Submitted:
01 November 2024
Posted:
04 November 2024
You are already at the latest version
Abstract
Prostate cancer (PCa) is the most prevalent malignancy and the second leading cause of can-cer-related death in men. Although current therapies can effectively manage the primary tumor, most patients with late-stage disease manifest with metastasis in different organs. From surgery to treatment intensification (TI), several combinations of therapies are administered to improve prognosis of patients with metastatic PCa. Due to the high frequency of mutation during the metastatic phase, the Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genetic engineering tool can accelerate the effects of TI by enhancing targeted gene therapy or immunotherapy. This review describes the genetic backgrounds of metastatic PCa and how CRISPR/Cas9 technology can contribute to the field of PCa treatment development. It also dis-cusses current limitations of conventional PCa therapy and the potential of CRISPR-based-PCa therapy.
Keywords:
1. Introduction
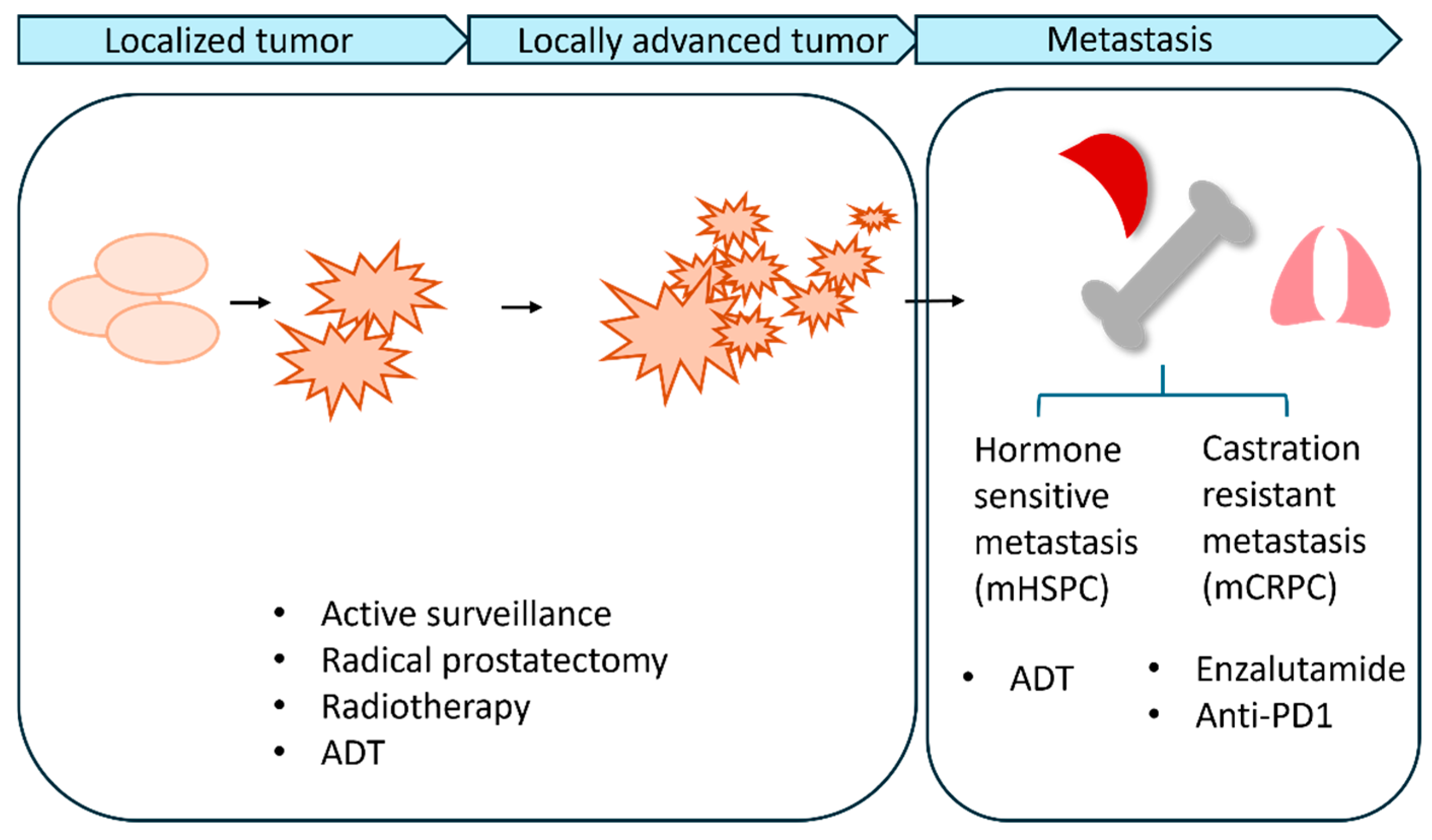
2. Biology of Metastatic PCa
3. Genetics of Metastatic PCa
| Somatic mutations | Localized (n=333) [28] | Metastatic castration sensitive (n=140) [29] | Metastatic castration resistant (n=444) [30] and (n=101) [31] |
