF-diseased lung fibroblast, as well as between lung and dermal fibroblasts. These findings highlight the heterogeneity of fibroblasts and tissue-specific differences.
2. Materials and Methods
2.1. Cell Culture
Primary human lung fibroblasts from Idiopathic Pulmonary Fibrosis-diseased (DHLF), healthy (NHLF) donors and normal primary adult human dermal fibroblasts (NHDF) were obtained from Lonza (Cat. # CC-2512, CC-7231, CC-2511 respectively, Basel, Switzerland). Primary cells were cultured in T-75 cm
2 flasks containing fibroblast growth basal media (FBM) (CC-3131) supplemented with insulin 0.5ml, gentamicin sulfate 0.5ml, human fibroblast growth factor-B (hFGF-B) 0.5ml and fetal bovine serum 10ml at 37°C in 5% CO
2 as recommended by the manufacturer. For serum starve media, we used FBM without supplements. In addition, the same donor-specific lots were used in all the experiments (till passage 4) to ensure reliability and consistency. All the lots were commercially validated and tested positive for smooth-muscle alpha actin expression and negative for epithelial cell-marker cytokeratin 14 (CK14), cytokeratin 18 (CK18) and cytokeratin 19 (CK19) expression ascertaining the purity of cultured fibroblasts. Healthy NHLF and DHLF were obtained from 5 different donors, while NHDF were obtained from 3 different donors. Clinical characteristics of the patient derived cell lines are listed in
Table 1. Commercially acquired primary cells were expanded and multiple frozen aliquots at early passage were cryopreserved. Cells were passaged a maximum of 3-4 times after which another parent aliquot of the same lot was used.
2.2. Treatment
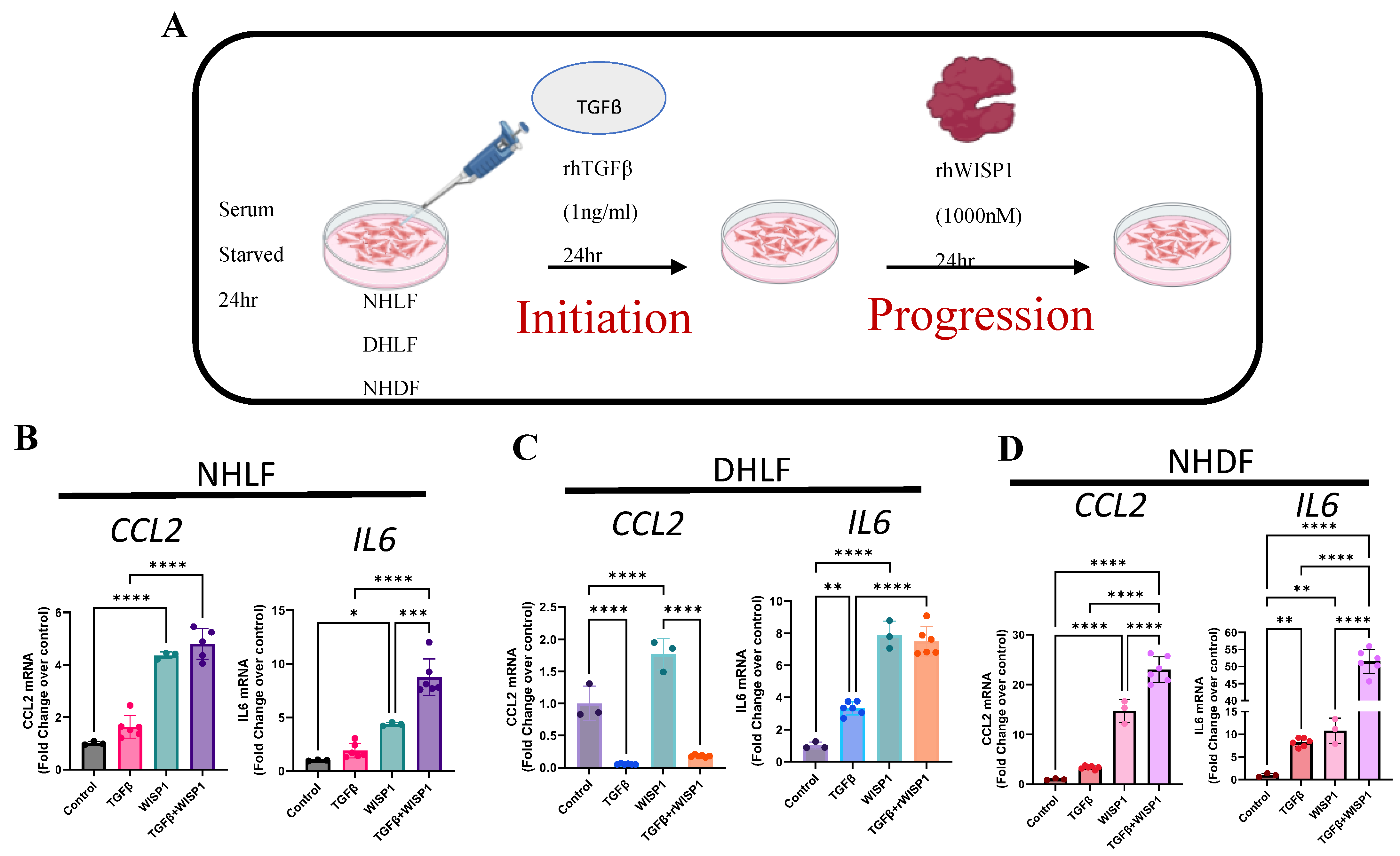
NHLF, DHLF and NHDF (4 ×105 cells/well) cells were seeded in 6-well plate and allowed to adhere overnight and then serum starved for 24 h prior to treatment with either TGFβ (1ng/ml) or recombinant human WISP1 (1000nM) in serum starve media for 24 h. To study progression, cells were incubated with TGFβ1 (7754-BH; R&D) (1ng/ml) for 24 h prior to stimulation with WISP1 (1000nM) for another 24 h. Following treatment, cell culture supernatant and RNA for collected for analysis. Recombinant human WISP1 was generated in-house. HEK293E cells were transiently transfected using polyethyleneimine hydrochloride (PEI MAX; 24765 (100)) in 2L shake flask at 37°C, 5-8% CO2 at 160rpm. Cells were cultured for 5 days and spined at 7000xg for 20min prior to protein-extraction using 5ml Hi trap His-excel column and 50ml Q-FF anion-exchange chromatography column.
2.3. Quantitative Real-Time PCR
Total RNA for cultured cells was extracted using PureLink RNA mini kit (Cat#12183018A; Invitrogen, Carlsbad, CA) and quantified using Nanodrop Eight Spectrophotometer (Thermo Scientific, Waltham, MA). Complementary DNAs were synthesized by reverse transcription with 100ng of total RNA using SuperScript III First strand (Cat#18080-051; Invitrogen). The following commercially available TaqMan probes and primers were utilized: human Col1A1 (Hs00164004_m1), Col3A1 (Hs00943809_m1), CCL2 (Hs00234140_m1), IL6 (Hs00985639_m1), ACTA2 (Hs05005341_m1), FN1 (Hs01549976_m1), CCN4/ WISP1 (Hs00987448_m1) and 18S (4310893E) from Applied Biosystems. cDNAs (1:20 dilution ratio) was added along with TaqMan Fast-advanced Mastermix in total 10µl reaction volume in 384-well plate as per manufacturer’s instructions and the plate was read using Quant Studio 7 Flex (Applied Biosystems, Waltham, MA). All the genes were normalized to endogenous 18S. Gene expression was determined using the 2ΔΔCt method. Briefly, ΔCt was calculated by subtracting sample Ct value from Ct value of 18S reference gene. ΔΔCt was calculated by subtracting the average ΔCt value of unstimulated control from sample ΔCt value. Relative quantification of gene expression was calculated by 2-ΔΔCt. Each reaction was performed in triplicates.
2.4. ELISA
Cell culture supernatants were collected post-treatment and stored in -20C until ready to analyze. Supernatants were thawed at room temperature and diluted 1:10 fold in PBS. WISP1 (DY1627), IL-6 (D6050) and CCL-2 (DCP00) protein content was assessed using enzyme-linked immunosorbent assay (ELISA) kit from R&D systems as per manufacturer’s recommendations. Protein standard and samples from three biological replicates were run in duplicate. Optical density was determined at 450nm with wavelength correction set to 540nm by SpectraMax® i3x (Molecular Devices, San Jose, CA) microplate reader.
2.5. Cell Proliferation Assay
5 x 103 cells were seeded per well in a 96 well black-clear bottom plate in total 100µl growth media and culture overnight. Cells were serum starved for 96 h prior to treatment with CCN4 or PDGF-BB (positive control) for 24 h. Equal volume of 4% paraformaldehyde solution was added per well and incubated for 15 min at room temperature. Post-fixation, cells were washed three-times with 1X PBS solution and blocked in 2% normal goat serum (in PBS) for 1 h at room temperature with gentle rocking. Cells were incubated with 1:1000 pRb primary antibody (Rabbit anti-phospho-Rb (Ser807/811) -D20B12) AF488 Ab used at 1:2000 dilution (Cell Signaling Tech Cat. #4277S) and Nuc Blue live cell stain (Cat. #R37605) in PBS containing 0.1% triton-X 100 overnight at 4 °C. Cells were washed three-times with 1X PBS solution and images were acquired using Phenix Opera microscope (Revvity, Waltham, MA). %pRb positive cells were analyzed using Harmony high-content imaging software.
2.6. NanoString® Gene Analysis
nCounter Human Fibrosis V2 panel (NanoString® Technologies, Inc., Seattle, WA) comprised of 770 genes was utilized to assess fibrotic gene signature. 100ng of total RNA isolated from cells were hybridized with the capture and reporter probe set at 65 °C for 20 h in PCRmax Alpha Cycler 4 thermal cycler (Cole-Parmer, Vernon Hills, Il) following manufacturer’s instructions. The samples were processed on the nCounter MAX/FLEX system followed by cartridge scanning by digital analyzer. The. rcc files obtained were uploaded as the input data and normalized using housekeeping genes included in the panel using NanoString nSolver analysis software version 4.0 (nSolver 4.0, NanoString Technologies, Inc., Seattle, WA). The background thresholding count was set to 20 and pairwise ratios were generated to compare different groups. Each sample was performed at least in triplicates.
2.7. Data Analysis
All the statistical analysis were performed and graphed using GraphPad Prism 10 (San Diego, CA) and RStudio. Data are represented as mean ± SD and experiments were performed at least three independent times in triplicates as noted in figure legends. Statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey’s post-hoc analysis with ninety-five percent confidence interval and statistical significance is represented as *p<0.5, **p<0.01, ***p<0.001, ****p<0.0001 as described in the figure legends.
