Submitted:
30 October 2024
Posted:
01 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Perturb-seq as standard strategy
1.2. Resolving affected biological pathways by interpreting high-throughput data
2. Methods
2.1. Data and filtering
2.2. Comparing variants
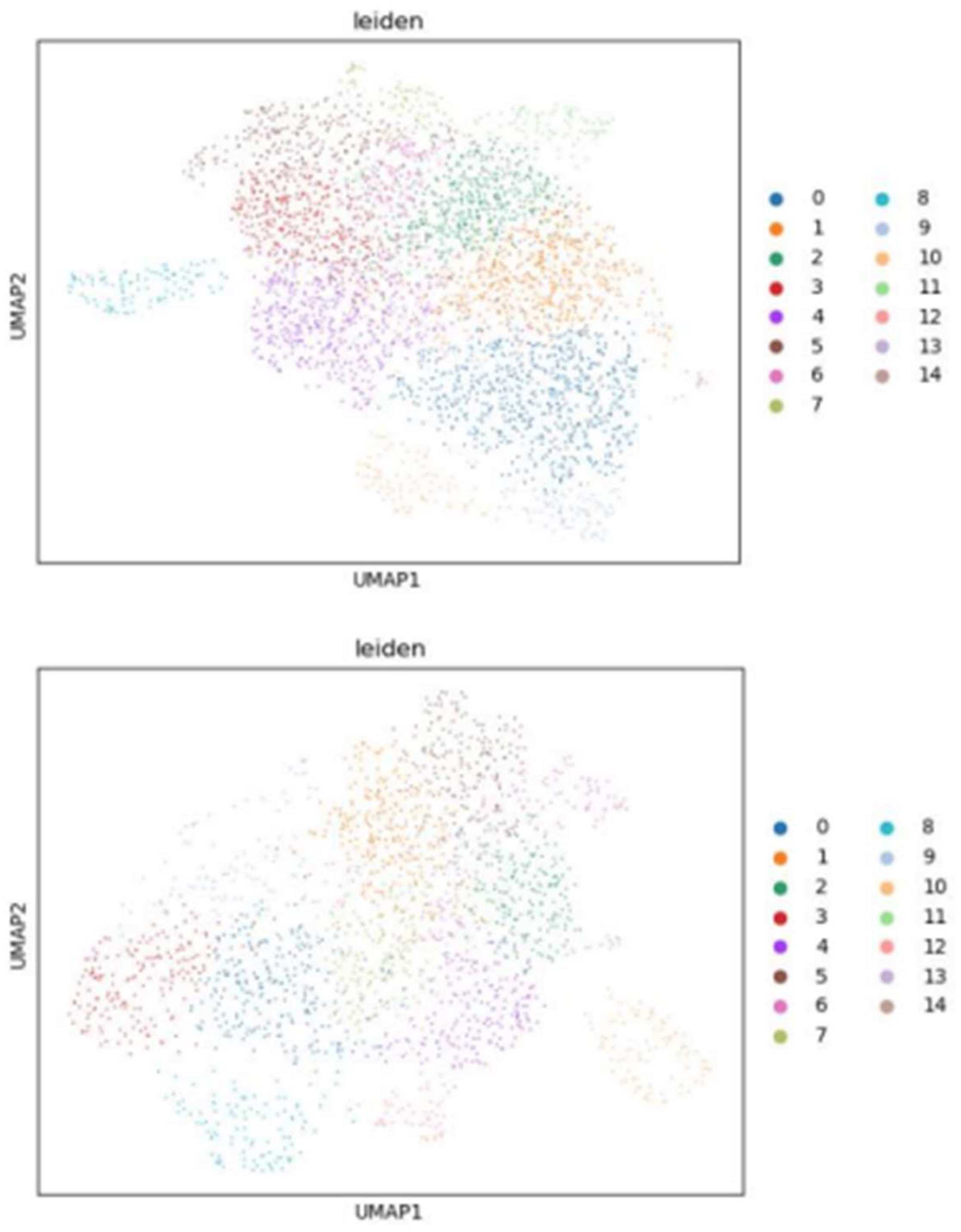
2.3. Leiden clustering