|---|---|---|---|
| TMPRSS2-ERG fusion | 46.0% | Not reported | 41.0% and 43.0% |
| Other ETS family gene fusions | 14.0% | Not reported | 10.0% and 15.0% |
| SPOP mutation | 11.0% | 11.0% | 5.0% and 6.0% |
| CHD1 deletion | 7.0% | 6.0% | 23.0% and 33.0% |
| FOXA1 mutation | 4.0% | 10.0% | 9.0% and 19.0% |
| PTEN deletion | 17.0% | 17.0% | 32.0% and 45.0% |
| TP53 mutation or deletion | 8.0% | 30.0% | 40.0% and 57.0% |
| RB1 deletion | 1.0% | 7.0% | 12.0% and 13.0% |
| PI3K mutation | 3.0% | 5.0% | 5.0% and 5.0% |
| AKT mutation | 1.0% | 2.0% | 1.0% and 2.0% |
| BRCA1 mutation or deletion | 1.0% | 1.0% | 1.0% and 2.0% |
| BRCA2 mutation or deletion | 3.0% | 7.0% | 10.0% and 11.0% |
| ATM mutation | 1.0% | 2.0% | 1.0% and 2.0% |
| CDK12 mutation | 2.0% | 6.0% | 3.0% and 7.0% |
| Mismatch repair mutation | 5.0% | 5.0% | 4.0% and 5.0% |
| APC deletion | 5.0% | 13.0% | 8.0% and 9.0% |
| CTNNB1 mutation | 2.0% | 6.0% | 4.0% and 6.0% |
| MYC gain-of-function | 7.0% | 6.0% | 23.0% and 33.0% |
| AR amplification or mutation | 1.0% | 4.0% | 59.0% and 70.0% |
| Germline mutations | Localized (n=499) [32] | Metastatic* (n=692) [32] |
|---|---|---|
| BRCA1 | 0.6% | 0.9% |
| BRCA2 | 0.2% | 5.3% |
| ATM | 1.0% | 1.6% |
| CHEK2 | 0.4% | 1.9% |
| PALB2 | 0.4% | 0.4% |