3. Results
3.1. WISP1 mRNA and Protein Are Upregulated in IPF Lung Fibroblasts
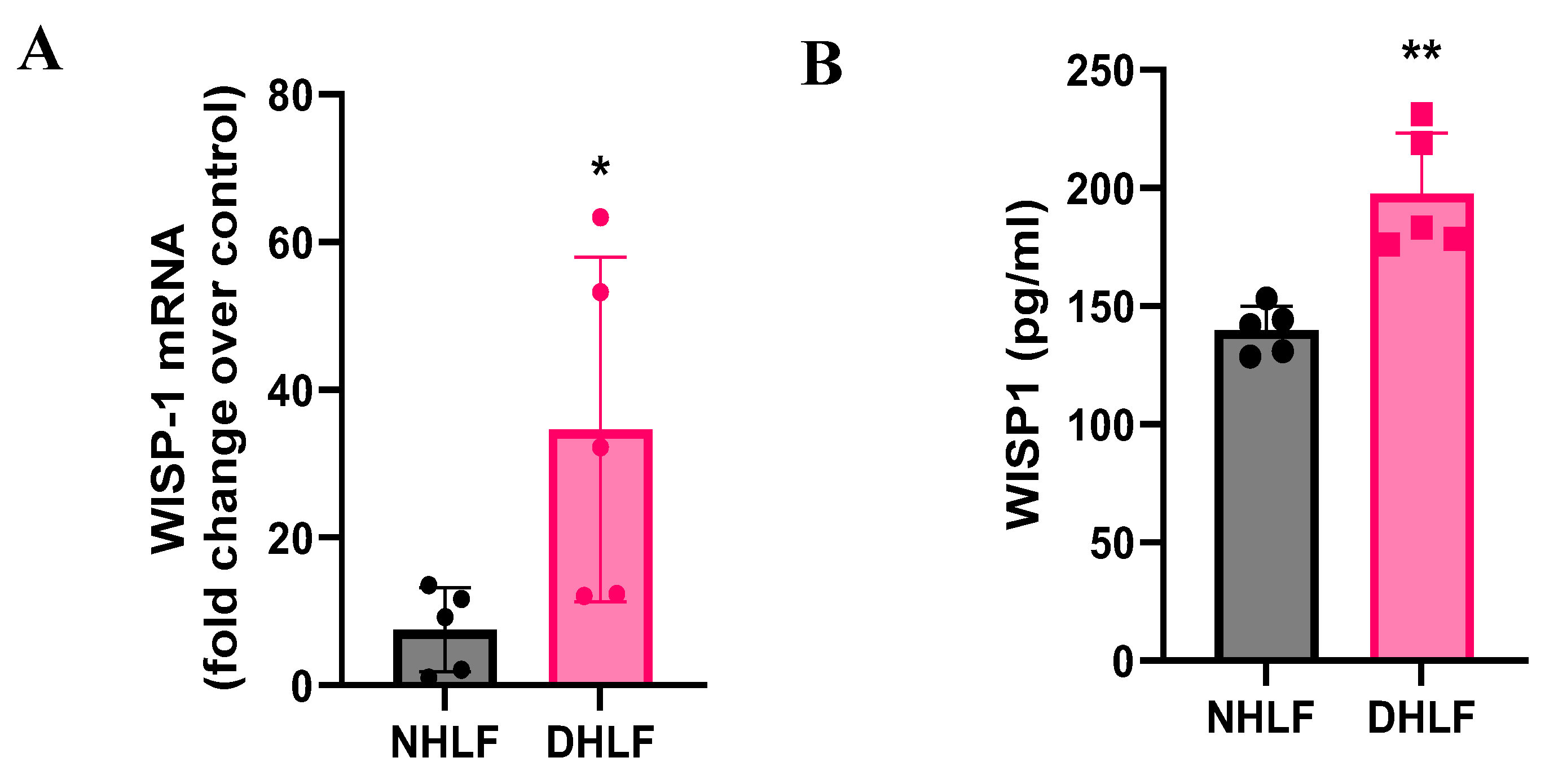
To investigate the role of WISP1 in initiating fibrosis, we first examined and compared both basal mRNA expression along with secreted WISP1 protein levels in healthy control (NHLF) and IPF-derived (DHLF) primary human lung fibroblasts obtained from 5 donors each. WISP1 is highly upregulated at both transcript (
Figure 1A) and protein levels (
Figure 1B) in DHLF as compared to NHLF. Results also demonstrate variability in endogenous WISP1 expression among DHLF patients, which may be attributed to distinct underlying co-morbidities and patient history (
Table 1). Similar expression profile has been shown by others in diverse pathophysiological conditions, such as cancer [
8,
9,
10,
11,
12,
13], fibrosis [
14,
15,
16], metabolic disorders [
17,
18,
19], and arthritis [
20,
21,
22,
23,
24,
25,
26].
3.2. TGFβ1 Stimulation Displays Differential WISP1 Expression Profile in Primary NHLF and DHLF
Transforming growth factor-β (TGFβ1) is the most potent, master regulator of fibrosis and inflammation [
27,
28]. However, due to its pleiotropic role in maintaining homeostasis, TGFβ1 blocking therapeutic modalities have caused severe adverse effects in clinical trials [
29,
30]. Given the challenges of targeting TGFβ1 for fibrogenesis, we aimed to assess the relationship between TGFβ1 and WISP1. Stimulation with TGFβ1 (1ng/ml) yielded a time-dependent increase in profibrotic gene signature with statistically significant effects noted at 24h in both NHLF and DHLF (
Figure S1A and S1B). Furthermore, the effect of TGFβ1 was apparent on WISP1 gene expression as early as 2h in DHLF (1.9-fold) and continued to increase till 24h for both NHLF (8.3-fold) and DHLF (3.2-fold) (
Figure S1A and S1B).
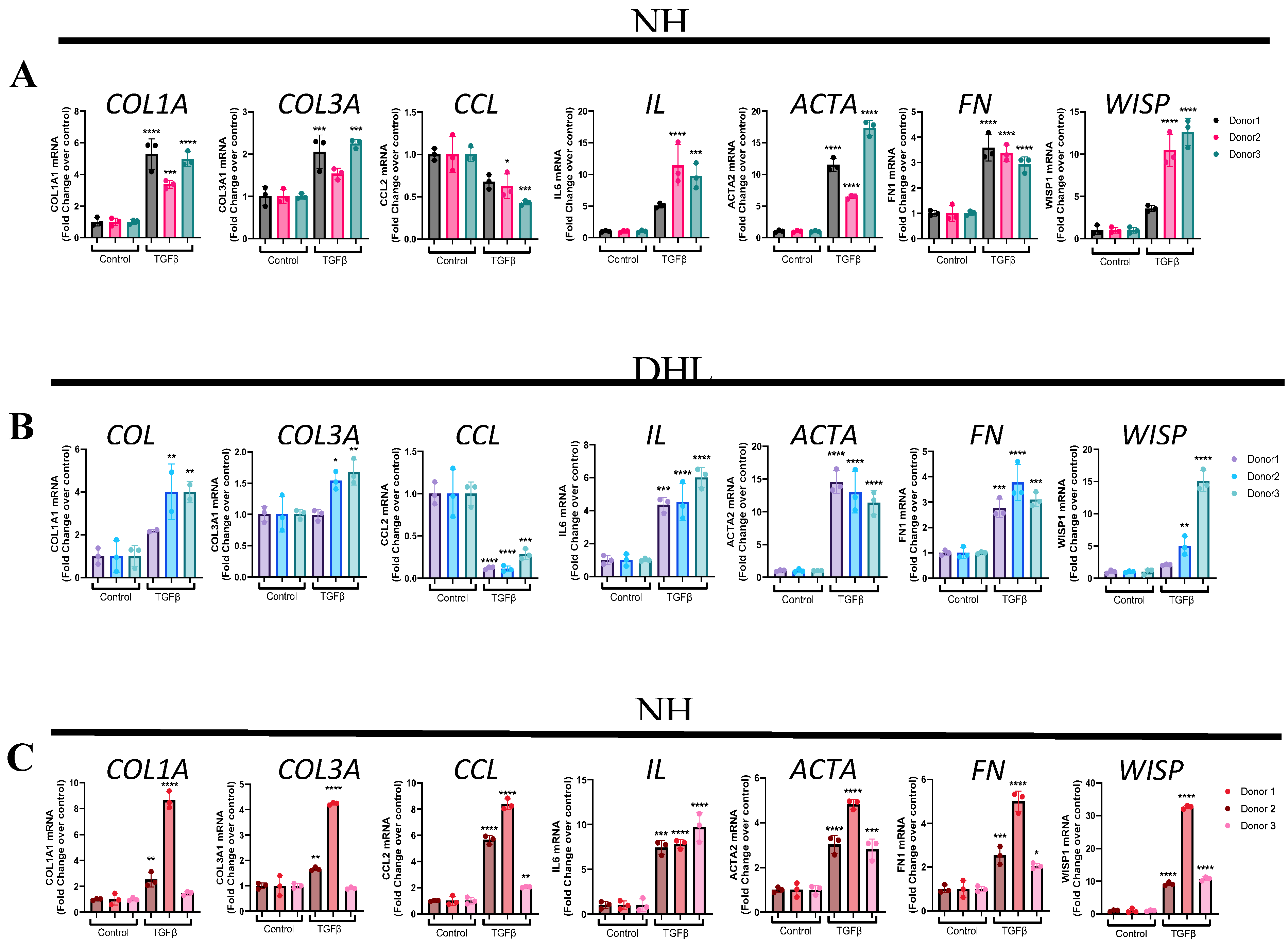
After we established that 24h time point induces fibroblast-to-myofibroblast transition (FMT) gene signature, we next wanted to determine if this is also true for multiple donor-derived primary fibroblast cell lines (n=3/each) to gain confidence with respect to utilizing TGFβ1 as positive control. Results from these experiments reveal that TGFβ1 stimulation significantly elevated pro-fibrotic genes such as collagen type 1 α1 chain (COL1A1), collagen type 3 α1 chain (COL3A1), α smooth muscle actin (ACTA2/ αSMA), fibronectin (FN1) and inflammatory gene such as interleukin-6 (IL6) in all 3-donor cell lines, with expected donor-to-donor variability for NHLF (
Figure 2A), DHLF (
Figure 2B) and NHDF (
Figure 2C). However, to our surprise, chemokine ligand 2/ monocyte chemoattractant protein-1 (CCL2/ MCP1) was significantly downregulated in NHLF (
Figure 2A) and DHLF (
Figure 2B), but not in NHDF (
Figure 2C), a striking difference between lung versus dermal fibroblast response to TGFβ1. Furthermore, TGFβ1 time course studies in NHLF and DHLF confirm these findings and reveal a biphasic response to CCL2 gene expression which increases until 4 h, followed by a clear decline until 24h (
Figure S1A and S1B). Overall, these results suggest that WISP1 operates downstream of TGFβ. Given that TGFβ levels are markedly elevated in the fibrotic disease state, as shown by others [
31,
32], this could partly explain the significant elevation of WISP1 observed in DHLF (
Figure 1A). Interestingly, similar findings have been reported in hepatic stellate cell lines [
16].
3.3. WISP1 Promotes Fibroblast Cell Proliferation
Fibroblast cell proliferation is a key mechanism in the induction and progression of fibrosis. Given our results showing that WISP1 is upregulated in IPF, next, we wanted to study the role of WISP1 in fibroblast proliferation. Retinoblastoma protein (Rb) is a tumor suppressor protein that regulates cell proliferation and cell cycle. CDK dependent phosphorylation of Rb (pRb) inactivates the protein and facilitates the entry into S-phase of cell cycle, thereby promoting cell proliferation [
33,
34]. Immunofluorescence technique was utilized to detect and visualize pRb positive cells as a surrogate marker for cell proliferation (
Figure 3A). Our results show that treatment with recombinant human WISP1 significantly increases cell proliferation in a concentration-dependent manner, with a maximum response peaking at approximately 30% pRb positive cells at the highest concentration compared to unstimulated control in primary human dermal fibroblast (
Figure 3B). Platelet-derived growth factor (PDGF) served as internal positive control in all the experiments and yields a robust fluorescent signal indicative of saturated cell-proliferation response at 100nM with approximately 40% pRb positive cells (p< 0.0001 via One-Way ANOVA) (
Figure S2A). Although, TGFβ1 treatment for 24h did not increase cell proliferation at higher doses but at lower doses (0.0625ng/ml) it significantly increased pRb positive cells (p< 0.0001 via One-Way ANOVA) (
Figure S2B and S2C).