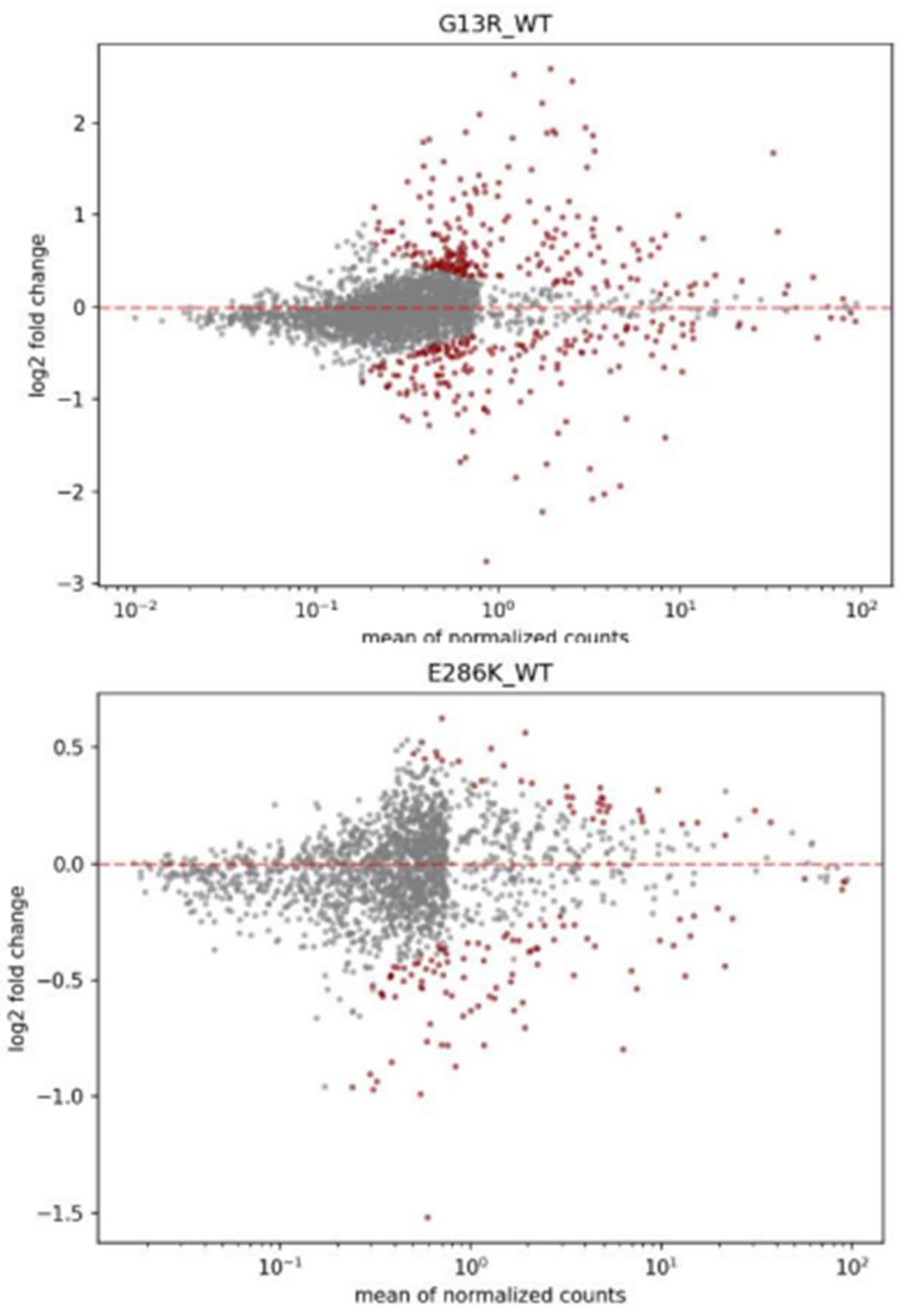
2.4. Differential expression análisis (DEA)
2.5. The STRING database
2.6. Adaptation of Hierarchical Hotnet