| RAD51D | 0.4% | 0.4% |
| Mismatch repair (Lynch syndrome) | 0.6% | 0.6% |
4. Current Standard Treatments
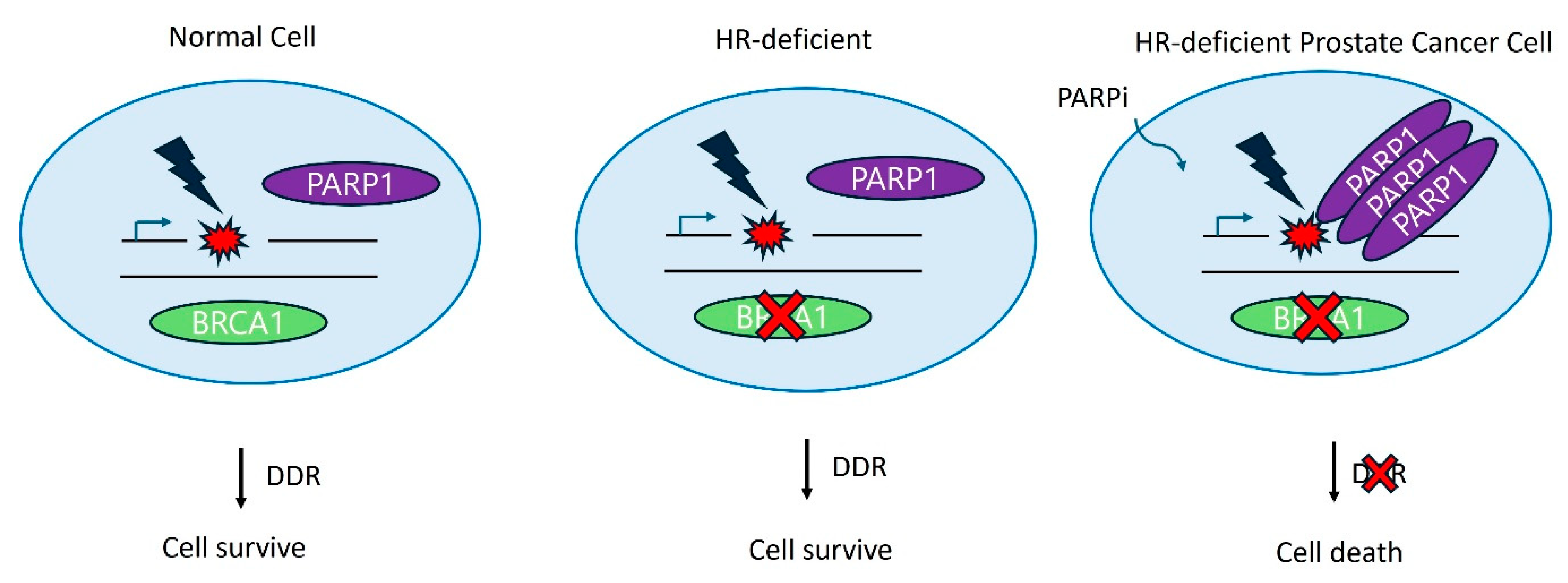
5. DNA Repair Inhibition/Targeted Therapy
5.1. Ly Results in Cancer Cell Death
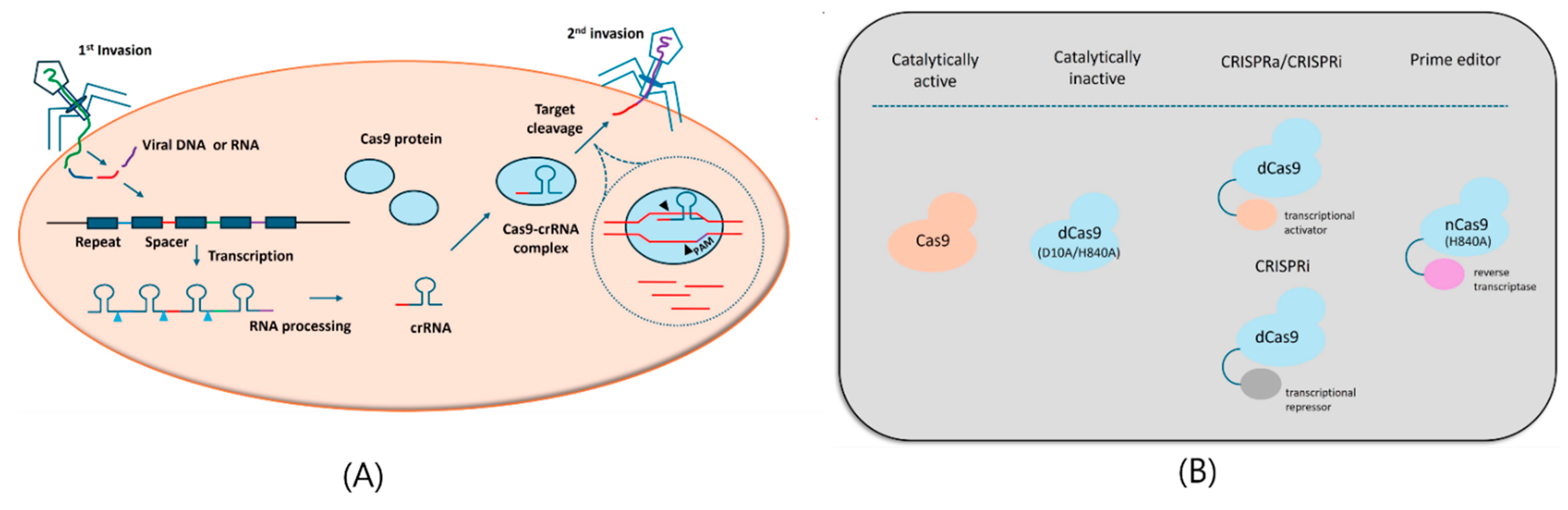
6. CRISPR Technology for mPCa Therapeutics
6.1. Drug Resistance
6.2. Metastasis
6.3. Treatment
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.; Madaan, S. What Is New in the Management of High-Risk Localized Prostate Cancer? J Clin Med 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. Ca-Cancer J Clin 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Wasim, S.; Lee, S.Y.; Kim, J. Complexities of Prostate Cancer. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat Rev Dis Primers 2021, 7, 9. [Google Scholar] [CrossRef]
- Kinsella, N.; Helleman, J.; Bruinsma, S.; Carlsson, S.; Cahill, D.; Brown, C.; Van Hemelrijck, M. Active surveillance for prostate cancer: a systematic review of contemporary worldwide practices. Transl Androl Urol 2018, 7, 83–97. [Google Scholar] [CrossRef]
- Prevention., C.f.D.C.a. U.S. Cancer Statistics Prostate Cancer Stat Bite.US Department of Health and Human Services; 2023. Available online: https://www.cdc.gov/cancer/uscs/about/stat-bites/stat-bite-prostate.htm (accessed on.