3.4. WISP1 Drives Initiation of Pro-Inflammatory Cytokines Induction
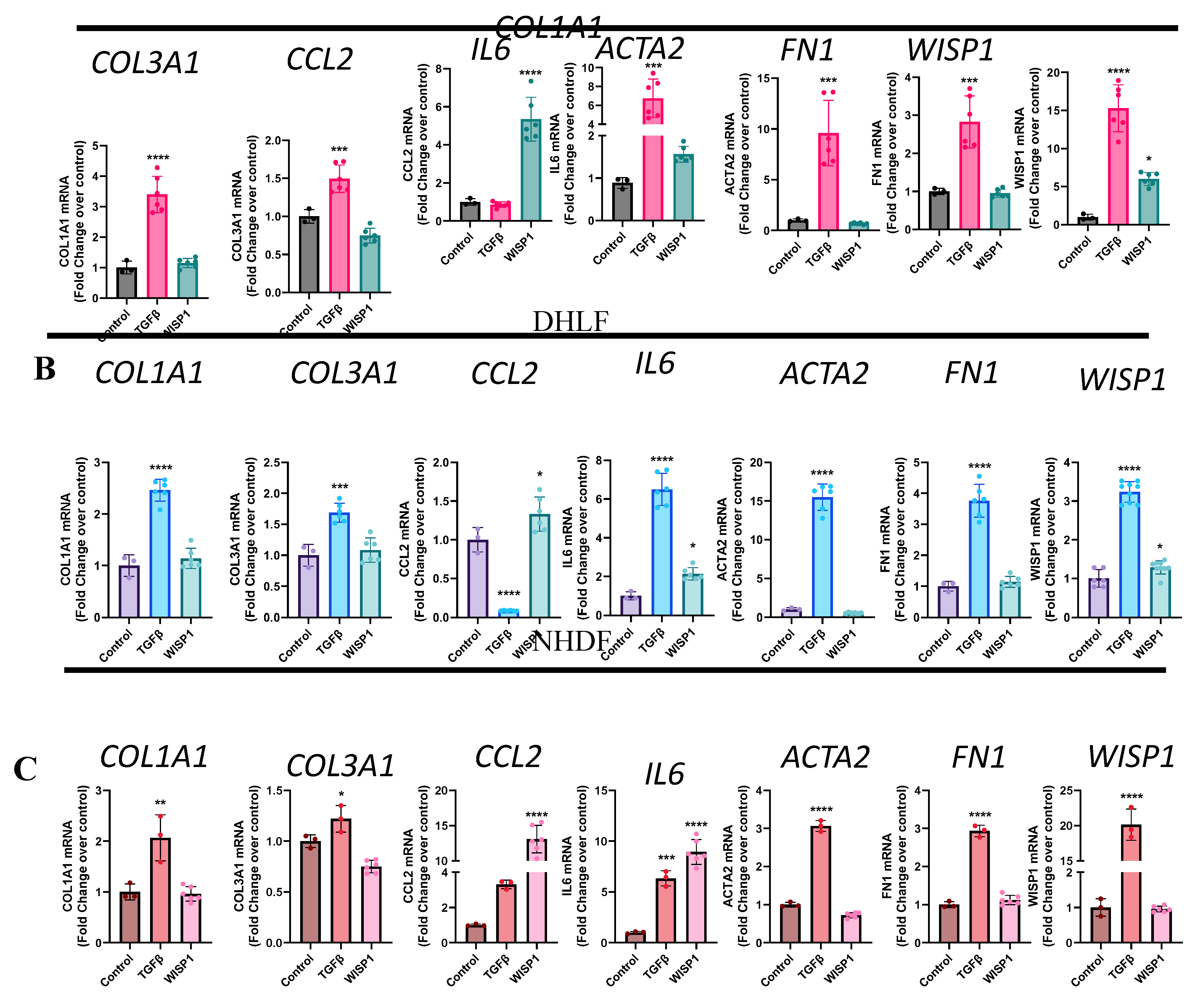
To study whether WISP1 effect on fibrogenesis is direct or indirect we first examined its role in fibrosis initiation by assessing profibrotic gene signature, similar to TGFβ1 stimulation studies, shown previously (
Figure 2). TGFβ1 (1ng/ml) served as a positive control as stimulation for 24h robustly initiated fibrotic gene induction (
Figure 2). To our surprise and contrary to what others have reported [
35], we did not observe any direct effect of WISP1(1000nM) treatment in initiating the induction of pro-fibrotic genes increase particularly collagens such as COL1A1, COL3A1, and ACTA2 and FN1 in NHLF (
Figure 4A), IPF-DHLF (
Figure 4B) and NHDF (
Figure 4C). However, interestingly stimulation of NHLF, and DHLF with WISP1 significantly elevated pro-inflammatory cytokines particularly IL6 (1.5 and 2.1-fold, respectively) and CCL2/MCP1 (5 and 1.3-fold, respectively) as compared to control (
Figure 4A and 4B). Among the three main fibroblast cell lines, WISP1 had the most significant impact on IL6 and CCL2 in NHDF, showing approximately 8.9-fold and 13-fold increases compared to the unstimulated control (
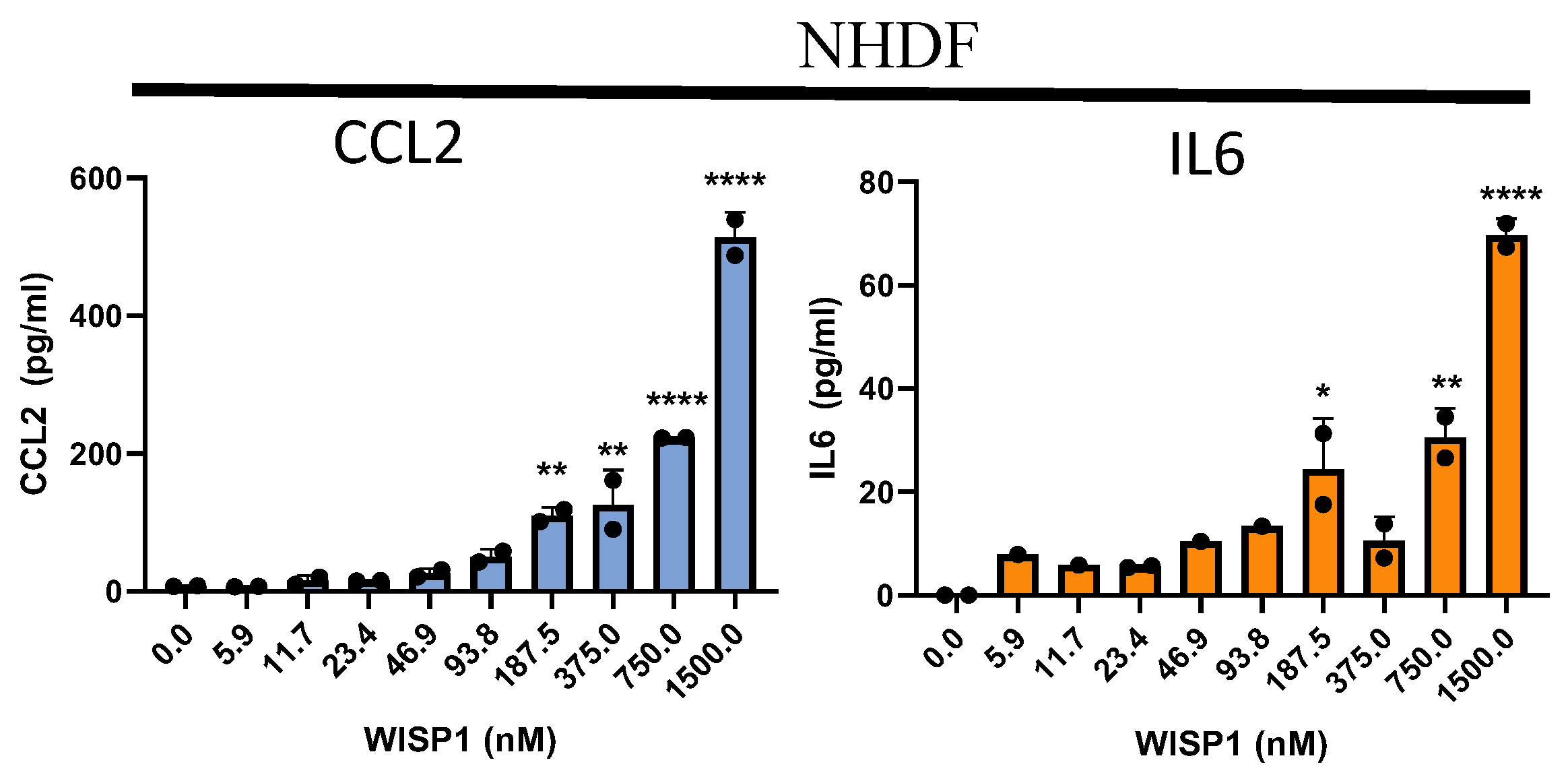
Figure 4C). The levels of IL6 and CCL2 in the cell-culture supernatant increased in a concentration-dependent fashion when compared to the control (
Figure 4D). In addition, these results also highlight that gene-expression analysis (
Figure 4C) confirms and aligns well with the observed secreted protein levels (
Figure 4D).
Notably, our findings reveal that, like TGFβ1, WISP1 stimulation also enhances its own expression in NHLF (6-fold) and DHLF (1.3-fold) (
Figure 4 A and 4B), indicating a possible positive auto-regulatory mechanism. This autocrine effect of WISP1 was not observed in NHDF (
Figure 4C), highlighting the self-regulatory mechanistic disparity amongst different-organ derived fibroblast subpopulations.
3.5. WISP1 with TGFβ1 Synergistically Induced Pro-Inflammatory Cytokines
Although WISP1 did not directly initiate fibrosis, we aimed to investigate its potential role in disease progression, similar to findings observed in liver fibrosis in vivo [
7]. We reasoned that establishing a simple in-vitro system would be beneficial. We first treated cells with TGFβ1 to initiate profibrotic gene expression and mimic pre-existing fibrotic environment, and then followed by WISP1 stimulation, to further assess whether WISP1 works in concert with TGFβ1 to facilitate disease progression in human fibroblasts (
Figure 5A). Our results show that gene expression of pro-inflammatory cytokines, particularly IL6 was synergistically upregulated when NHLF (
Figure 5B) and NHDF (
Figure 5D) were treated with TGFβ1+ WISP1 as compared to either TGFβ1 or WISP1 only. Data also reveals an additive effect of TGFβ1+ WISP1 treatment condition on CCL2 gene expression in NHLF (
Figure 5B) and NHDF (
Figure 5D), while similar effects were not observed in DHLF (
Figure 5C). Surprisingly, similar to our previous findings, TGFβ1 significantly reduced, while on the contrary WISP1 increased CCL2 gene expression (
Figure 2B and 4B). For IL6, although either TGFβ1 or WISP1 alone facilitated IL6 induction, no additive effects were observed when treated in combination in DHLF (
Figure 5C).
We also wished to assess the compound effect of TGFβ1+ WISP1 on other structural and extracellular genes, given their direct role in disease pathophysiology. To our surprise, the combination of TGFβ1+ WISP1 also did not show any significant alterations in the profibrotic gene expression, such as COL1A1, COLl3A1, ACTA2 and FN1 in all three primary human fibroblast cell lines (
Figure S3A, S3B and S3C). Further, given the positive-self regulatory function of WISP1 on itself, in conjugation with the TGFβ1-mediated WISP1 expression seen earlier in our studies (
Figure 4 A and 4B), WISP1 expression was notably upregulated upon combinatorial treatment in both NHLF (
Figure S3A) and NHDF (
Figure S3C) but not in DHLF (
Figure S3B). To our knowledge, these findings are the first to demonstrate the cooperative interaction between TGFβ1 and WISP1 in vitro, inducing inflammatory responses and showing differences between healthy and diseased fibroblasts, as well as between lung and skin fibroblasts, indicating organ-specific fibroblast heterogeneity. Given the highly dynamic and multifarious nature of fibroblast, we next utilized NanoString® for the transcriptional profiling of human lung and dermal fibroblast to better understand the relationship between WISP1 and fibrogenesis by identifying novel WISP1-mediated pro-fibrotic and inflammatory targets.