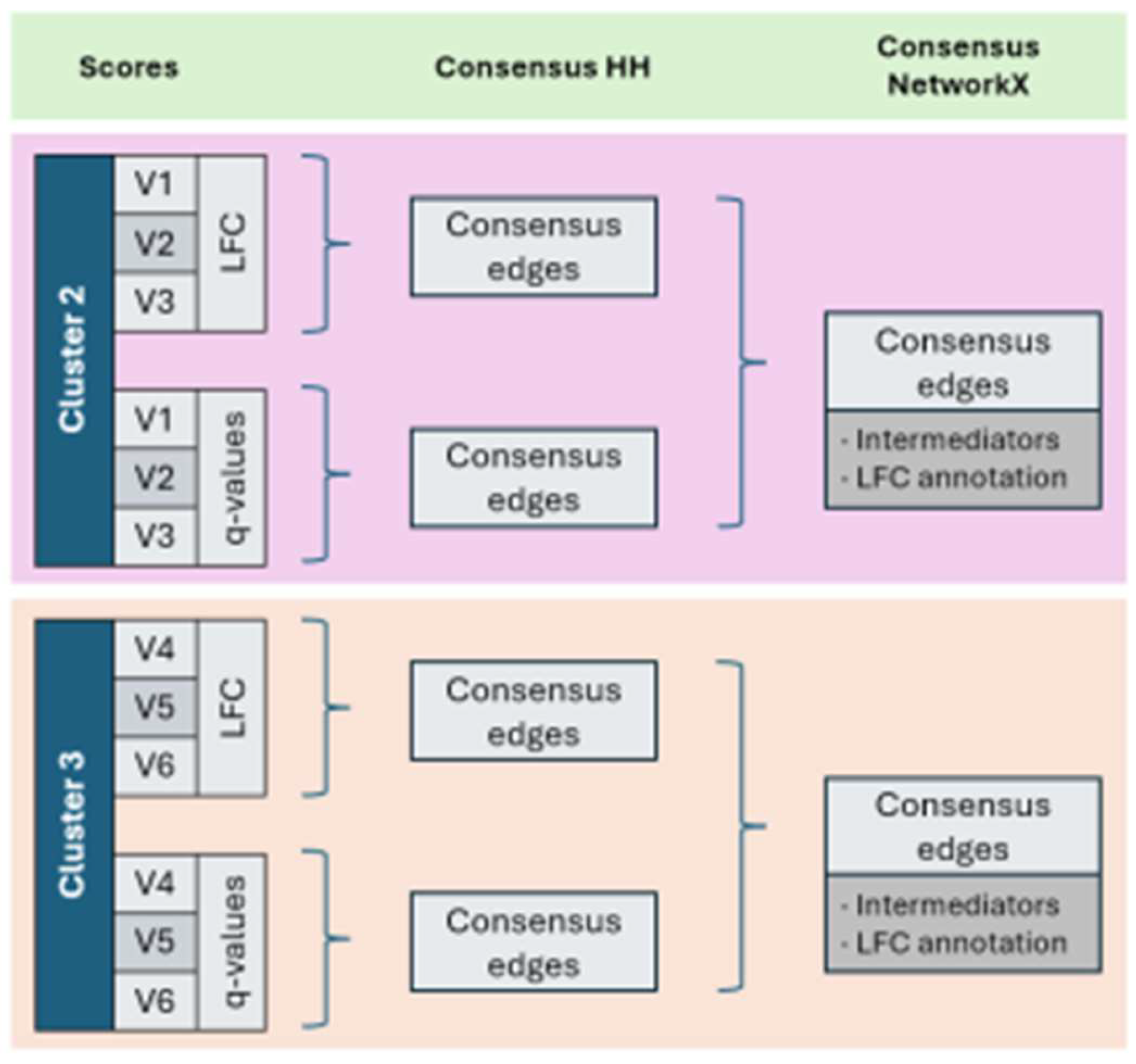
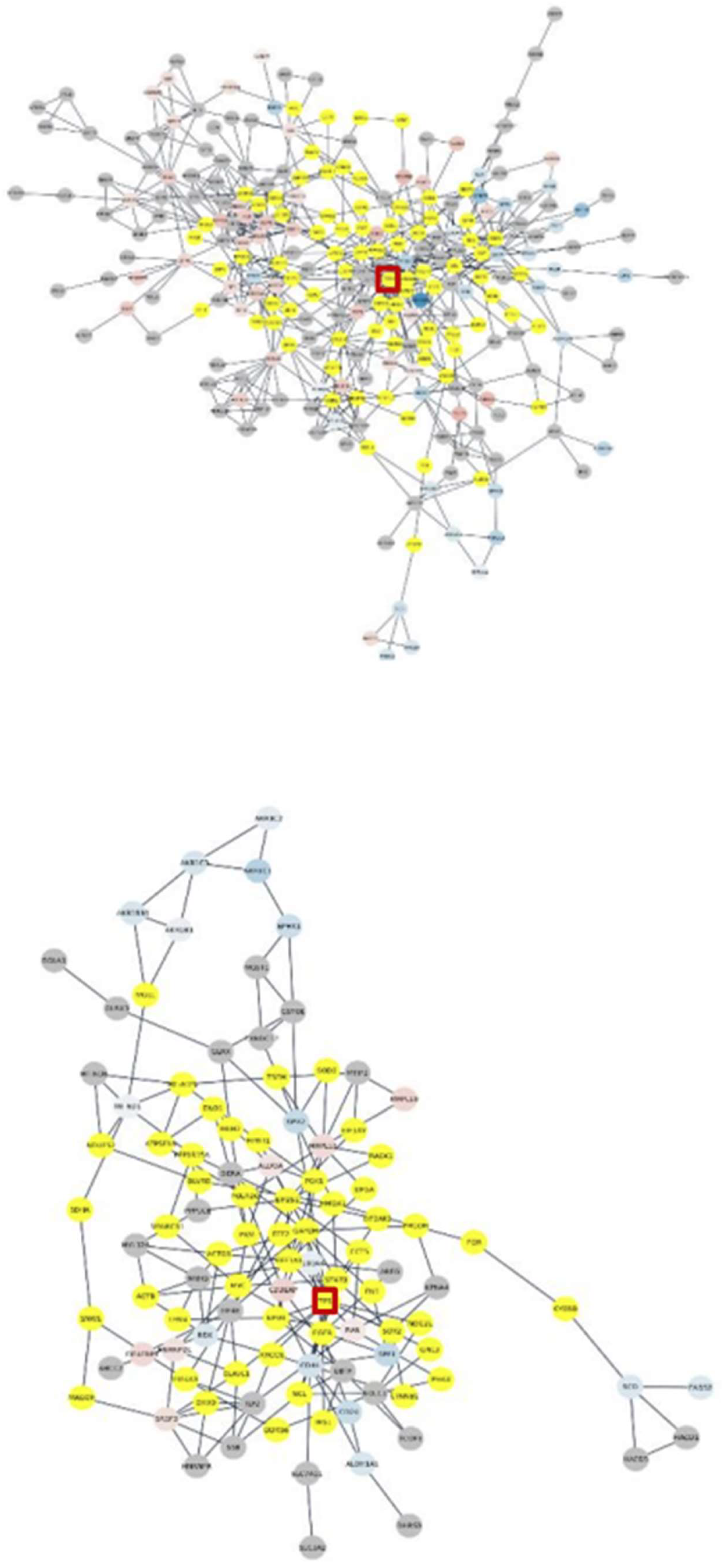
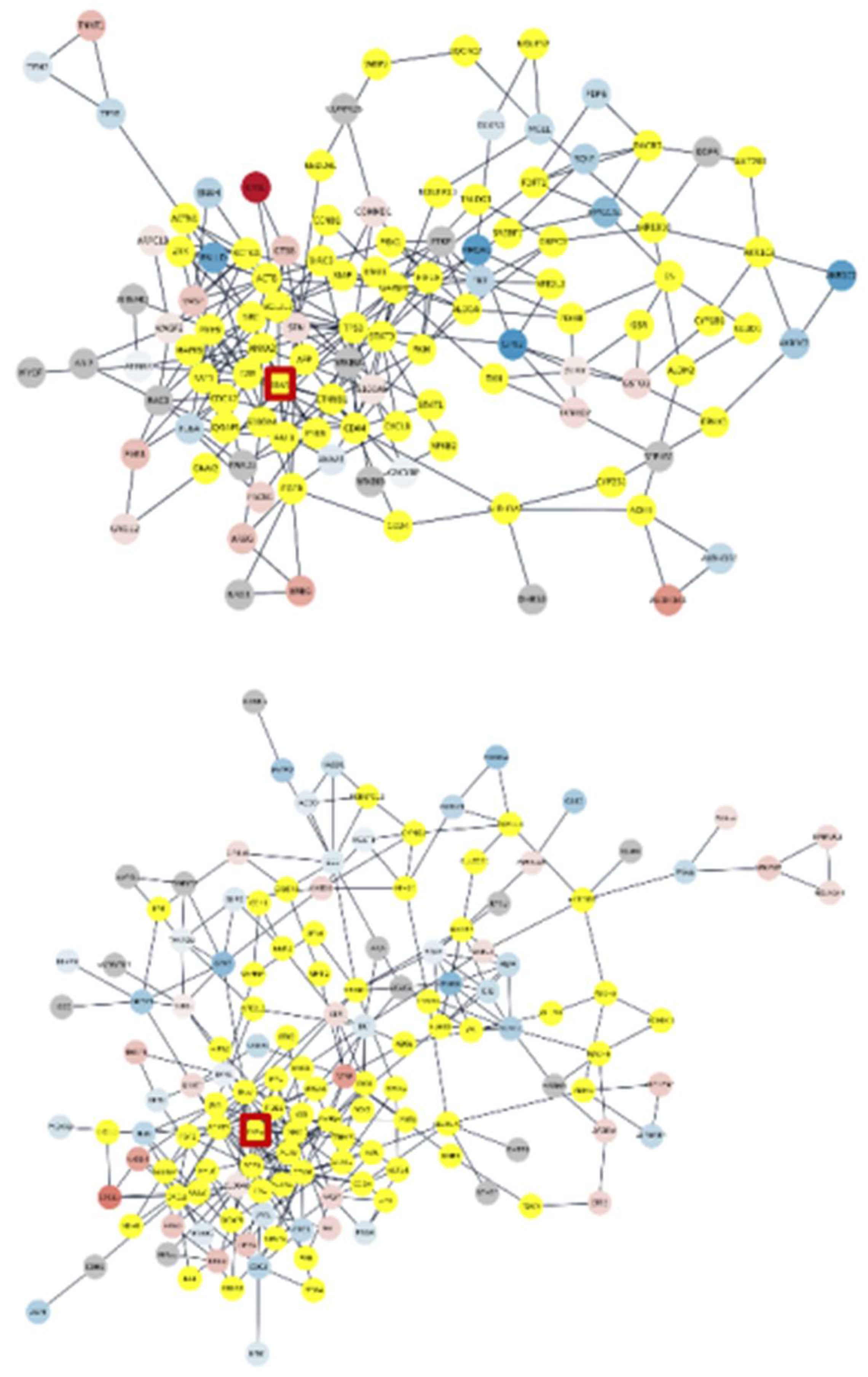
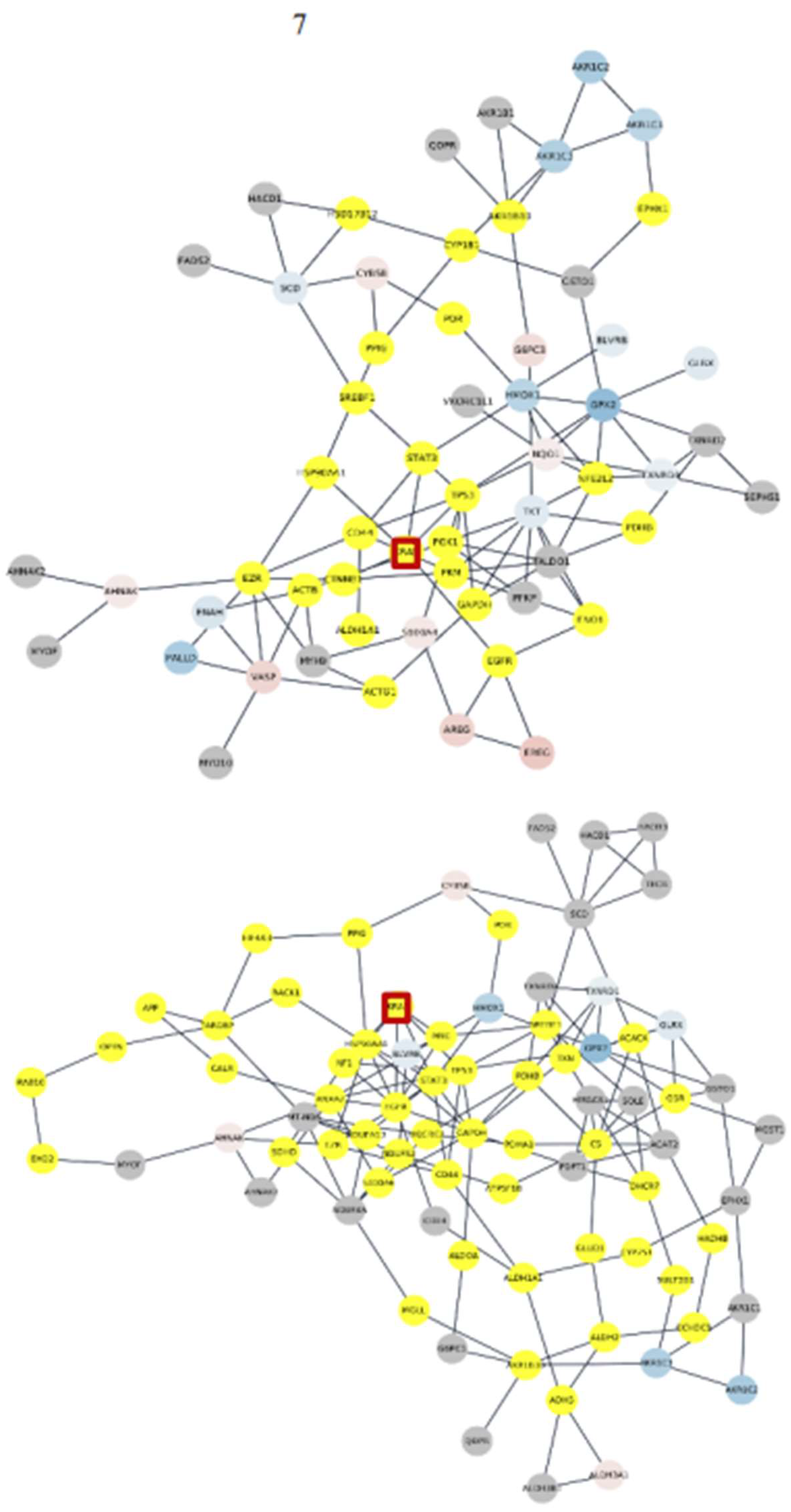