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef]
- Desai, M.M.; Cacciamani, G.E.; Gill, K.; Zhang, J.; Liu, L.; Abreu, A.; Gill, I.S. Trends in Incidence of Metastatic Prostate Cancer in the US. JAMA Netw Open 2022, 5, e222246. [Google Scholar] [CrossRef]
- Cuzick, J.; Thorat, M.A.; Andriole, G.; Brawley, O.W.; Brown, P.H.; Culig, Z.; Eeles, R.A.; Ford, L.G.; Hamdy, F.C.; Holmberg, L.; et al. Prevention and early detection of prostate cancer. Lancet Oncol 2014, 15, e484–e492. [Google Scholar] [CrossRef]
- Cheville, J.C.; Tindall, D.; Boelter, C.; Jenkins, R.; Lohse, C.M.; Pankratz, V.S.; Sebo, T.J.; Davis, B.; Blute, M.L. Metastatic prostate carcinoma to bone: clinical and pathologic features associated with cancer-specific survival. Cancer 2002, 95, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.; Vogl, F.D.; Naume, B.; Janni, W.; Osborne, M.P.; Coombes, R.C.; Schlimok, G.; Diel, I.J.; Gerber, B.; Gebauer, G.; et al. A pooled analysis of bone marrow micrometastasis in breast cancer. New Engl J Med 2005, 353, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Niu, C.; Ye, L.; Huang, H.Y.; He, X.; Tong, W.G.; Ross, J.; Haug, J.; Johnson, T.; Feng, J.Q.; et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 2003, 425, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, Y.; Pienta, K.J.; Taichman, R.S. Hematopoietic Stem Cell Niche Is a Potential Therapeutic Target for Bone Metastatic Tumors. Clin Cancer Res 2011, 17, 5553–5558. [Google Scholar] [CrossRef]
- Shiozawa, Y.; Berry, J.E.; Eber, M.R.; Jung, Y.H.; Yumoto, K.; Cackowski, F.C.; Yoon, H.J.; Parsana, P.; Mehra, R.; Wang, J.C.; et al. The marrow niche controls the cancer stem cell phenotype of disseminated prostate cancer. Oncotarget 2016, 7, 41217–41232. [Google Scholar] [CrossRef]
- Kobayashi, A.; Okuda, H.; Xing, F.; Pandey, P.R.; Watabe, M.; Hirota, S.; Pai, S.K.; Liu, W.; Fukuda, K.; Chambers, C.; et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med 2011, 208, 2641–2655. [Google Scholar] [CrossRef]
- Chéry, L.; Lam, H.M.; Coleman, I.; Lakely, B.; Coleman, R.; Larson, S.; Aguirre-Ghiso, J.A.; Xia, J.; Gulati, R.; Nelson, P.S.; et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget 2014, 5, 9939–9951. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017, 9, 34. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Laxman, B.; Dhanasekaran, S.M.; Helgeson, B.E.; Cao, X.; Morris, D.S.; Menon, A.; Jing, X.; Cao, Q.; Han, B.; et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007, 448, 595–599. [Google Scholar] [CrossRef]
- Hagglof, C.; Hammarsten, P.; Stromvall, K.; Egevad, L.; Josefsson, A.; Stattin, P.; Granfors, T.; Bergh, A. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS One 2014, 9, e86824. [Google Scholar] [CrossRef]
- Song, C.; Chen, H. Predictive significance of TMRPSS2-ERG fusion in prostate cancer: a meta-analysis. Cancer Cell Int 2018, 18, 177. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, A.; Graff, R.E.; Bauer, S.R.; Pitt, M.J.; Lis, R.T.; Stack, E.C.; Martin, N.E.; Kunz, L.; Penney, K.L.; Ligon, A.H.; et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev 2012, 21, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.P.; Chen, R.; Zhang, C.L.; Zhang, W.; Xiao, G.A.; Wang, F.B.; Ta, N.; Gao, X.; Sun, Y.H. Prevalence and clinical application of TMPRSS2-ERG fusion in Asian prostate cancer patients: a large-sample study in Chinese people and a systematic review. Asian J Androl 2020, 22, 200–207. [Google Scholar] [CrossRef]
- Li, Y.; Yang, R.; Henzler, C.M.; Ho, Y.; Passow, C.; Auch, B.; Carreira, S.; Nava Rodrigues, D.; Bertan, C.; Hwang, T.H.; et al. Diverse AR Gene Rearrangements Mediate Resistance to Androgen Receptor Inhibitors in Metastatic Prostate Cancer. Clin Cancer Res 2020, 26, 1965–1976. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Bangma, C.H.; Bjartell, A.; Catto, J.W.; Culig, Z.; Gronberg, H.; Luo, J.; Visakorpi, T.; Rubin, M.A. The mutational landscape of prostate cancer. Eur Urol 2013, 64, 567–576. [Google Scholar] [CrossRef]