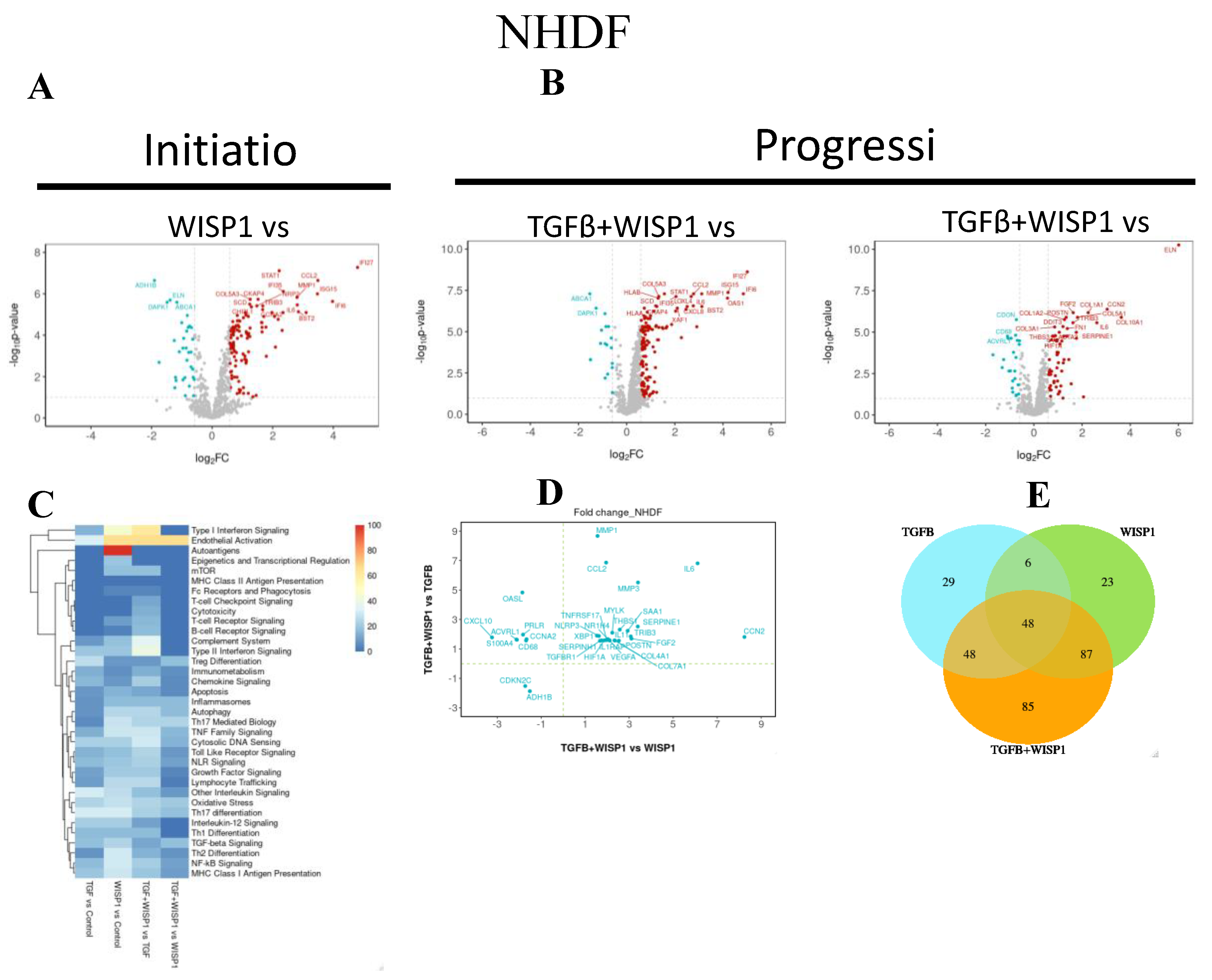
3.6. WISP1 Mediated Novel-Fibrotic Gene Signature in Primary Human Dermal Fibroblasts
Although we did not observe any direct effect of WISP1 on pro-fibrotic genes, we next employed NanoString nCounter® technology, to further understand the role of WISP1 in initiation and progression of fibrogenesis by treating primary human lung and dermal fibroblasts with either WISP1 (initiation) or in combination with TGFβ1 (progression).
NanoString® transcriptional profiling analysis revealed novel WISP1 induced gene targets. Amongst these, 117 genes were significantly upregulated while 34 genes were downregulated (|FC| >1.5; adj. P value <0.1) upon WISP1 (1000nM) stimulation for 24h in NHDF (
Table S1). Of the 117 upregulated genes, our results demonstrated highly interconnected group of inflammatory genes such as interferon α (IFNα) inducible protein, IF127, IFI6 and IFI35, ISG15 and XAF1 (
Figure 6A). Furthermore, WISP1 also induced the expression of another cluster of interleukins related inflammatory markers such as IL12RB2, IL18R1, IL1R1, IL1RAP, IRAK3, IL6 and IL6ST. In addition, genes involved in chemokine signaling such as CCL2, CXCL2, CXCL8, CXCL10, and CCR4 were amongst the most significantly associated genes, suggesting the inflammatory axis of WISP1 biology. Cellular adhesion molecules, like ICAM1 and VCAM1 that are crucial for leukocyte infiltration and inflammation were also notably upregulated. Similar to these findings, others have also shown WISP1-mediated upregulation of VCAM1 [
10,
36,
37] and ICAM1 [
10,
38] are crucial for cancer cell migration, invasion, and wound healing. In addition, integrins have been identified as receptors for most WISP1 biological functions [
5,
39]. For the first time, our results here demonstrate that WISP1 can in turn regulate the expression of integrins particularly α4 (ITGA4), α5 (ITGA5) and ILK (
Table S1). Overall, our NanoString® analysis indicates that WISP1 significantly promotes inflammation by activating genes associated with inflammatory signaling pathways, thus contributing to the disease pathology.
Interestingly, similar to what others have shown, WISP1 drives matrix metalloprotease expression, particularly MMP1 and MMP3 expression, which could further modulate fibrosis [
5]. Other potential genes involved in cell proliferation and cell cycle regulation such as E2F2, CCNA2 along with MAPK11 were markedly elevated by WISP1 in dermal fibroblast. These results further confirm and strengthen our previous results (
Figure 3A and 3B) by providing a molecular basis for WISP1-mediated cellular proliferation in NHDF. In addition to the genes mentioned above, a list of all WISP1-induced genes, with their respective fold change (FC) value, Log2FC, Adj. p-value and other statistical parameters is provided in the
Supplementary Materials (
Table S1).
To further gain insight into the cooperative interaction of TGFβ1 + WISP1 in disease progression, we next sought to determine other synergistically regulated fibrotic-transcriptional signature, elicited by these two mediators in NHDF. We identified 24 synergistically upregulated genes, while 2 genes were synergistically downregulated (|FC| >1.5; adj. P value <0.1) in TGFβ1 + WISP1 condition as compared to either TGFβ1 only or WISP1 only (
Figure 6B, 6D and
Table S2). In addition to IL6 and CCL2, the gene set comprises transcripts associated with growth factors, extracellular matrix (ECM), inflammation and other fibrotic targets, such as collagen (COL4A1, COL7A1), growth factors (FGF2, VEGFA, CCN2), matrix metalloproteases (MMP1, MMP3), interleukins (IL1RAP, IL11), along with TGFβ receptor 1 (TGFBR1), TNF receptor superfamily 17 (TNFRSF17), hypoxia inducible factor 1-α (HIF1α), serpin family E member 1 (SERPINE1) and serpin family H member 1 (SERPINH1) amongst others listed in the
Supplementary Materials (
Table S2). While WISP1 alone did not directly increase structural proteins like collagen, their levels were significantly upregulated in the presence of TGFβ1. This suggests an interdependency between WISP1 and TGFβ1 to produce a coordinated fibrogenic effect. Cyclin dependent kinase inhibitor 2C (CDKN2C) was markedly downregulated in TGFβ1+ WISP1 condition suggesting another parallel yet indirect mechanism promoting WISP1-mediated cell proliferation. Furthermore, we also highlighted unique and overlapping set of genes (
Figure 6E) along with their associated pathways induced by different treatment paradigms (
Figure 6C).
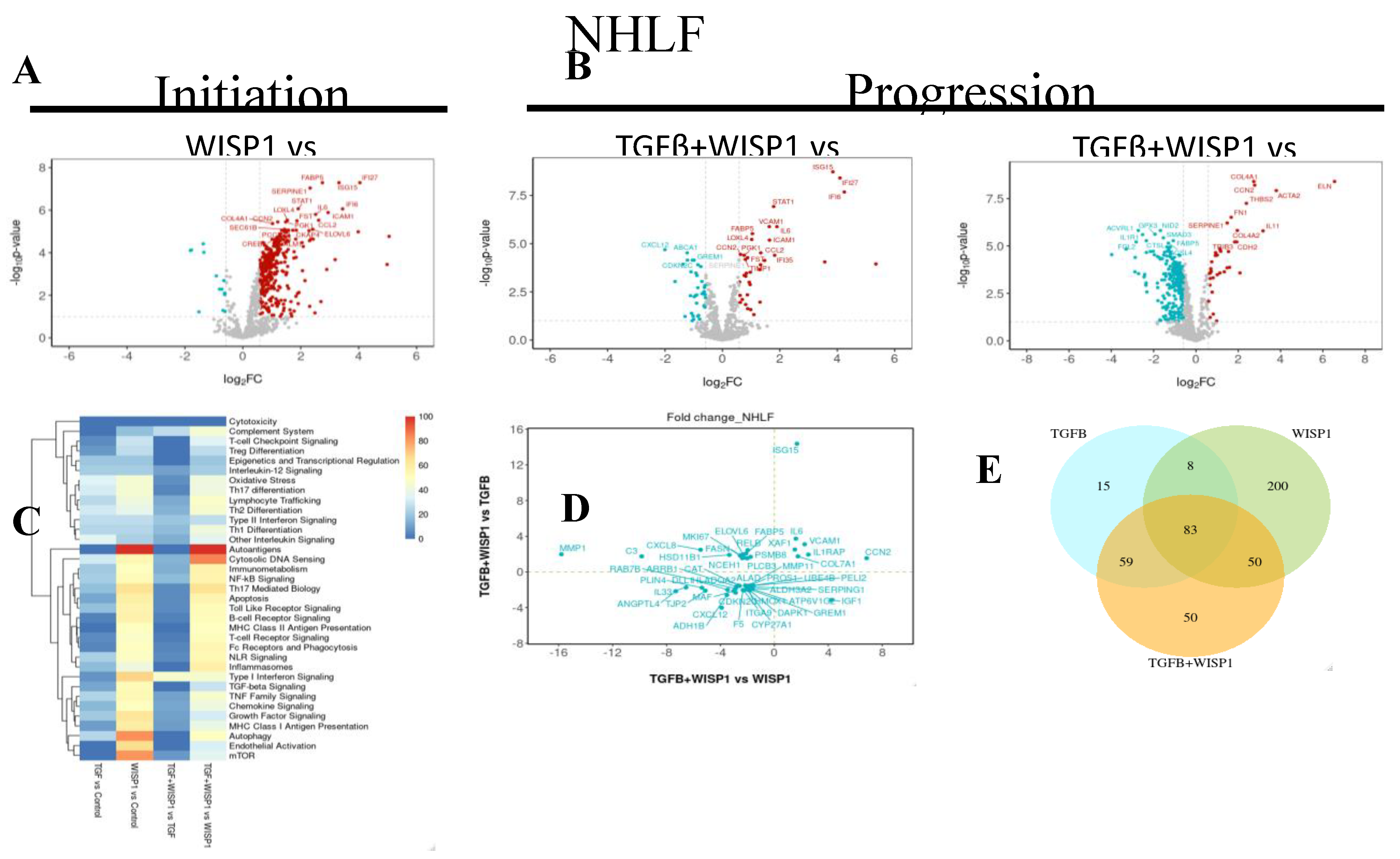
3.7. WISP1 Mediated Fibrotic Gene Signature in Primary Human Lung Fibroblasts
NanoString® analysis revealed WISP1 induced novel gene targets. Among these genes, 331 were significantly upregulated while 13 were downregulated (|FC| >1.5; adj. P value <0.1) in NHLF (
Figure 7A and
Table S3). Amongst these, interleukin related inflammatory markers such as IL6, IL4, IL21R, IL6ST, IL1RAP and IRAK3 were most notably upregulated. Similar to dermal fibroblast, chemokine signaling associated genes such as CCL2, CXCL2, CXCL8, CXCL10 were also upregulated in lung fibroblast as well, with the addition of CCL13 and CXCL11 in NHLF. Furthermore, our results also demonstrate a remarkably high degree of similarity in normal lung and dermal fibroblast with respect to IFNα inducible protein related genes upon WISP1 stimulation, such as IFI6, IFI27, IFI35, ISG15 and ISG20 (
Figure 7A and
Table S3). We observed a significant association of WISP1 with proliferative genes, such as MAPK1, MAPK9, MAPK11, MAP2K1, MK167, CDK4 and CCNA2 along with transcriptional factor, like E2F4, S100A4 suggesting a direct role of WISP1 in promoting fibroblast cell proliferation, an important part of fibrosis (
Figure 3). Surprisingly, and notably a significant fold-change increase in 9 different collagen associated genes, such as COL1A1, COL4A1, COL5A1, COL5A3, COL6A3, COL6A5, COL7A1, COL10A1 and COL16A1, and FN1 was observed in contrast to our previous qPCR data (
Table S3) suggesting higher sensitivity of NanoString® based multiplex technique over conventional PCR and a potential direct action of WISP1 on fibroblast in promoting fibrosis. Other profibrotic related genes directly modulated by WISP1 includes growth factors (VEGFA, EGFR, NEGF, FGF2, FLT1/ VEGFR1, CCN2), cellular hypoxia inducible factors (HIF1A, HIF3A), cellular adhesion molecules (VCAM1, ICAM1) and serine protease inhibitor family members (SERPINE1, SERPINH1) (
Table S3). Recent study shows that WISP1 phosphorylates and activates EGFR to activate MIF, which in turn drives lung inflammation and remodeling [
40].