3. Results
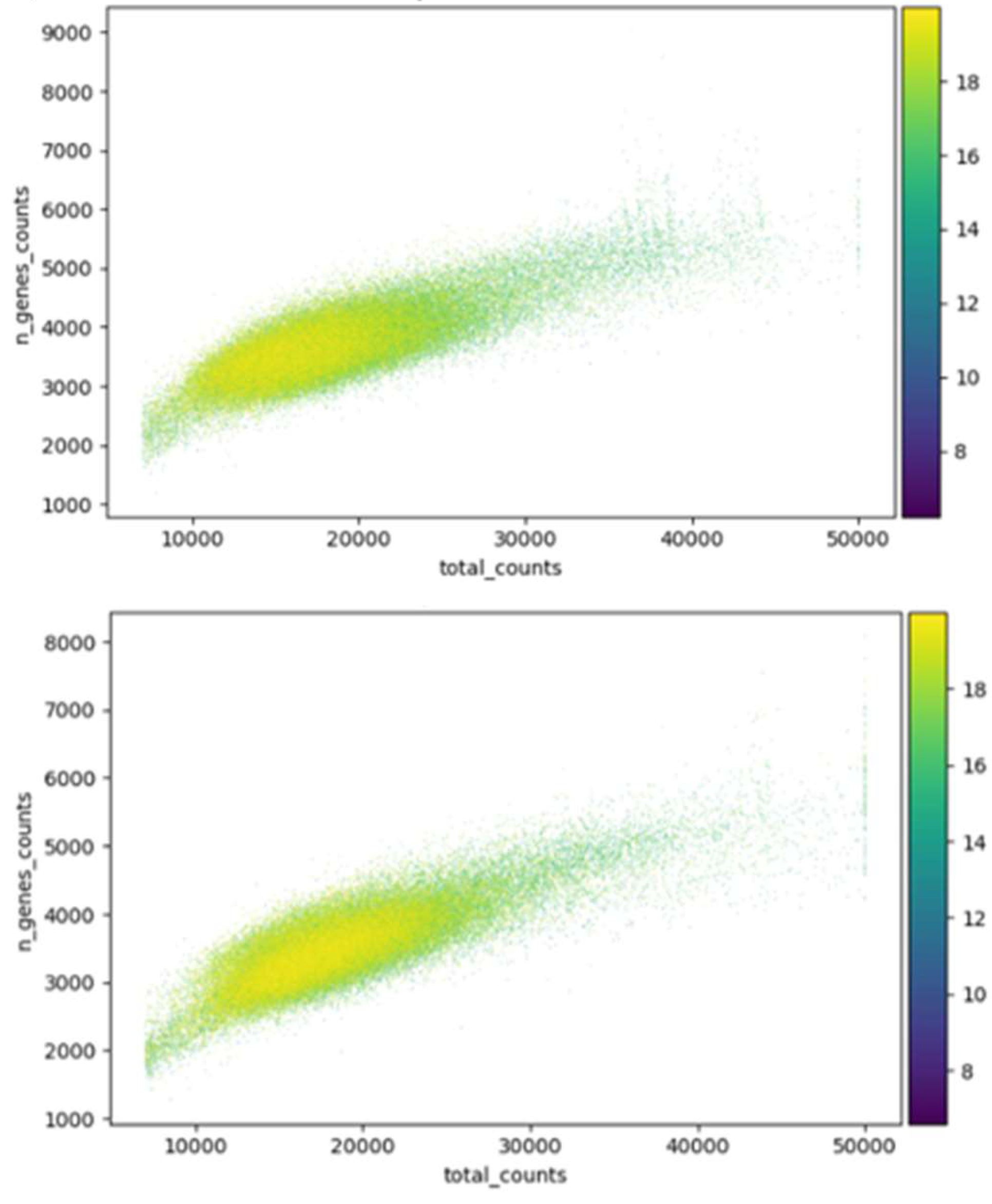
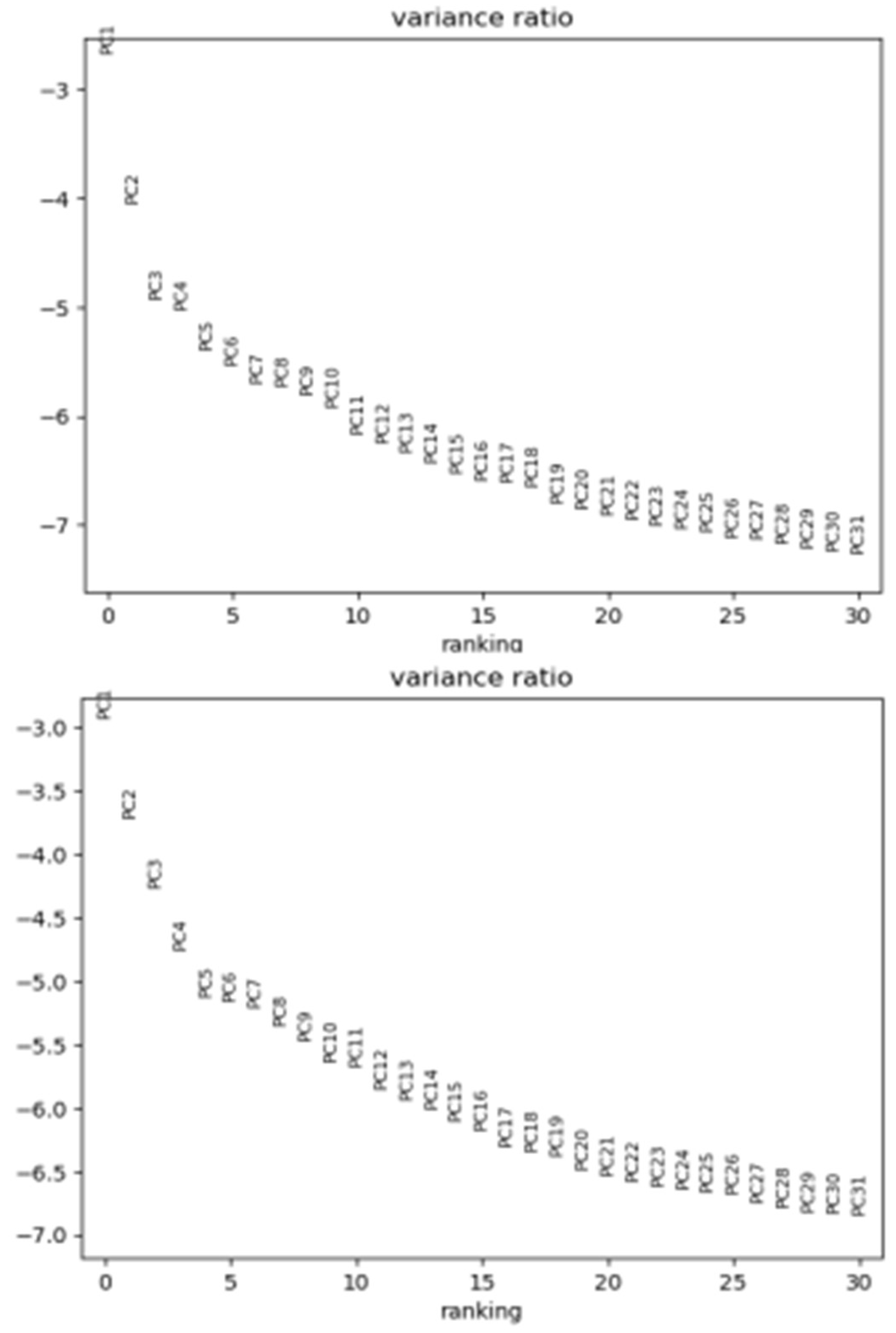
3.1. Overview of data processing
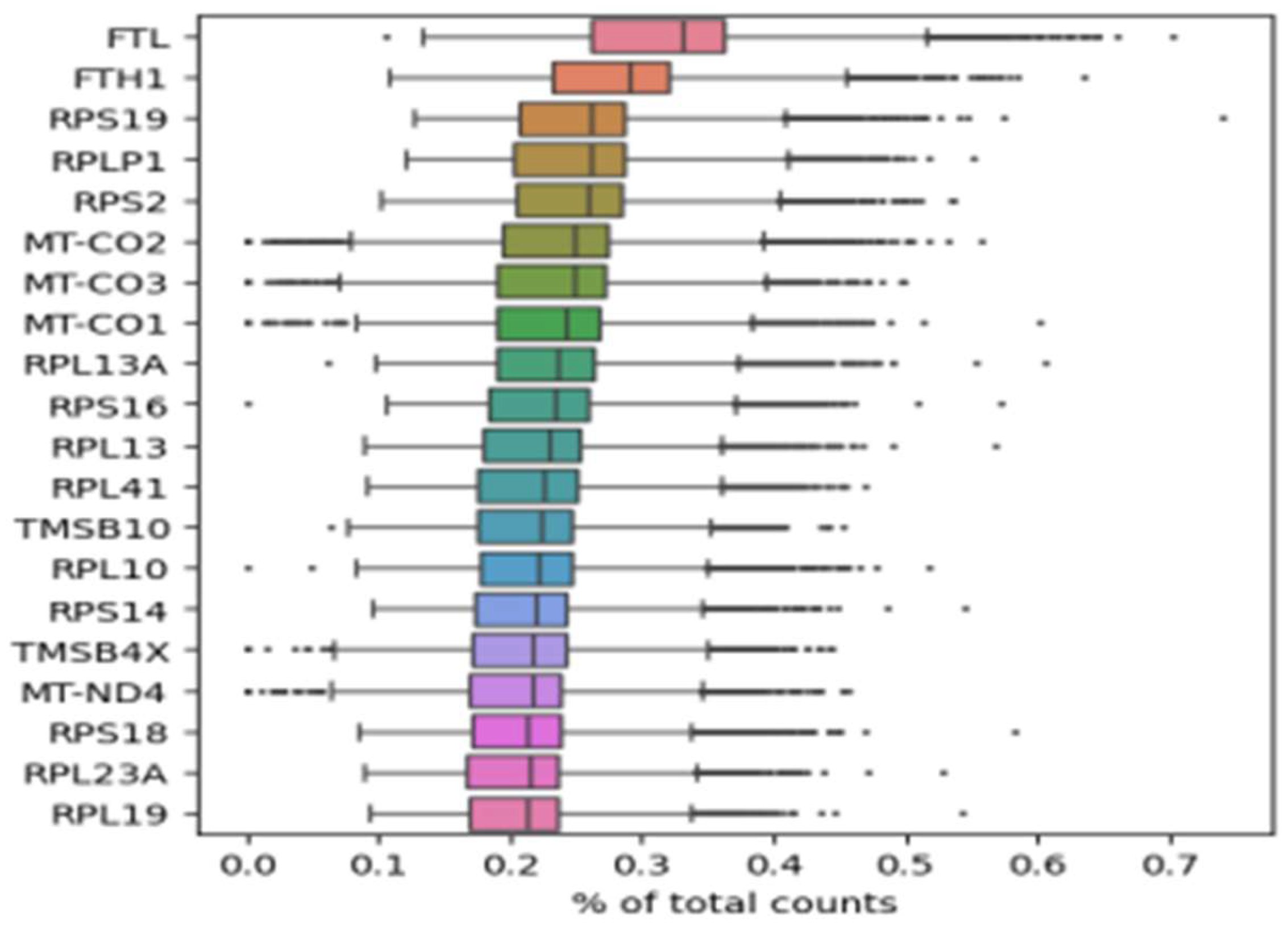