- Li, J.; Xu, C.L.; Lee, H.J.; Ren, S.C.; Zi, X.Y.; Zhang, Z.M.; Wang, H.F.; Yu, Y.W.; Yang, C.H.; Gao, X.F.; et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020, 580, 93. [Google Scholar] [CrossRef]
- Abeshouse, A.; Ahn, J.; Akbani, R.; Ally, A.; Amin, S.; Andry, C.D.; Annala, M.; Aprikian, A.; Armenia, J.; Arora, A.; et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Abida, W.; Armenia, J.; Gopalan, A.; Brennan, R.; Walsh, M.; Barron, D.; Danila, D.; Rathkopf, D.; Morris, M.; Slovin, S.; et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017, 2017. [Google Scholar] [CrossRef]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019, 116, 11428–11436. [Google Scholar] [CrossRef]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018, 174, 758–769 e759. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Dai, X.; Lunardi, A.; Li, Z.; Inuzuka, H.; Liu, P.; Varmeh, S.; Zhang, J.; Cheng, L.; Sun, Y.; et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell 2015, 59, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Blattner, M.; Liu, D.; Robinson, B.D.; Huang, D.; Poliakov, A.; Gao, D.; Nataraj, S.; Deonarine, L.D.; Augello, M.A.; Sailer, V.; et al. SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell 2017, 31, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Gan, W.; Li, X.; Wang, S.; Zhang, W.; Huang, L.; Liu, S.; Zhong, Q.; Guo, J.; Zhang, J.; et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med 2017, 23, 1063–1071. [Google Scholar] [CrossRef]
- Boysen, G.; Barbieri, C.E.; Prandi, D.; Blattner, M.; Chae, S.S.; Dahija, A.; Nataraj, S.; Huang, D.; Marotz, C.; Xu, L.; et al. SPOP mutation leads to genomic instability in prostate cancer. Elife 2015, 4. [Google Scholar] [CrossRef]
- Wang, S.; Gao, J.; Lei, Q.; Rozengurt, N.; Pritchard, C.; Jiao, J.; Thomas, G.V.; Li, G.; Roy-Burman, P.; Nelson, P.S.; et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003, 4, 209–221. [Google Scholar] [CrossRef]
- Wu, Y.M.; Cieslik, M.; Lonigro, R.J.; Vats, P.; Reimers, M.A.; Cao, X.; Ning, Y.; Wang, L.; Kunju, L.P.; de Sarkar, N.; et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 2018, 173, 1770–1782 e1714. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 2011, 25, 2158–2172. [Google Scholar] [CrossRef]
- Rescigno, P.; Gurel, B.; Pereira, R.; Crespo, M.; Rekowski, J.; Rediti, M.; Barrero, M.; Mateo, J.; Bianchini, D.; Messina, C.; et al. Characterizing CDK12-Mutated Prostate Cancers. Clin Cancer Res 2021, 27, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018, 50, 645-+. [Google Scholar] [CrossRef] [PubMed]
- Eeles, R.; Goh, C.; Castro, E.; Bancroft, E.; Guy, M.; Al Olama, A.A.; Easton, D.; Kote-Jarai, Z. The genetic epidemiology of prostate cancer and its clinical implications. Nature Reviews Urology 2014, 11, 18–31. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Abida, W.; Patnaik, A.; Campbell, D.; Shapiro, J.; Bryce, A.H.; McDermott, R.; Sautois, B.; Vogelzang, N.J.; Bambury, R.M.; Voog, E.; et al. Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a or Gene Alteration. J Clin Oncol 2020, 38, 3763-+. [Google Scholar] [CrossRef]
- Carter, H.B.; Helfand, B.; Mamawala, M.; Wu, Y.; Landis, P.; Yu, H.; Wiley, K.; Na, R.; Shi, Z.; Petkewicz, J.; et al. Germline Mutations in ATM and BRCA1/2 Are Associated with Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur Urol 2019, 75, 743–749. [Google Scholar] [CrossRef]
- Castro, E.; Goh, C.; Leongamornlert, D.; Saunders, E.; Tymrakiewicz, M.; Dadaev, T.; Govindasami, K.; Guy, M.; Ellis, S.; Frost, D.; et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur Urol 2015, 68, 186–193. [Google Scholar] [CrossRef]