Our previous data reveals that WISP1 is downstream of TGFβ1 (
Figure 2A), however, another interesting observation from the transcriptomics data reveal that WISP1 can also modulate TGFβ signaling by increasing TGFβ-receptor TGFBR1, TGFBR2 and TGFB1I1 expression in primary lung fibroblast (
Table S3). However, similar results were not observed with conventional qPCR gene expression analysis (data not shown), again highlighting the tool-specific disparity with respect to assay sensitivity and specificity. WISP1 can further self-regulate itself via augmenting its integrin receptor expression (ITGB1, ITGB5, ITGA4, ILK) or by promoting MMP expression (MMP1, MMP7, MMP10, MMP13, MMP16) in NHLF to drive cell migration via distinct mechanisms, which can further enhance fibrogenesis [
5]. A detailed list of all the genes is provided in
Supplementary Materials (
Table S3), with their respective FC value, Log2FC, Adj. p-value and other statistical parameters.
To further examine the synergistically regulated fibrotic-gene profiling, we identified a total 47 differentially regulated genes, out of which 7 of them were synergistically upregulated, while 27 were synergistically downregulated (|FC| >1.5; adj. P value <0.1) in TGFβ1+ WISP1 condition upon comparison to both, TGFβ1 only and WISP1 only (
Figure 7B, 7D and
Table S4). We found 4 out of the 7 upregulated genes stimulated in NHLF were exactly similar to what NHDF gene-signature, including transcripts associated to growth factor (CCN2/ CTGF), collagen (COL7A1) and interleukins (IL1RAP, IL6), highlighting high degree of congruency with respect to collective TGFβ1+ WISP1 induced signaling in non-diseased lung and dermal primary human fibroblast. Other synergistically upregulated genes were ISG15, VCAM1 and XAF1. As a detailed interpretation of all the genes is beyond the scope of this paper, a list of all the significantly modulated genes along with fold change and p-values is provided in
Supplementary Materials (
Table S4). Out of the unique gene, tight junction protein 2 (TJP2), catalase (CAT), death associated protein kinase (DAPK1) and CDKN2C were amongst the unique set of genes that are significantly downregulated (
Table S4). Chronic inflammatory lung diseases are characterized by oxidative stress due to the dysregulation in oxidant and antioxidant system, that can not only drive disease progression but can also determine therapeutic outcomes [
41,
42]. Catalase acts as an endogenous antioxidant enzyme that helps catalyze breakdown of hydrogen peroxide into water and oxygen. Decreased catalase expression at both mRNA and protein levels has been reported in humans and mouse-bleomycin fibrosis model [
43], and our results highlight the combined role of WISP1 and TGFβ1 in declined catalase activity, during fibrogenesis. In addition, TJP2 gene encodes for a tight junction associated protein, namely zonula occludens-2 (ZO-2) and dysregulation in TJ protein leads to barrier dysfunction with compromised integrity, a common feature of IPF [
44]. Furthermore, we also highlighted unique and overlapping set of genes (
Figure 7E) along with their associated pathways induced by different treatment paradigms (
Figure 7C).
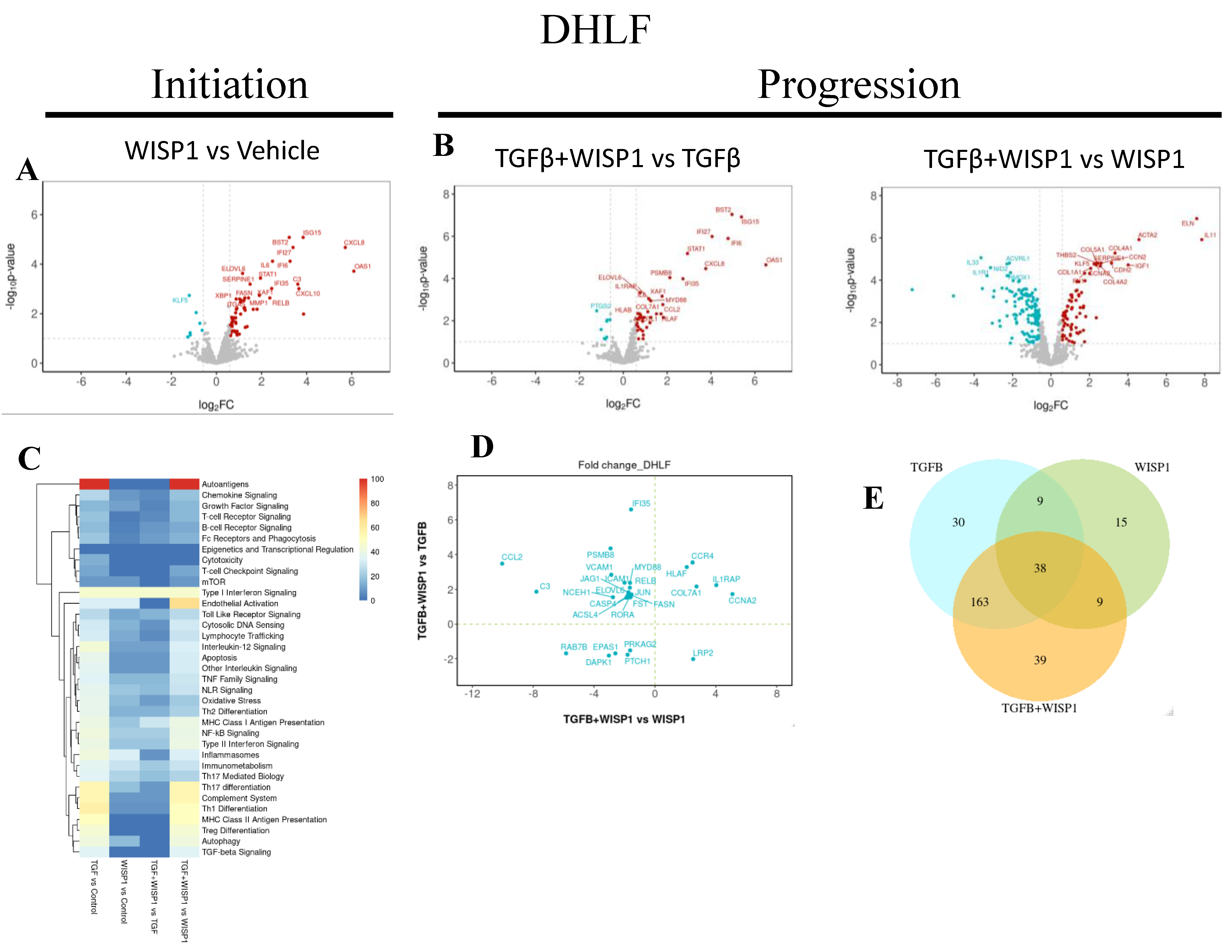
3.8. WISP1 Mediated Fibrotic Gene Signature in Primary Human Diseased (IPF) Lung Fibroblasts
To better understand the role of WISP1 in fibrogenesis and uncovering associated pathways, we also sought to identify gene transcripts in diseased (IPF) primary lung fibroblast. Our NanoString® based transcriptomics analysis revealed novel molecular signature, amongst which 64 genes were significantly upregulated while 7 genes were downregulated (
Figure 8A and
Table S5). These genes filtered and trimmed down based on our stringent inclusion criteria (|FC| >1.5; adj. P value <0.1) to only represent highly relevant effects in diseased human lung fibroblasts. Amongst these, IFNα inducible protein related genes, including IFI6, IFI27, IFI35 and ISG15 were also modulated in diseased human lung fibroblasts, analogous to both lung and dermal fibroblasts. In addition, our results also highlight high degree of similarity in WISP1-mediated gene signature, for instance, chemokine signaling related genes expression signature (CCL2, CCL13, CXCL2, CXCL8, CXCL10, CXCL11) in DHLF (
Table S5) is exactly similar to what was observed in NHLF (
Table S3) Other highly correlated genes were integrins (ITGA4, ITGA5), cellular adhesion molecules (VCAM1, ICAM1), serine protease inhibitor family members (SERPINE1, SERPINH1) and hypoxia inducible factors (HIF1A). However, fewer interleukin (IL6, IL1RAP), MMPs (MMP1) and growth factor (FLT4/ VEGFR3) related genes were observed in diseased state (
Table S5) as compared to non-diseased lung fibroblast (
Table S3), which could potentially be due to increased WISP1 leading to desensitization of Wnt-1 associated downstream signaling pathway in IPF-diseased state. Furthermore, consistent with our findings in lung-fibroblast, the expression of profibrotic genes, particularly collagen associated genes, such as COL1A1, COL3A1, COL4A1, COL5A1 and COL5A3 was strongly associated with WISP1 in IPF-diseased fibroblasts but not in dermal fibroblast, suggesting tissue-specific differences and heterogeneity in primary human fibroblasts (
Table S5).
In addition to the fibroblast-lineage associated disparity in molecular signature, we also found a common cluster of 31 highly significant core gene set which were highly correlated in both diseased and non-diseased lung and dermal fibroblasts (
Table S7). These set of genes comprises of transcript associated with interferon-induced genes, such as 2'-5'-oligoadenylate synthase1 (OAS1) and X-linked inhibitor of apoptosis (XIAP) associated factor 1 (XAF1) or NF-κB pathway associated members, including transcription factor RelB (RELB) and tribbles pseudokinase 3 (TRIB3). Transcription factor RELB is a member of NF-κB family which has been utilized not only as a promising diagnostic and monitoring biomarker in renal fibrosis but has also been attributed to promote inflammatory cytokines expression to drive disease progression [
45,
46]. Accumulating evidences from the literature also points towards the crucial role of stress response protein TRIB3 in pulmonary and renal fibrogenesis by promoting ECM deposition [
47,
48,
49,
50,
51]. In contrast, another study reports that TRIB3 expression is markedly downregulated in IPF patients both at transcript and protein level, and TRIB3 overexpression inhibits fibroblast activation and ECM deposition [
52]. In addition, signaling pathways of a broad range of proinflammatory and profibrotic cytokines (such as IL6), interferon as well as growth factors (such as VEGF, FGF) by canonical or non-canonical pathways, culminates downstream into JAK/STAT pathway [
53,
54], and given that our data supports direct modulation of these upstream modulator by WISP1, imperatively, downstream transcription factor, such as STAT1 was also found to be significantly upregulated, further validating our findings. Furthermore, the role of JAK/STAT pathway in fibrotic diseases has been extensively documented in literature and small-molecule JAK/STAT inhibitors alleviate excessive inflammation and provide anti-fibrotic effects both in-vitro and in-vivo [
54]. Here, our results reveal that WISP1 activates STAT1, which at least in part could contribute to heightened inflammation, influencing fibrosis pathophysiology. From the identified gene cluster, some unconventional fatty acid metabolism related genes such as fatty acid elongase 6 (ELOV6), fatty acid binding protein 5 (FABP5) and stearoyl-CoA desaturase (SCD) were also significantly associated with WISP1 in all the three lung and dermal primary fibroblast cell lines (
Table S7). Alteration in fatty acid composition and dysregulation in fatty acid metabolism has been identified in both preclinical bleomycin-model and in IPF lung tissue, and plays a critical role in macrophage polarization, epithelial-to-myofibroblast transition (EMT) and fibroblast activation, suggesting key role in disease pathophysiology [
55]. FABP5 gene encodes for fatty acid binding protein that is responsible for lipid trafficking, metabolism and has been shown to aggravate pulmonary fibrosis through Wnt-β-catenin pathway [
56]. On the contrary and to our surprise, SCD1 and ELOVL6 expression was downregulated in IPF-lungs tissues [
57,
58]. Although decreased ELOVL6 expression was specifically localized in alveolar type II (AT2) epithelial cells, our findings primarily focus on fibroblasts, indicating cell-type specific differences. Furthermore, SCD mRNA expression was significantly upregulated in hepatic stellate cells (HSCs) that activates Wnt-pathway by stabilizing Frizzled /low-density lipoprotein (LDL) receptor related protein (LRP) 5/6, promoting liver fibrosis [
59]. In addition, although the role of fatty acid metabolism in IPF remains controversial, unsaturated fatty acids were found to be significantly upregulated in bleomycin-animal model as compared to control. This aligns with our results, as increased SCD-catalyzed fatty acid desaturation leads to increased unsaturated lipids [
60]. Overall, our findings highlight the previously underappreciated role of dysregulated lipid metabolism and reveal its link with WISP1 in the pathogenesis of pulmonary fibrosis.