3.2. Characterization of variant profiles
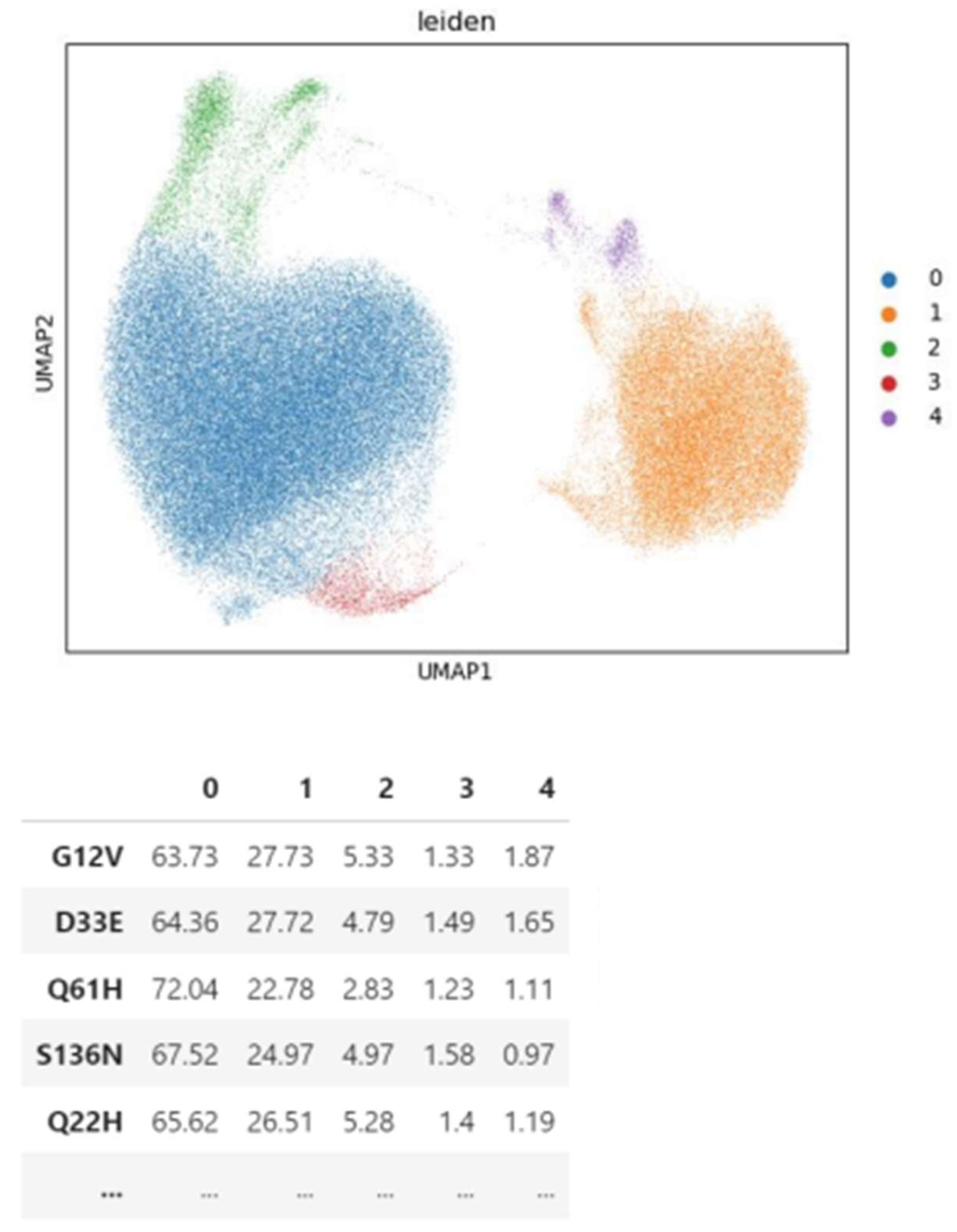
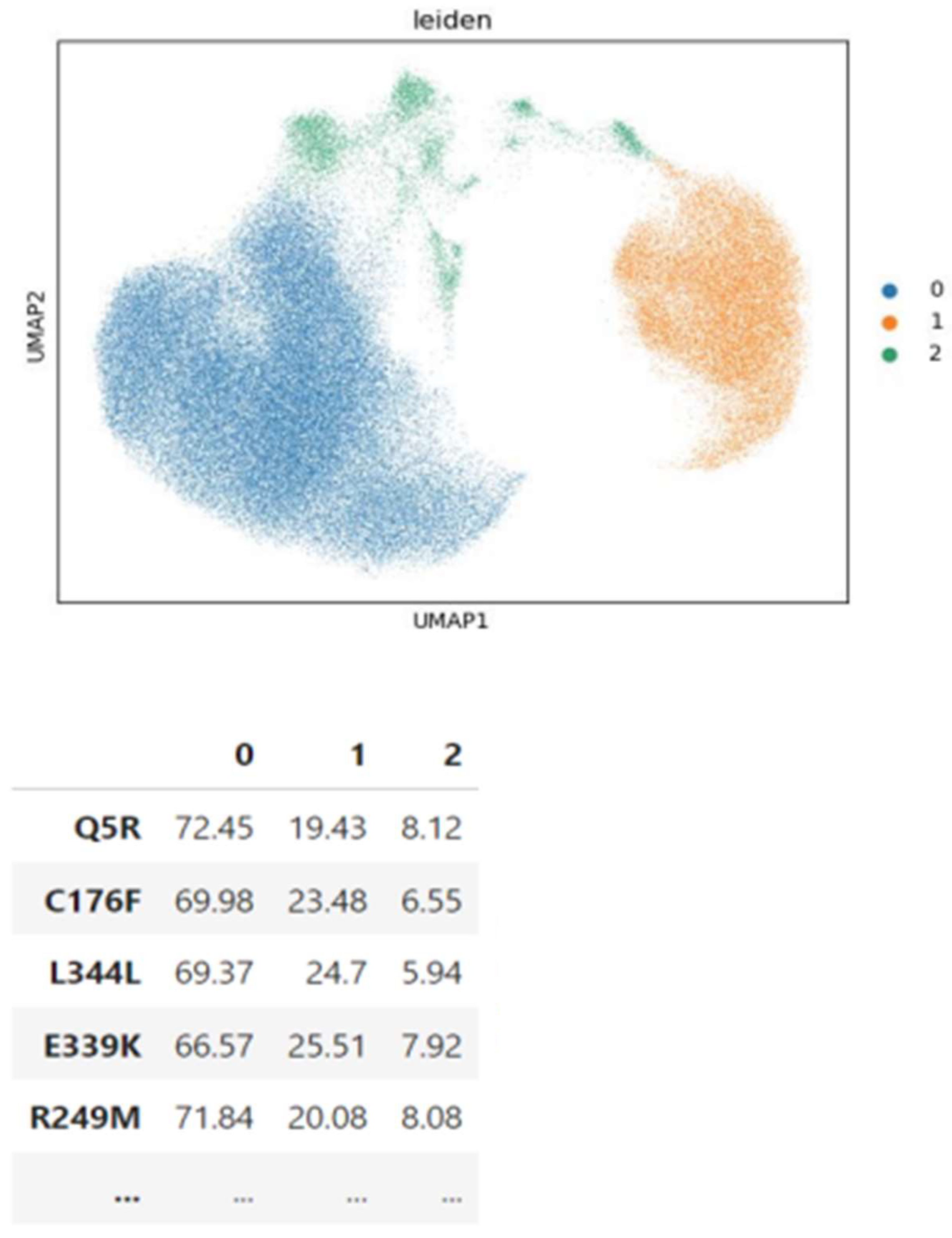
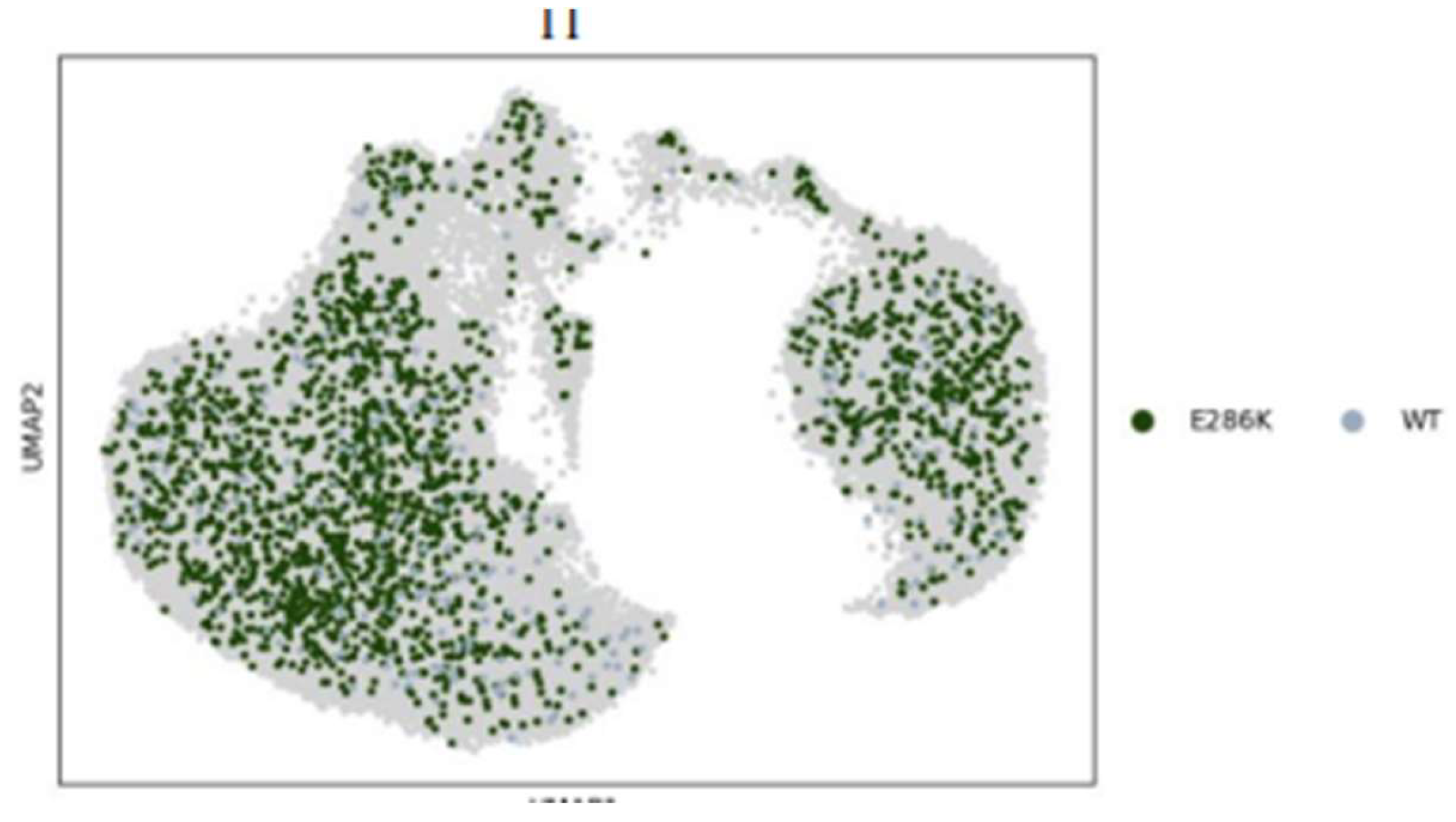