- Taylor, R.A.; Fraser, M.; Livingstone, J.; Espiritu, S.M.G.; Thorne, H.; Huang, V.; Lo, W.; Shiah, Y.J.; Yamaguchi, T.N.; Sliwinski, A.; et al. Germline mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun 2017, 8. [Google Scholar] [CrossRef]
- Taylor, R.A.; Fraser, M.; Rebello, R.J.; Boutros, P.C.; Murphy, D.G.; Bristow, R.G.; Risbridger, G.P. The influence of mutation on localized prostate cancer. Nature Reviews Urology 2019, 16, 281–290. [Google Scholar] [CrossRef]
- Huggins, C.; Hodges, C.V. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin 1972, 22, 232–240. [Google Scholar] [CrossRef]
- Sartor, O. Androgen deprivation therapy in prostate cancer: new findings and questions for the future. Lancet Oncol 2019, 20, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.D.; Martin, A.J.; Stockler, M.R.; Begbie, S.; Chi, K.N.; Chowdhury, S.; Coskinas, X.; Frydenberg, M.; Hague, W.E.; Horvath, L.G.; et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 2019, 381, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Fizazi, K.; Saad, F.; Rathenborg, P.; Shore, N.; Ferreira, U.; Ivashchenko, P.; Demirhan, E.; Modelska, K.; Phung, D.; et al. Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 2018, 378, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.K.; Schenkein, E.; Murali, R.; Subudhi, S.K.; Wongvipat, J.; Balbas, M.D.; Shah, N.; Cai, L.; Efstathiou, E.; Logothetis, C.; et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013, 155, 1309–1322. [Google Scholar] [CrossRef]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016, 22, 298–305. [Google Scholar] [CrossRef]
- Alumkal, J.J.; Sun, D.; Lu, E.; Beer, T.M.; Thomas, G.V.; Latour, E.; Aggarwal, R.; Cetnar, J.; Ryan, C.J.; Tabatabaei, S.; et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci U S A 2020, 117, 12315–12323. [Google Scholar] [CrossRef]
- Manna, F.; Karkampouna, S.; Zoni, E.; De Menna, M.; Hensel, J.; Thalmann, G.N.; Kruithof-de Julio, M. Metastases in Prostate Cancer. Cold Spring Harb Perspect Med 2019, 9. [Google Scholar] [CrossRef]
- Maitland, N.J.; Collins, A.T. Prostate cancer stem cells: a new target for therapy. J Clin Oncol 2008, 26, 2862–2870. [Google Scholar] [CrossRef]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005, 65, 10946–10951. [Google Scholar] [CrossRef]
- Colombel, M.; Eaton, C.L.; Hamdy, F.; Ricci, E.; van der Pluijm, G.; Cecchini, M.; Mege-Lechevallier, F.; Clezardin, P.; Thalmann, G. Increased expression of putative cancer stem cell markers in primary prostate cancer is associated with progression of bone metastases. Prostate 2012, 72, 713–720. [Google Scholar] [CrossRef]
- Li, S.; Topatana, W.; Juengpanich, S.; Cao, J.; Hu, J.; Zhang, B.; Ma, D.; Cai, X.; Chen, M. Development of synthetic lethality in cancer: molecular and cellular classification. Signal Transduct Target Ther 2020, 5, 241. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Sumanasuriya, S.; De Bono, J. Treatment of Advanced Prostate Cancer-A Review of Current Therapies and Future Promise. Csh Perspect Med 2018, 8. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Offit, K.; Nelson, P.S. DNA-Repair Gene Mutations in Metastatic Prostate Cancer Reply. New Engl J Med 2016, 375, 1804–1805. [Google Scholar] [CrossRef]
- Kim, C.; Chen, C.; Yu, Y. Avoid the trap: Targeting PARP1 beyond human malignancy. Cell Chem Biol 2021, 28, 456–462. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015, 373, 1697–1708. [Google Scholar] [CrossRef]
- Hussain, M.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer. N Engl J Med 2020, 383, 2345–2357. [Google Scholar] [CrossRef]
- Mulholland, D.J.; Kobayashi, N.; Ruscetti, M.; Zhi, A.; Tran, L.M.; Huang, J.; Gleave, M.; Wu, H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 2012, 72, 1878–1889. [Google Scholar] [CrossRef]