To gain better insight into the role of WISP1 in progression, we identified total 30 differentially modulated genes out of which, 7 genes were synergistically upregulated, while 5 genes were synergistically downregulated (|FC| >1.5; adj. P value <0.1) in TGFβ1+ WISP1 condition upon comparison to both, TGFβ1 only and WISP1 only in IPF-diseased fibroblast (
Figure 8B, 8D and
Table S6). Among these genes IL11, IL1RAP, ISG15, HLAF, CCR4, CCNA2 and COL7A1 were amongst the most significantly associated genes by conjoint TGFβ1+ WISP1 in IPF. In addition, DAPK1, endothelial PAS domain protein 1 (EPAS1), protein kinase AMP-activated non-catalytic subunit gamma 2 (PRKAG2), protein patched 1 (PTCH1) and RAS oncogene (RAB7B) were significantly downregulated (
Table S6). Patched-1 protein plays a critical role in fibrogenesis and has been shown to be significantly downregulated both in-vitro in hepatic stellate cells and in-vivo in CCl4-induced rat fibrogenesis model, due PTCH1 hypermethylation [
61]. Furthermore, DAPK1 is an interferon-induced serine/threonine kinase involved in programmed cell death and was significantly decreased by TGFβ1+ WISP1 treatment, making fibroblast resistant to apoptosis, which further contributes to disease pathophysiology and fibrogenesis in both non-diseased and IPF- primary lung fibroblast. Furthermore, we also highlighted unique and overlapping set of genes (
Figure 8E) along with their associated pathways induced by different treatment paradigms (
Figure 8C).
4. Discussion
CCN family members have drawn considerable attention due to their pleiotropic role in human health and diseases. Despite the extensive research and increased attention, WISP1 still remains one of the understudied members of the CCN family. Thus far, majority of the research efforts were directed towards CCN2/ CTGF, but the discontinuation of phase-3 ZEPHYRUS-1 study of Pamrevlumab for IPF, shifted the spotlight towards other promising CCN members as therapeutic targets. Over the past two decades, many studies have suggested that WISP1 plays a profibrotic role by driving disease progression rather than being involved in the initiation of fibrosis. Accordingly, as therapeutic strategies targeting WISP1 are currently at Phase 1 clinical trials, our results, for the first time, uncover the molecular signature of WISP1 profibrotic biology in a cell and disease-specific context i.e., dermal vs lung fibroblasts and between healthy vs. IPF-diseased states. Importantly, our current study indicates that WISP1 is upregulated at both transcript and protein level in IPF-diseased primary lung fibroblasts compared to non-diseased primary lung. TGFβ is the master regulator of fibrosis, and our results also highlight a feedback-loop between TGFβ-WISP1, where the two proteins positively regulate each other.
Furthermore, fibroblast proliferation is one of the key mechanisms involved in fibrogenesis and our study demonstrates WISP1 markedly induced human fibroblast cell proliferation. In addition, our transcriptomics data reveals mitogenic and cell-cycle associated molecular signature, including MAPKs, CDK4, CCNA2 marker of proliferation Ki-67 (MK167), along with transcriptional factor, like E2F4, further corroborating WISP1 induced fibroblast proliferation. Results from the current study also indicate that inflammatory gene signature exhibits high sensitivity to WISP1 as it positively regulates chemokine signaling, interleukin related inflammatory markers and interferon-stimulated gene in human fibroblasts.
Given the biological complexity and aggressive nature of IPF, it is not surprising that narrowly targeting one molecule fails to meet the clinical endpoint. Multimodal therapeutic strategy targeting multiple molecular pathways may yield better patient outcomes. Our study reveals the molecular underpinnings of the multifaceted role of WISP1 in driving various cellular processes ranging from cell proliferation, migration and inflammation. Therefore, targeting WISP1 could yield positive outcomes. Future research and experiments are underway in our laboratory to delve deeper into the downstream mechanisms of action and extend this work in-vivo, further testing and demonstrating translatability.
While the current study aims at unraveling WISP1 biology under IPF setting focusing on human fibroblast, our study has certain limitations. Firstly, it is critical to acknowledge that fibrosis is a complex disease with spatial and temporal heterogeneity due to the highly dynamic cellular state. Here, we are studying primary fibroblasts in isolation, which fails to capture the paracrine effects of WISP1 and crosstalk between fibroblasts and tissue-resident cells including but not limited to epithelial cells, endothelial cells, as well as other immune cells, such as macrophages and neutrophils. We are currently establishing co-culture systems that would shed more light on both autocrine and paracrine effects of WISP1. Second, as these results represent only the initial investigation of the WISP1-induced molecular signature in primary dermal and lung fibroblasts, future work aims to increase the translatability of these findings by utilizing preclinical animal models to better recapitulate disease biology. Furthermore, future work in our lab will also focus on uncovering gene-maps based on the cell type in native animal tissues by utilizing spatial transcriptomics. Nonetheless, to our knowledge these are the first results that delineate molecular underpinnings of WISP1 mediated fibrosis initiation and progression in primary human fibroblasts. In addition, our transcriptomics results must also be validated at the protein level to better correlate these findings. However, the IL6 and CCL2 message at the transcript levels positively corelate with secreted protein levels, suggesting no disconnect between mRNA and protein levels. While here we only focused on lung and dermal fibroblasts, to better understand whether WISP1 has a direct or indirect role in promoting fibrosis, more studies are underway to assess its role in other organ derived fibroblasts, particularly hepatic stellate cells, cardiac fibroblasts, and renal fibroblasts. Furthermore, biomanufacturing of active macromolecules, such as recombinant proteins and antibodies, has significantly advanced over time and is a key process in pharmaceutical industry. However, some proteins remain difficult to express in the conventional CHO or HEK cell-based systems due to several factors, including improper folding, degradation, oligomerization from overproduction, and post-translational modification. These issues can result in low yields or sometimes inactive products. WISP1 is one such ‘difficult-to-express’ protein in cell-based systems, often resulting in inconsistent yield and activity. To address these challenges, we are utilizing lentivirus-mediated approach to mimic endogenously upregulated WISP1 expression seen in diseased states. This method not only facilitates WISP1 production but also relies on cellular machinery to apply the necessary post-translational modifications for proper cellular functions, thereby bypassing the need for recombinant protein. Furthermore, this lentivirus-mediated approach will provide us with the flexibility to explore the domain-specific functional effects of WISP1, as producing domain-specific recombinant proteins can be challenging. Overall, the current study underscores the identification of a novel gene-map to gain better insights into WISP1 profibrotic function and provides a foundation for future investigation, enabling the development of therapeutic strategies for fibrosis. It is critical to emphasize that further research is essential to delve deeper into the mechanism of action, particularly in understanding the diverse functional roles of WISP1 in organ specific inflammation and fibrogenesis.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, K.S and S.S.O.; methodology, K.S; validation, K.S; formal analysis, K.S.; investigation, K.S; writing—original draft preparation, K.S.; writing—review and editing, S.S.O., M.W., K.T., J.B.; supervision, S.S.O.; project administration, S.S.O.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Eli Lilly and Company
Informed Consent Statement
Not applicable
Data Availability Statement
All the relevant data are within the article and
Supplementary Materials. For questions and queries can be directed to the corresponding author.
Acknowledgments
We acknowledge the support of the Department of Biotherapeutics Enabling Biology. The authors would also like to thank Charlie Changzhi Hu and Jacquelyn Mc Entire for their support with the project and Zhe Sun for helping with statistical data analysis.
Conflicts of Interest
The authors declare no conflicts of interest related to the work reported in this paper.
References
- Mei, Q.; Liu, Z.; Zuo, H.; Yang, Z.; Qu, J. Idiopathic Pulmonary Fibrosis: An Update on Pathogenesis. Front. Pharmacol. 2022, 12, 797292. [Google Scholar] [CrossRef] [PubMed]
- Kendall, R.T.; Feghali-Bostwick, C.A. Fibroblasts in fibrosis: novel roles and mediators. Front. Pharmacol. 2014, 5, 123. [Google Scholar] [CrossRef] [PubMed]
- Abuserewa, S.T.; Duff, R.; Becker, G. Treatment of Idiopathic Pulmonary Fibrosis. Cureus 2021, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Proesmans, V.L.J.; Drent, M.; Elfferich, M.D.P.; Wijnen, P.A.H.M.; Jessurun, N.T.; Bast, A. Self-reported Gastrointestinal Side Effects of Antifibrotic Drugs in Dutch Idiopathic Pulmonary Fibrosis patients. Lung 2019, 197, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Oladipupo, S.S. An overview of CCN4 (WISP1) role in human diseases. J. Transl. Med. 2024, 22, 1–24. [Google Scholar] [CrossRef]
- Maiese, K. WISP1: Clinical Insights for a Proliferative and Restorative Member of the CCN Family. Curr. Neurovascular Res. 2014, 11, 378–389. [Google Scholar] [CrossRef]
- Xi, Y.; LaCanna, R.; Ma, H.-Y.; N’diaye, E.-N.; Gierke, S.; Caplazi, P.; Sagolla, M.; Huang, Z.; Lucio, L.; Arlantico, A.; et al. A WISP1 antibody inhibits MRTF signaling to prevent the progression of established liver fibrosis. Cell Metab. 2022, 34, 1377–1393.e8. [Google Scholar] [CrossRef]
- Chen, P.-P.; Li, W.-J.; Wang, Y.; Zhao, S.; Li, D.-Y.; Feng, L.-Y.; Shi, X.-L.; Koeffler, H.P.; Tong, X.-J.; Xie, D. Expression of Cyr61, CTGF, and WISP-1 Correlates with Clinical Features of Lung Cancer. PLOS ONE 2007, 2, e534. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Liu, T.; Lv, N.; Yuan, X.; Li, P. WISP1 induces ovarian cancer via the IGF1/αvβ3/Wnt axis. J Ovarian Res. 2022. [Google Scholar] [CrossRef]
- Wu, J.; Long, Z.; Cai, H.; Du, C.; Liu, X.; Yu, S.; Wang, Y. High expression of WISP1 in colon cancer is associated with apoptosis, invasion and poor prognosis. Oncotarget 2016, 7, 49834–49847. [Google Scholar] [CrossRef]
- Chiang, K.-C.; Yeh, C.-N.; Chung, L.-C.; Feng, T.-H.; Sun, C.-C.; Chen, M.-F.; Jan, Y.-Y.; Yeh, T.-S.; Chen, S.-C.; Juang, H.-H. WNT-1 inducible signaling pathway protein-1 enhances growth and tumorigenesis in human breast cancer. Sci. Rep. 2015, 5, srep08686. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, W.; Yang, Y.; Li, Y.; Zhao, Y. WISP1 indicates poor prognosis and regulates cell proliferation and apoptosis in gastric cancer via targeting AKT/mTOR signaling pathway. Am J Transl Res. 2020, 12, 7297–7311. [Google Scholar] [PubMed]