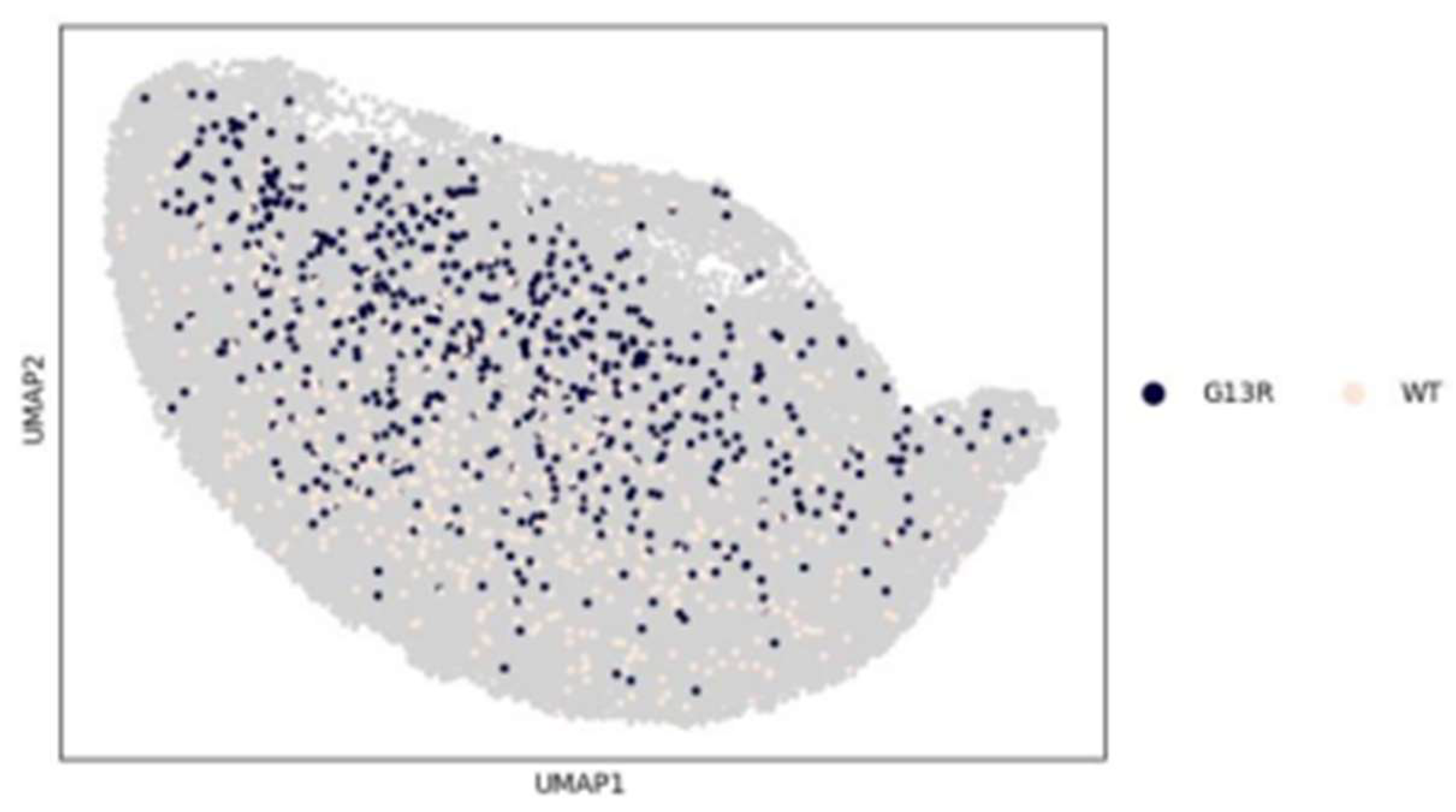
3.3. Insights from Leiden clustering
3.4. DEA insights
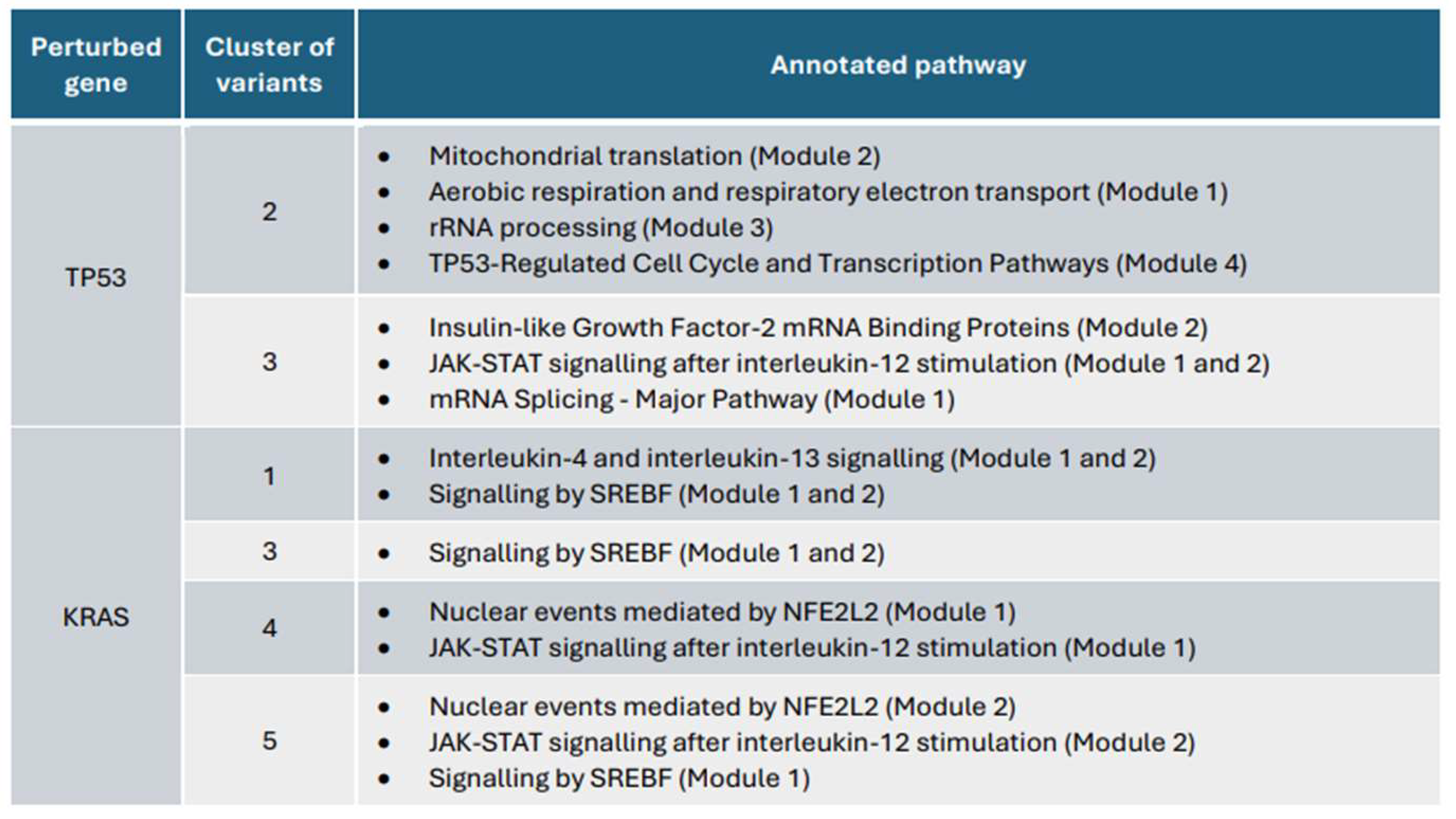