- Zhao, D.; Lu, X.; Wang, G.; Lan, Z.; Liao, W.; Li, J.; Liang, X.; Chen, J.R.; Shah, S.; Shang, X.; et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 2017, 542, 484–488. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Witz, A.; Dardare, J.; Francois, A.; Husson, M.; Rouyer, M.; Demange, J.; Merlin, J.L.; Gilson, P.; Harle, A. CRISPR/Cas9-mediated knock-in of BRCA1/2 mutations restores response to olaparib in pancreatic cancer cell lines. Sci Rep 2023, 13, 18741. [Google Scholar] [CrossRef]
- Tsujino, T.; Takai, T.; Hinohara, K.; Gui, F.; Tsutsumi, T.; Bai, X.; Miao, C.; Feng, C.; Gui, B.; Sztupinszki, Z.; et al. CRISPR screens reveal genetic determinants of PARP inhibitor sensitivity and resistance in prostate cancer. Nat Commun 2023, 14, 252. [Google Scholar] [CrossRef]
- Lei, H.Q.; Wang, Z.F.; Jiang, D.G.; Liu, F.; Liu, M.L.; Lei, X.X.; Yang, Y.F.; He, B.; Yan, M.; Huang, H.; et al. CRISPR screening identifies CDK12 as a conservative vulnerability of prostate cancer. Cell Death Dis 2021, 12. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Y.; He, D.; Jones, K.M.; Tang, S.; Allison, D.B.; Zhang, Y.; Chen, J.; Zhang, Q.; Wang, X.; et al. A kinome-wide CRISPR screen identifies CK1alpha as a target to overcome enzalutamide resistance of prostate cancer. Cell Rep Med 2023, 4, 101015. [Google Scholar] [CrossRef]
- Cai, H.; Zhang, B.; Ahrenfeldt, J.; Joseph, J.V.; Riedel, M.; Gao, Z.; Thomsen, S.K.; Christensen, D.S.; Bak, R.O.; Hager, H.; et al. CRISPR/Cas9 model of prostate cancer identifies Kmt2c deficiency as a metastatic driver by Odam/Cabs1 gene cluster expression. Nat Commun 2024, 15, 2088. [Google Scholar] [CrossRef]
- Arriaga, J.M.; Ronaldson-Bouchard, K.; Picech, F.; Nunes de Almeida, F.; Afari, S.; Chhouri, H.; Vunjak-Novakovic, G.; Abate-Shen, C. In vivo genome-wide CRISPR screening identifies CITED2 as a driver of prostate cancer bone metastasis. Oncogene 2024, 43, 1303–1315. [Google Scholar] [CrossRef]
- Camargo, J.A.; Viana, N.I.; Pimenta, R.; Guimaraes, V.R.; dos Santos, G.A.; Candido, P.; Ghazarian, V.; Romao, P.; Silva, I.A.; Birbrair, A.; et al. The Effect of Gene Editing by CRISPR-Cas9 of miR-21 and the Indirect Target MMP9 in Metastatic Prostate Cancer. International Journal of Molecular Sciences 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, N.; Dong, B.; Guo, W.; Wei, H.; Chen, Q.; Yuan, H.; Han, Y.; Chang, H.; Kan, S.; et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J Clin Invest 2019, 129, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Gries, K.; Valimukhametova, A.R.; McKinney, R.L.; Gonzalez-Rodriguez, R.; Topkiran, U.C.; Coffer, J.; Akkaraju, G.R.; Naumov, A.V. In Vitro Prostate Cancer Treatment via CRISPR-Cas9 Gene Editing Facilitated by Polyethyleneimine-Derived Graphene Quantum Dots. Adv Funct Mater 2023, 33. [Google Scholar] [CrossRef] [PubMed]
- O'Donnell, L.; Panier, S.; Wildenhain, J.; Tkach, J.M.; Al-Hakim, A.; Landry, M.C.; Escribano-Diaz, C.; Szilard, R.K.; Young, J.T.; Munro, M.; et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 2010, 40, 619–631. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat Rev Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Zhang, P.; Lee, H.; Brunzelle, J.S.; Couture, J.F. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res 2012, 40, 4237–4246. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef]
- Shin, S.H.; Lee, G.Y.; Lee, M.; Kang, J.; Shin, H.W.; Chun, Y.S.; Park, J.W. Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat Commun 2018, 9, 4113. [Google Scholar] [CrossRef]
- Lau, W.M.; Weber, K.L.; Doucet, M.; Chou, Y.T.; Brady, K.; Kowalski, J.; Tsai, H.L.; Yang, J.; Kominsky, S.L. Identification of prospective factors promoting osteotropism in breast cancer: a potential role for CITED2. Int J Cancer 2010, 126, 876–884. [Google Scholar] [CrossRef]
- Gong, Y.X.; Chippada-Venkata, U.D.; Oh, W.K. Roles of Matrix Metalloproteinases and Their Natural Inhibitors in Prostate Cancer Progression. Cancers 2014, 6, 1298–1327. [Google Scholar] [CrossRef]