- Zhang, H.; Luo, H.; Hu, Z.; Peng, J.; Jiang, Z.; Song, T.; Wu, B.; Yue, J.; Zhou, R.; Xie, R.; et al. Targeting WISP1 to sensitize esophageal squamous cell carcinoma to irradiation. Oncotarget 2015, 6, 6218–6234. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lv, S.; Ma, W.; Yang, D.; Zhang, X. Effect of WISP1 on paraquat-induced EMT. Toxicol. Vitr. 2023, 93, 105693. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, Z.; Wang, M.; Huang, C.; Ren, Y.; Zhang, W.; Gao, F.; Cao, L.; Li, L.; Nie, S. Paraquat induces pulmonary fibrosis through Wnt/β-catenin signaling pathway and myofibroblast differentiation. Toxicol. Lett. 2020, 333, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.C.; Wang, J.J.; Dong, S.; Hu, J.W.; Hu, L.J.; Yang, G.M.; et al. Wnt-induced secreted protein 1/CCN4 in liver fibrosis both in vitro and in vivo. Clin Lab. 2014, 60, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Tacke, C.; Aleksandrova, K.; Rehfeldt, M.; Murahovschi, V.; Markova, M.; Kemper, M.; et al. Assessment of circulating Wnt1 inducible signalling pathway protein 1 (WISP-1)/CCN4 as a novel biomarker of obesity. J Cell Commun Signal. 2018, 12, 539–548. [Google Scholar] [CrossRef]
- Hörbelt, T.; Tacke, C.; Markova, M.; de Wiza, D.H.; Van de Velde, F.; Bekaert, M.; Van Nieuwenhove, Y.; Hornemann, S.; Rödiger, M.; Seebeck, N.; et al. The novel adipokine WISP1 associates with insulin resistance and impairs insulin action in human myotubes and mouse hepatocytes. Diabetologia 2018, 61, 2054–2065. [Google Scholar] [CrossRef]
- Wang A ru Yan X qin Zhang, C.; Du C qi Long W jun Zhan, D.; et al. Characterization of Wnt1-inducible Signaling Pathway Protein-1 in Obese Children and Adolescents. Curr Med Sci. 2018, 38, 868–874. [Google Scholar] [CrossRef]
- Blom, A.B.; Brockbank, S.M.; van Lent, P.L.; van Beuningen, H.M.; Geurts, J.; Takahashi, N.; van der Kraan, P.M.; van de Loo, F.A.; Schreurs, B.W.; Clements, K.; et al. Involvement of the Wnt signaling pathway in experimental and human osteoarthritis: Prominent role of Wnt-induced signaling protein 1. Arthritis Rheum. 2009, 60, 501–512. [Google Scholar] [CrossRef]
- Chou, C.-H.; Wu, C.-C.; Song, I.-W.; Chuang, H.-P.; Lu, L.-S.; Chang, J.-H.; Kuo, S.-Y.; Lee, C.-H.; Wu, J.-Y.; Chen, Y.-T.; et al. Genome-wide expression profiles of subchondral bone in osteoarthritis. Arthritis Res. Ther. 2013, 15, R190–R190. [Google Scholar] [CrossRef]
- Komatsu, M.; Nakamura, Y.; Maruyama, M.; Abe, K.; Watanapokasin, R.; Kato, H. Expression profiles of human CCN genes in patients with osteoarthritis or rheumatoid arthritis. J Orthop Sci. 2015, 20, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Macsai, C.E.; Georgiou, K.R.; Foster, B.K.; Zannettino, A.C.; Xian, C.J. Microarray expression analysis of genes and pathways involved in growth plate cartilage injury responses and bony repair. Bone 2012, 50, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Grässel, S.; Straub, R.H.; Schett, G.; Dinser, R.; Grifka, J.; et al. Differential transcriptome analysis of intraarticular lesional vs intact cartilage reveals new candidate genes in osteoarthritis pathophysiology. Osteoarthr Cartil. 2009, 17, 328–335. [Google Scholar] [CrossRef]
- Van Den Bosch, M.H.J.; Ramos, Y.F.M.; Den Hollander, W.; Bomer, N.; Nelissen, R.G.H.H.; Bovée, J.V.M.G.; et al. Increased WISP1 expression in human osteoarthritic articular cartilage is epigenetically regulated and decreases cartilage matrix production. Rheumatol (United Kingdom). 2019, 58, 1065–1074. [Google Scholar] [CrossRef]
- Chen, S.; Li, B. MiR-128-3p Post-Transcriptionally Inhibits WISP1 to Suppress Apoptosis and Inflammation in Human Articular Chondrocytes via the PI3K/AKT/NF-κB Signaling Pathway. Cell Transplant. 2020, 29, 1–13. [Google Scholar] [CrossRef]
- Meng, X.-M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Biernacka, A.; Dobaczewski, M.; Frangogiannis, N.G. TGF-beta signaling in fibrosis. Libert Pap. 2011, 6, 57. [Google Scholar]
- Sun, T.; Heiden, J.A.V.; Gao, X.; Yin, J.; Uttarwar, S.; Liang, W.-C.; Jia, G.; Yadav, R.; Huang, Z.; Mitra, M.; et al. Isoform-selective TGF-β3 inhibition for systemic sclerosis. Med 2024, 5, 132–147.e7. [Google Scholar] [CrossRef]
- Mitra, M.S.; Lancaster, K.; O Adedeji, A.; Palanisamy, G.S.; A Dave, R.; Zhong, F.; Holdren, M.S.; Turley, S.J.; Liang, W.-C.; Wu, Y.; et al. A Potent Pan-TGFβ Neutralizing Monoclonal Antibody Elicits Cardiovascular Toxicity in Mice and Cynomolgus Monkeys. Toxicol. Sci. 2020, 175, 24–34. [Google Scholar] [CrossRef]
- Frank, S.; Madlener, M.; Werner, S. Transforming Growth Factors β1, β2, and β3 and Their Receptors Are Differentially Regulated during Normal and Impaired Wound Healing. J. Biol. Chem. 1996, 271, 10188–10193. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Transforming growth factor–ß in tissue fibrosis. J Exp Med. 2020, 217, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Cancer cell cycles. Science (80-) 1996, 274, 1672–1674. [Google Scholar] [CrossRef] [PubMed]
- Rizzolio, F.; Lucchetti, C.; Caligiuri, I.; Marchesi, I.; Caputo, M.; Klein-Szanto, A.J.; Bagella, L.; Castronovo, M.; Giordano, A. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ. 2012, 19, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Königshoff, M.; Kramer, M.; Balsara, N.; Wilhelm, J.; Amarie, O.V.; Jahn, A.; Rose, F.; Fink, L.; Seeger, W.; Schaefer, L.; et al. WNT1-inducible signaling protein–1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Investig. 2009, 119, 772–787. [Google Scholar] [CrossRef]
- Chang, A.-C.; Chen, P.-C.; Lin, Y.-F.; Su, C.-M.; Liu, J.-F.; Lin, T.-H.; Chuang, S.-M.; Tang, C.-H. Osteoblast-secreted WISP-1 promotes adherence of prostate cancer cells to bone via the VCAM-1/integrin α4β1 system. Cancer Lett. 2018, 426, 47–56. [Google Scholar] [CrossRef]
- Tai, H.; Chang, A.; Yu, H.; Huang, C.; Lai, Y.; Sun, H.; et al. Osteoblast-derived WISP-1 increases VCAM-1 expression and enhances prostate cancer metastasis by down-regulating miR-126. Oncotarget. 2014, 5, 7589–7598. [Google Scholar] [CrossRef]
- Chuang, J.-Y.; Chang, A.-C.; Chiang, I.-P.; Tsai, M.-H.; Tang, C.-H. Apoptosis Signal-Regulating Kinase 1 Is Involved in WISP-1–Promoted Cell Motility in Human Oral Squamous Cell Carcinoma Cells. PLOS ONE 2013, 8, e78022. [Google Scholar] [CrossRef]
- Stephens, S.; Palmer, J.; Konstantinova, I.; Pearce, A.; Jarai, G.; Day, E. A functional analysis of Wnt inducible signalling pathway protein −1 (WISP-1/CCN4). J Cell Commun Signal. 2015, 9, 63–72. [Google Scholar] [CrossRef]
- Christopoulou, M.-E.; Skandalis, S.S.; Papakonstantinou, E.; Stolz, D.; Aletras, A.J. WISP1 induces the expression of macrophage migration inhibitory factor in human lung fibroblasts through Src kinases and EGFR-activated signaling pathways. Am. J. Physiol. Physiol. 2024, 326, C850–C865. [Google Scholar] [CrossRef]
- Singh, K.; Teyani, R.L.; Moniri, N.H. Agonists and hydrogen peroxide mediate hyperoxidation of β2-adrenergic receptor in airway epithelial cells: Implications for tachyphylaxis to β2-agonists in constrictive airway disorders. Biomed. Pharmacother. 2023, 168, 115763–115763. [Google Scholar] [CrossRef] [PubMed]
- Kinnula, V.L.; Fattman, C.L.; Tan, R.J.; Oury, T.D. Oxidative stress in pulmonary fibrosis: A possible role for redox modulatory therapy. Am J Respir Crit Care Med. 2005, 172, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Odajima, N.; Betsuyaku, T.; Nagai, K.; Moriyama, C.; Wang, D.H.; Takigawa, T.; et al. The Role of Catalase in Pulmonary Fibrosis. Respir Res [Internet]. 2010, 11 183. Available from: http://respiratory-research.com/content/11/1/183.
- Zou, J.; Li, Y.; Yu, J.; Dong, L.; Husain, A.N.; Shen, L.; Weber, C.R. Idiopathic pulmonary fibrosis is associated with tight junction protein alterations. Biochim. et Biophys. Acta (BBA) - Biomembr. 2020, 1862, 183205. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Xie, N.; Wang, X.; Wu, W.; Li, X.; Chen, X.; Qian, G.; Li, C.; Zhang, H.; Jiang, Y.; et al. Serum RelB is correlated with renal fibrosis and predicts chronic kidney disease progression. Clin. Transl. Med. 2021, 11, e362. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Huang, W.; Wei, J.; Zhang, J.; Liu, Z.; Ji, R.; Ge, S.; Xiao, M.; Fan, Y.; Lu, C. RelB promotes liver fibrosis via inducing the release of injury-associated inflammatory cytokines. J. Cell. Mol. Med. 2020, 24, 6008–6014. [Google Scholar] [CrossRef]
- Lv, X.; Liu, S.; Liu, C.; Li, Y.; Zhang, T.; Qi, J.; Li, K.; Hua, F.; Cui, B.; Zhang, X.; et al. TRIB3 promotes pulmonary fibrosis through inhibiting SLUG degradation by physically interacting with MDM2. Acta Pharm. Sin. B 2023, 13, 1631–1647. [Google Scholar] [CrossRef]
- Ding, W.-Y.; Li, W.-B.; Ti, Y.; Bi, X.-P.; Sun, H.; Wang, Z.-H.; Zhang, Y.; Zhang, W.; Zhong, M. Protection from renal fibrosis, putative role of TRIB3 gene silencing. Exp. Mol. Pathol. 2013, 96, 80–84. [Google Scholar] [CrossRef]
- Tomcik, M.; Palumbo-Zerr, K.; Zerr, P.; Sumova, B.; Avouac, J.; Dees, C.; Distler, A.; Becvar, R.; Distler, O.; Schett, G.; et al. Tribbles homologue 3 stimulates canonical TGF-β signalling to regulate fibroblast activation and tissue fibrosis. Ann. Rheum. Dis. 2015, 75, 609–616. [Google Scholar] [CrossRef]
- Liu, S.; Lv, X.; Wei, X.; Liu, C.; Li, Q.; Min, J.; Hua, F.; Zhang, X.; Li, K.; Li, P.; et al. TRIB3‒GSK-3β interaction promotes lung fibrosis and serves as a potential therapeutic target. Acta Pharm. Sin. B 2021, 11, 3105–3119. [Google Scholar] [CrossRef]
- Wang, W.; Sun, A.; Lv, W.; Cheng, J.; Lv, S.; Liu, X.; Guan, G.; Liu, G. TRB3, up-regulated in kidneys of rats with type1 diabetes, mediates extracellular matrix accumulation in vivo and in vitro. Diabetes Res. Clin. Pr. 2014, 106, 101–109. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, W.; Xia, C.; Li, Z.; Zhao, W.; Xu, K.; Wang, N.; Lian, H.; Rosas, I.O.; Yu, G. TRIB3 Mediates Fibroblast Activation and Fibrosis though Interaction with ATF4 in IPF. Int. J. Mol. Sci. 2022, 23, 15705. [Google Scholar] [CrossRef] [PubMed]
- Montero, P.; Milara, J.; Roger, I.; Cortijo, J. Role of JAK/STAT in Interstitial Lung Diseases; Molecular and Cellular Mechanisms. Int. J. Mol. Sci. 2021, 22, 6211. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Luo, F. The Role of JAK/STAT Pathway in Fibrotic Diseases: Molecular and Cellular Mechanisms. Biomolecules 2023, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Liu, Y.; Dai, H.; Wang, C. Fatty Acid Metabolism and Idiopathic Pulmonary Fibrosis. Front. Physiol. 2022, 12, 794629. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Yu, Z.; Li, H.; Cheng, J.; Wang, Y. Fatty acid-binding protein 5 aggravates pulmonary artery fibrosis in pulmonary hypertension secondary to left heart disease via activating wnt/β-catenin pathway. J. Adv. Res. 2021, 40, 197–206. [Google Scholar] [CrossRef]
- Sunaga, H.; Matsui, H.; Ueno, M.; Maeno, T.; Iso, T.; Syamsunarno, M.R.A.A.; Anjo, S.; Matsuzaka, T.; Shimano, H.; Yokoyama, T.; et al. Deranged fatty acid composition causes pulmonary fibrosis in Elovl6-deficient mice. Nat. Commun. 2013, 4, 2563. [Google Scholar] [CrossRef]
- Yang, F.; Ma, Z.; Li, W.; Kong, J.; Zong, Y.; Wendusu, B.; Wu, Q.; Li, Y.; Dong, G.; Zhao, X.; et al. Identification and immune characteristics of molecular subtypes related to fatty acid metabolism in idiopathic pulmonary fibrosis. Front. Nutr. 2022, 9, 992331. [Google Scholar] [CrossRef]
- Lai, K.K.; Kweon, S.-M.; Chi, F.; Hwang, E.; Kabe, Y.; Higashiyama, R.; Qin, L.; Yan, R.; Wu, R.P.; Lai, K.; et al. Stearoyl-CoA Desaturase Promotes Liver Fibrosis and Tumor Development in Mice via a Wnt Positive-Signaling Loop by Stabilization of Low-Density Lipoprotein-Receptor-Related Proteins 5 and 6. Gastroenterology 2017, 152, 1477–1491. [Google Scholar] [CrossRef]
- Swendsen, C.L.; Skita, V.; Thrall, R.S. Alterations in surfactant neutral lipid composition during the development of bleomycin-induced pulmonary fibrosis. Biochim. et Biophys. Acta (BBA) - Lipids Lipid Metab. 1996, 1301, 90–96. [Google Scholar] [CrossRef]
- Yang, J.-J.; Tao, H.; Huang, C.; Shi, K.-H.; Ma, T.-T.; Bian, E.-B.; Zhang, L.; Liu, L.-P.; Hu, W.; Lv, X.-W.; et al. DNA methylation and MeCP2 regulation of PTCH1 expression during rats hepatic fibrosis. Cell. Signal. 2013, 25, 1202–1211. [Google Scholar] [CrossRef]
Figure 1.
WISP1 is endogenously upregulated in primary human DHLF. WISP1 transcript and secreted protein are expressed in NHLF and DHLF. DHLF exhibit significantly higher (A) WISP1 mRNA transcript and (B) secreted WISP1 protein in the condition media, detected by RT-qPCR and ELISA, respectively. Each data point represents one donor-derived primary cell line for both NHLF (n=5 donors) and DHLF (n=5 donors), with each experiment performed in triplicates. Statistical analysis was performed using paired T-test and significance denoted as * p< 0.5 or ** p< 0.01 versus healthy NHLF, as shown.
Figure 1.
WISP1 is endogenously upregulated in primary human DHLF. WISP1 transcript and secreted protein are expressed in NHLF and DHLF. DHLF exhibit significantly higher (A) WISP1 mRNA transcript and (B) secreted WISP1 protein in the condition media, detected by RT-qPCR and ELISA, respectively. Each data point represents one donor-derived primary cell line for both NHLF (n=5 donors) and DHLF (n=5 donors), with each experiment performed in triplicates. Statistical analysis was performed using paired T-test and significance denoted as * p< 0.5 or ** p< 0.01 versus healthy NHLF, as shown.
Figure 2.
TGFβ induces WISP1 expression in primary lung and dermal fibroblast. Stimulation with TGFβ (1ng/ml) for 24 hr significantly increases Col1a1, Col3a1, Il6, Acta2, Fn1 and Wisp1 mRNA as compared to control in (A) NHLF, (B) DHLF and (C) NHDF for 3 respective donors (n=3/each) analyzed by RT-qPCR. Notably, TGFβ decreased CCL2 in NHLF and DHLF, except in NHDF. Each experiment was independently performed in triplicates with three biological replicates (n=3) for each donor. Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the unstimulated control, as shown.
Figure 2.
TGFβ induces WISP1 expression in primary lung and dermal fibroblast. Stimulation with TGFβ (1ng/ml) for 24 hr significantly increases Col1a1, Col3a1, Il6, Acta2, Fn1 and Wisp1 mRNA as compared to control in (A) NHLF, (B) DHLF and (C) NHDF for 3 respective donors (n=3/each) analyzed by RT-qPCR. Notably, TGFβ decreased CCL2 in NHLF and DHLF, except in NHDF. Each experiment was independently performed in triplicates with three biological replicates (n=3) for each donor. Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the unstimulated control, as shown.
Figure 3.
WISP1 promotes a striking increase in primary human dermal fibroblast cell proliferation.
(A) Treatment with WISP1 (1000nM) shows a striking increase in cell proliferation (p<0.0001 via One-way ANOVA with Tukey’s post-hoc analysis) detected by increases pRb positive cells as compared to control in NHDF. Representative 10X images were captured with the top image representing Hoechst nuclear stain, while the bottom panel representing pRb green fluorescence. PDGF-BB (100nM) was used as an internal positive control to ensure assay reliability, PDGF-BB dose response curve is shown in
Supplementary Materials.
(B) % pRb positive cells were calculated by dividing pRb positive cells over total number of cells calculated by nuclear Hoechst staining. Three independent experiments (n=3) were performed in triplicates. Statistical analysis was performed using One-way ANOVA with Tukey’s post-hoc analysis versus the vehicle-control condition and significance denoted as *p<0.5, **p<0.01, ***p<0.001, ****p<0.0001.
Figure 3.
WISP1 promotes a striking increase in primary human dermal fibroblast cell proliferation.
(A) Treatment with WISP1 (1000nM) shows a striking increase in cell proliferation (p<0.0001 via One-way ANOVA with Tukey’s post-hoc analysis) detected by increases pRb positive cells as compared to control in NHDF. Representative 10X images were captured with the top image representing Hoechst nuclear stain, while the bottom panel representing pRb green fluorescence. PDGF-BB (100nM) was used as an internal positive control to ensure assay reliability, PDGF-BB dose response curve is shown in
Supplementary Materials.
(B) % pRb positive cells were calculated by dividing pRb positive cells over total number of cells calculated by nuclear Hoechst staining. Three independent experiments (n=3) were performed in triplicates. Statistical analysis was performed using One-way ANOVA with Tukey’s post-hoc analysis versus the vehicle-control condition and significance denoted as *p<0.5, **p<0.01, ***p<0.001, ****p<0.0001.

Figure 4.
WISP1 increases CCL2 and IL6 in primary fibroblasts. Stimulation with WISP1 (1000nM) for 24hr significantly increases CCL2 and IL6 gene expression in (A) NHLF, (B) DHLF and (C) NHDF as compared to control. TGFβ (1ng/ml) for 24 hr served as a positive control and significantly increases Col1a1, Col3a1, Il6, Acta2, Fn1 and Wisp1 versus vehicle control. (D) Stimulation with increasing WISP1 concentrations for 24 hr initiates concentration-dependent increase in CCL2 and IL6 protein levels, detected via ELISA in cell-supernatant of NHDF. Each experiment performed in duplicates with three biological replicates (n=3). Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the vehicle control, as shown.
Figure 4.
WISP1 increases CCL2 and IL6 in primary fibroblasts. Stimulation with WISP1 (1000nM) for 24hr significantly increases CCL2 and IL6 gene expression in (A) NHLF, (B) DHLF and (C) NHDF as compared to control. TGFβ (1ng/ml) for 24 hr served as a positive control and significantly increases Col1a1, Col3a1, Il6, Acta2, Fn1 and Wisp1 versus vehicle control. (D) Stimulation with increasing WISP1 concentrations for 24 hr initiates concentration-dependent increase in CCL2 and IL6 protein levels, detected via ELISA in cell-supernatant of NHDF. Each experiment performed in duplicates with three biological replicates (n=3). Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the vehicle control, as shown.
Figure 5.
Recombinant WISP1 synergistically induces inflammatory markers along with TGFβ. (A) Primary human fibroblasts were stimulated with TGFβ (1ng/ml) for 24 hr for initiation, followed by WISP1 (1000nM) for another 24hr as per the experimental layout. CCL2 and IL6 mRNA expression was significantly increased in (B) non-diseased lung and (D) dermal fibroblasts, but not in (C) IPF-diseased lung fibroblasts. Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the unstimulated control, for three biological replicates (n=3).
Figure 5.
Recombinant WISP1 synergistically induces inflammatory markers along with TGFβ. (A) Primary human fibroblasts were stimulated with TGFβ (1ng/ml) for 24 hr for initiation, followed by WISP1 (1000nM) for another 24hr as per the experimental layout. CCL2 and IL6 mRNA expression was significantly increased in (B) non-diseased lung and (D) dermal fibroblasts, but not in (C) IPF-diseased lung fibroblasts. Statistical analysis was performed using One-way ANOVA with Tukey’s Post-hoc analysis vs vehicle control for respective donors. * denotes p< 0.5, ** denotes p< 0.01, *** denotes p< 0.001, **** denotes p< 0.0001 versus the unstimulated control, for three biological replicates (n=3).
Figure 6.
WISP1 Fibrotic gene signature in dermal fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis)
(E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Figure 6.
WISP1 Fibrotic gene signature in dermal fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis)
(E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Figure 7.
WISP1 Fibrotic gene signature in lung fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis)
(E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Figure 7.
WISP1 Fibrotic gene signature in lung fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis)
(E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Figure 8.
WISP1 Fibrotic gene signature in IPF-diseased fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis) (E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Figure 8.
WISP1 Fibrotic gene signature in IPF-diseased fibroblast. The volcano plot illustrates fibroblast genes upregulated (red) and downregulated (blue) upon stimulation with either
(A) WISP1 alone or
(B) in conjugation with TGFβ as compared to vehicle control (PBS) in NHDF (n=3). The plots represent statistically significant gene with |FC| > 1.5 and Adj Pval < 0.1 are plotted with the x-axis representing log2Fold change (FC) versus the y-axis –log10 p-value.
(C) The heatmap represent pathways significantly associated with corresponding treatment condition with
(D) Scatterplot showing log2 fold change for the differentially expressed genes when comparing WISP1+TGFβ treatment with WISP1 (x-axis) versus TGFβ (y-axis) (E) Venn diagram represent unique and overlapping set of genes between WISP1, TGFβ and WISP1+TGFβ treatment paradigm. Comprehensive lists of genes are provided in the
Supplementary Materials.
Table 1.
Clinical Characteristics of donor.
Table 1.
Clinical Characteristics of donor.
| Characteristics: Mean ± SD (range) |
NHLF (N=5) |
DHLF (N=5) |
NHDF(N=3) |
| Age (years) |
57.3 ± 14.3 (45-73) |
65.6 ± 15.8 (52-83) |
38.3 ± 9.1 |
| Height (Inches) |
61.3 ± 7.1 (55-70) |
68 ± 0 (68-68) |
62.3 ± 3.21 (60-66) |
| Weight (Kg) |
80.1 ± 17.3 (60.7-108.4) |
81.2 ± 6.6 (74.8-88) |
67.8 ± 5.9 (61.2-69.9) |
| Sex (M/F) |
2/3 |
5/0 |
0/3 |
| Race |
C (5) |
C (5) |
C (1), B(1), H(1) |
| Diabetes |
0 |
2 |
0 |
| Heart Disease |
0 |
0 |
0 |
| Hypertension |
2 |
1 |
0 |
| Alcohol |
1 |
2 |
0 |
| Smoke |
0 |
1 |
0 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).