- Kim, K.; Kim, H.H.; Lee, C.H.; Kim, S.; Cheon, G.J.; Kang, K.W.; Chung, J.K.; Youn, H. Therapeutic efficacy of modified anti-miR21 in metastatic prostate cancer. Biochem Biophys Res Commun 2020, 529, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Coppola, V.; Musumeci, M.; Patrizii, M.; Cannistraci, A.; Addario, A.; Maugeri-Sacca, M.; Biffoni, M.; Francescangeli, F.; Cordenonsi, M.; Piccolo, S.; et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 2013, 32, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun 2009, 383, 280–285. [Google Scholar] [CrossRef]
- Park, J.H.; Riviere, I.; Gonen, M.; Wang, X.; Senechal, B.; Curran, K.J.; Sauter, C.; Wang, Y.; Santomasso, B.; Mead, E.; et al. Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N Engl J Med 2018, 378, 449–459. [Google Scholar] [CrossRef]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014, 371, 1507–1517. [Google Scholar] [CrossRef]
- Dorff, T.B.; Blanchard, M.S.; Adkins, L.N.; Luebbert, L.; Leggett, N.; Shishido, S.N.; Macias, A.; Del Real, M.M.; Dhapola, G.; Egelston, C.; et al. PSCA-CAR T cell therapy in metastatic castration-resistant prostate cancer: a phase 1 trial. Nat Med 2024, 30, 1636–1644. [Google Scholar] [CrossRef]
- Porter, L.H.; Zhu, J.J.; Lister, N.L.; Harrison, S.G.; Keerthikumar, S.; Goode, D.L.; Urban, R.Q.; Byrne, D.J.; Azad, A.; Vela, I.; et al. Low-dose carboplatin modifies the tumor microenvironment to augment CAR T cell efficacy in human prostate cancer models. Nat Commun 2023, 14, 5346. [Google Scholar] [CrossRef]
- Wei, W.; Chen, Z.N.; Wang, K. CRISPR/Cas9: A Powerful Strategy to Improve CAR-T Cell Persistence. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Nishimura, H.; Minato, N.; Nakano, T.; Honjo, T. Immunological studies on PD-1 deficient mice: implication of PD-1 as a negative regulator for B cell responses. Int Immunol 1998, 10, 1563–1572. [Google Scholar] [CrossRef]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef]
- Dötsch, S.; Svec, M.; Schober, K.; Hammel, M.; Wanisch, A.; Gökmen, F.; Jarosch, S.; Warmuth, L.; Barton, J.; Cicin-Sain, L.; et al. Long-term persistence and functionality of adoptively transferred antigen-specific T cells with genetically ablated PD-1 expression. P Natl Acad Sci USA 2023, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Zhang, X.Y.; Cheng, C.; Mu, W.; Liu, X.J.; Li, N.; Wei, X.F.; Liu, X.; Xia, C.Q.; Wang, H.Y. CRISPR-Cas9 mediated disruption in CAR-T cells. Front Med-Prc 2017, 11, 554–562. [Google Scholar] [CrossRef] [PubMed]
| Subject | Organism | Target | Methods | Genetic factors | Ref. | |
|---|---|---|---|---|---|---|
| Drug resistance | in-vitro, in-vivo | PARP inhibitor sensitivity and resistance | CRISPR KO library | MMS22L KO | CHEK2 KO | Tsujino et al [77] (2023) |
| Increasement of sensitivity to PARPi | Increasement of resistance to PARPi | |||||
| in-vitro | AR inhibitor resistance | CRISPR KO library | CDK12 KO | Lei et al [78] (2021) | ||
| Synergistic effect with ARi | ||||||
| in-vitro, in-vivo | AR inhibitor resistance | CRISPR KO library | CK1α KO | Liu et al [79] (2023) | ||
| Increasement of sensitivity to ARi | ||||||
| Metastasis | in-vivo | Lung metastasis | CRISPR KO library | KMT2C | Cai et al [80] (2024) | |
| Driver of lung metastasis | ||||||
| in-vivo | Bone metastasis | CRISPRa/CRISPRi library | CTIED2 | Arriaga et a [81]l (2024) | ||
| Driver of bone metastasis | ||||||
| in-vitro | Cancer cell proliferation and migration | CRISPR KO library | MMP9, miR-21 | Camargo et al [82] (2023) | ||
| Driver of metastasis | ||||||
| Treatment | in-vivo | Synthetic lethal target dentification | CRISPR KO library | BRG1 KO | Ding et al [83] (2019) | |
| Inhibition of PTEN-deficient Pca progression | ||||||
| in-vitro | Nanotherapeutics Correction of oncogene TP53 |
PEI-GQD/CRISPR RNP | TP53 KI | Lee et al [84] (2023) | ||
| Induction of apoptotic cell death of prostate cancer cell | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