3.4. Subnetworks identification and annotation
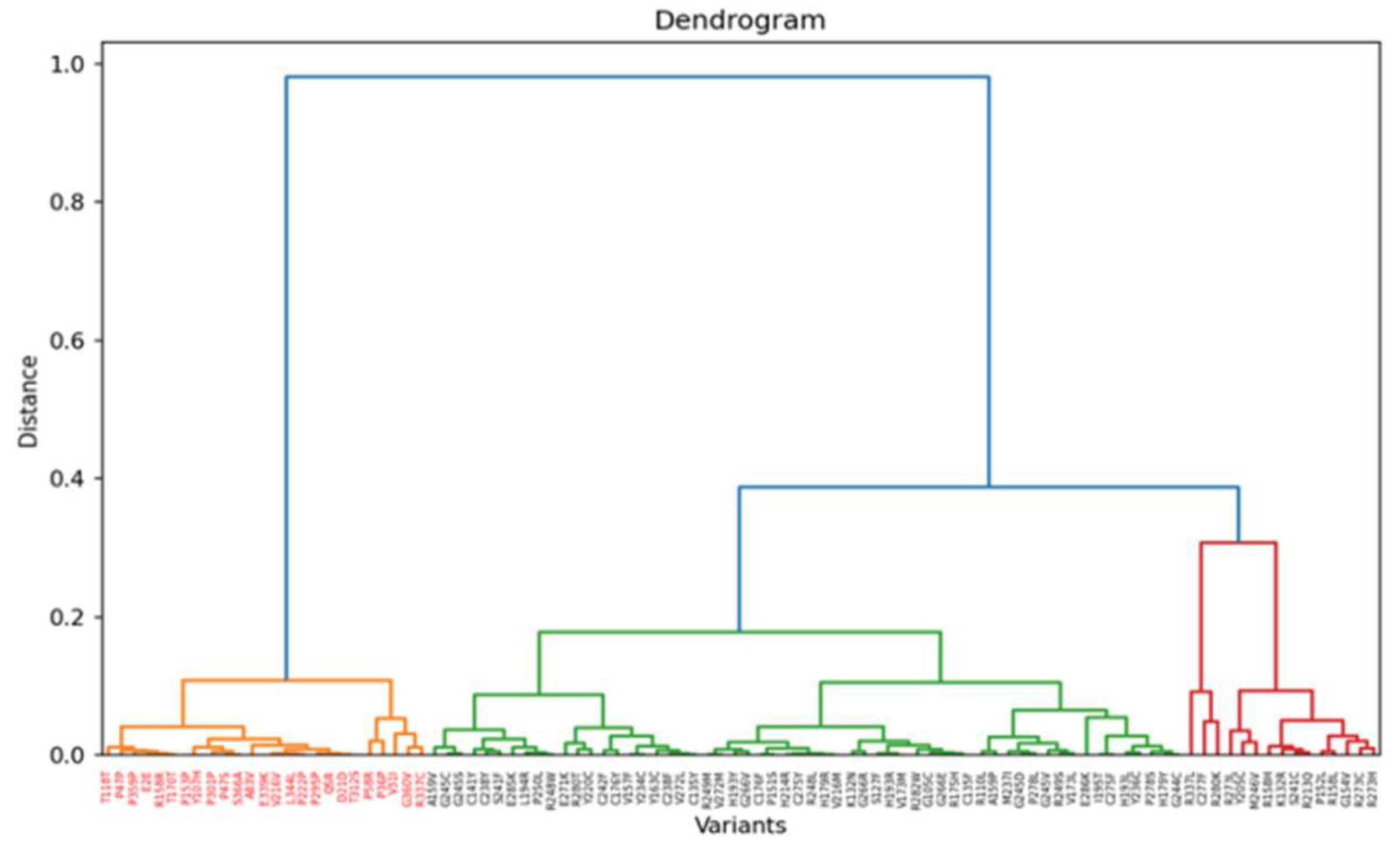
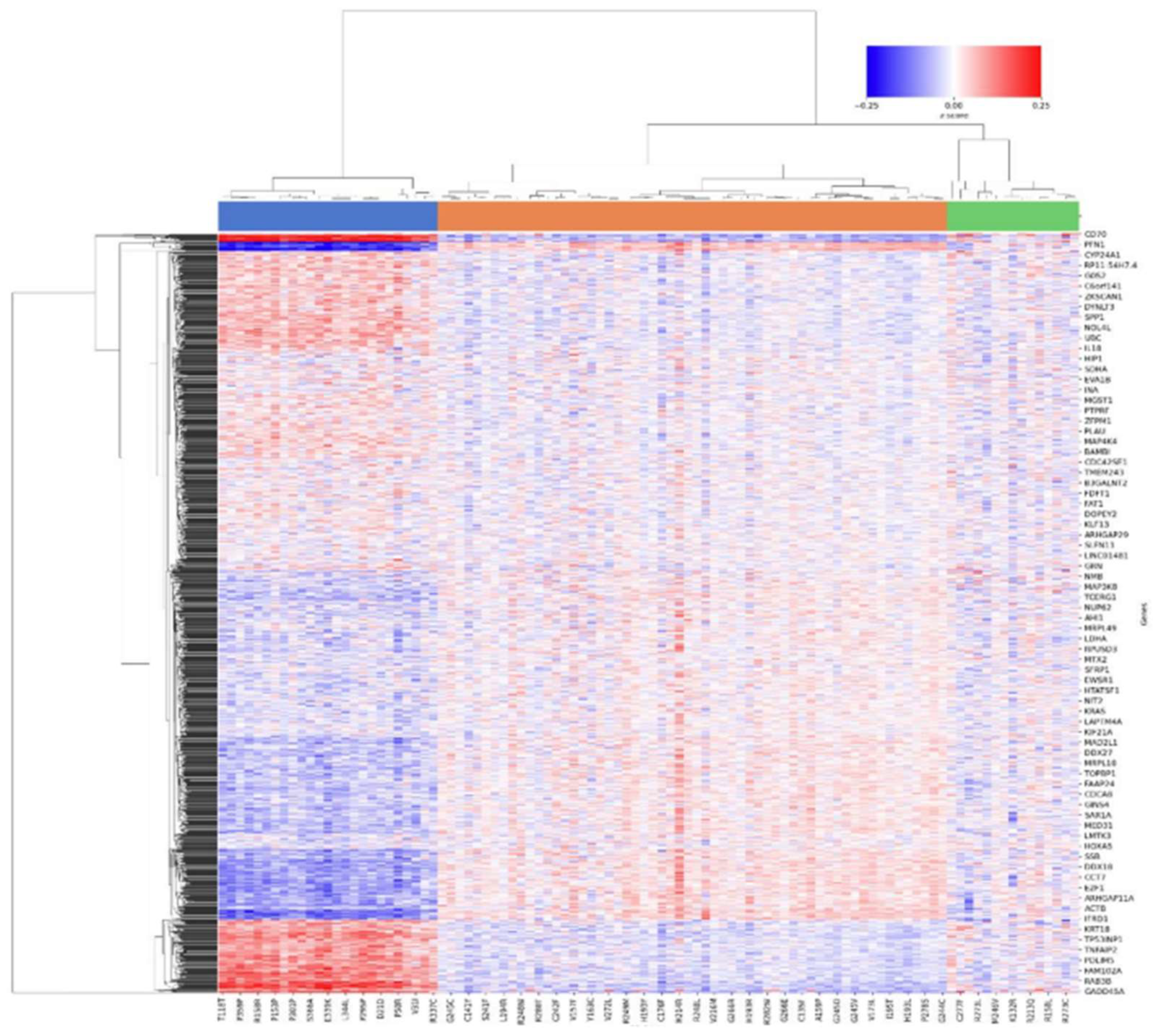
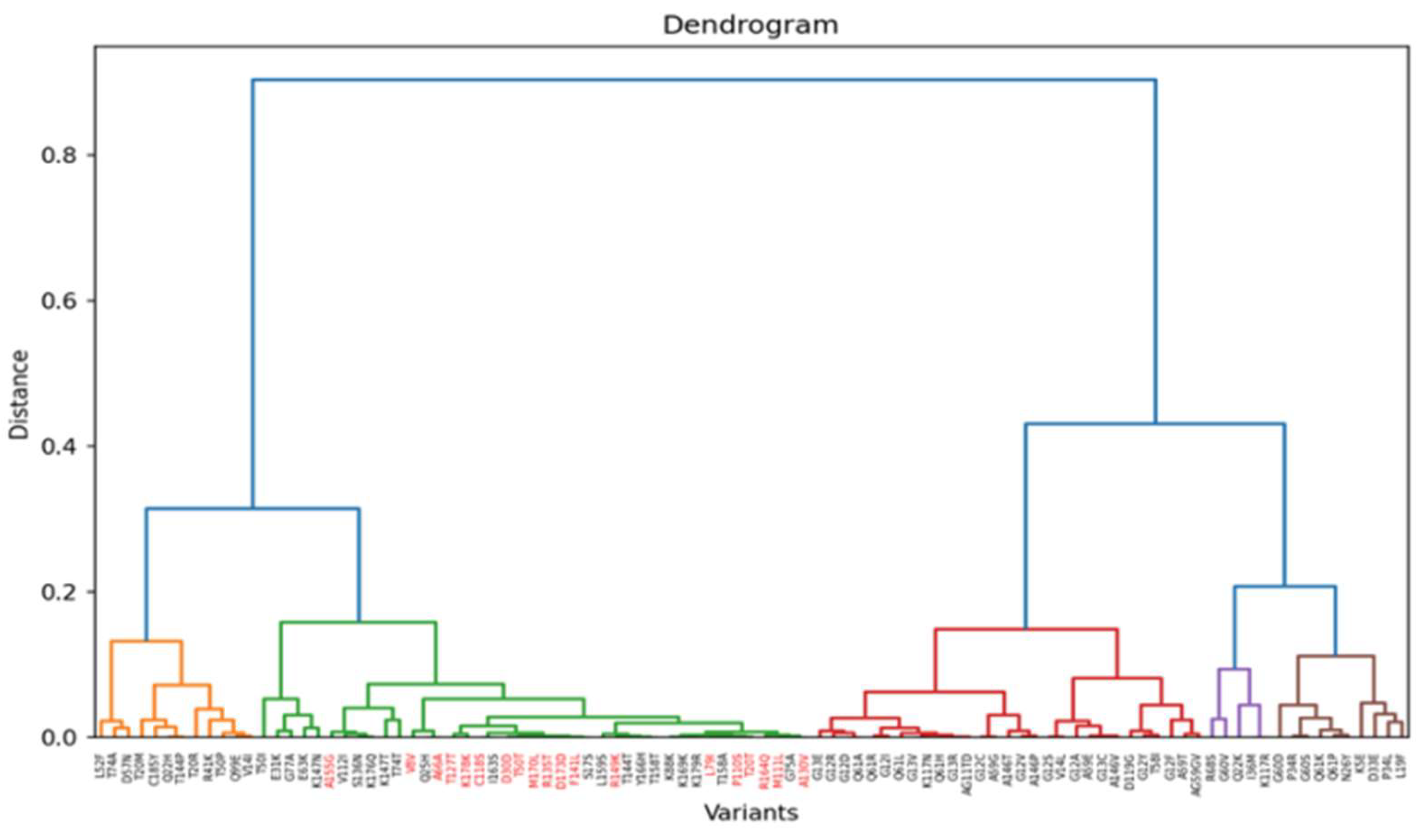
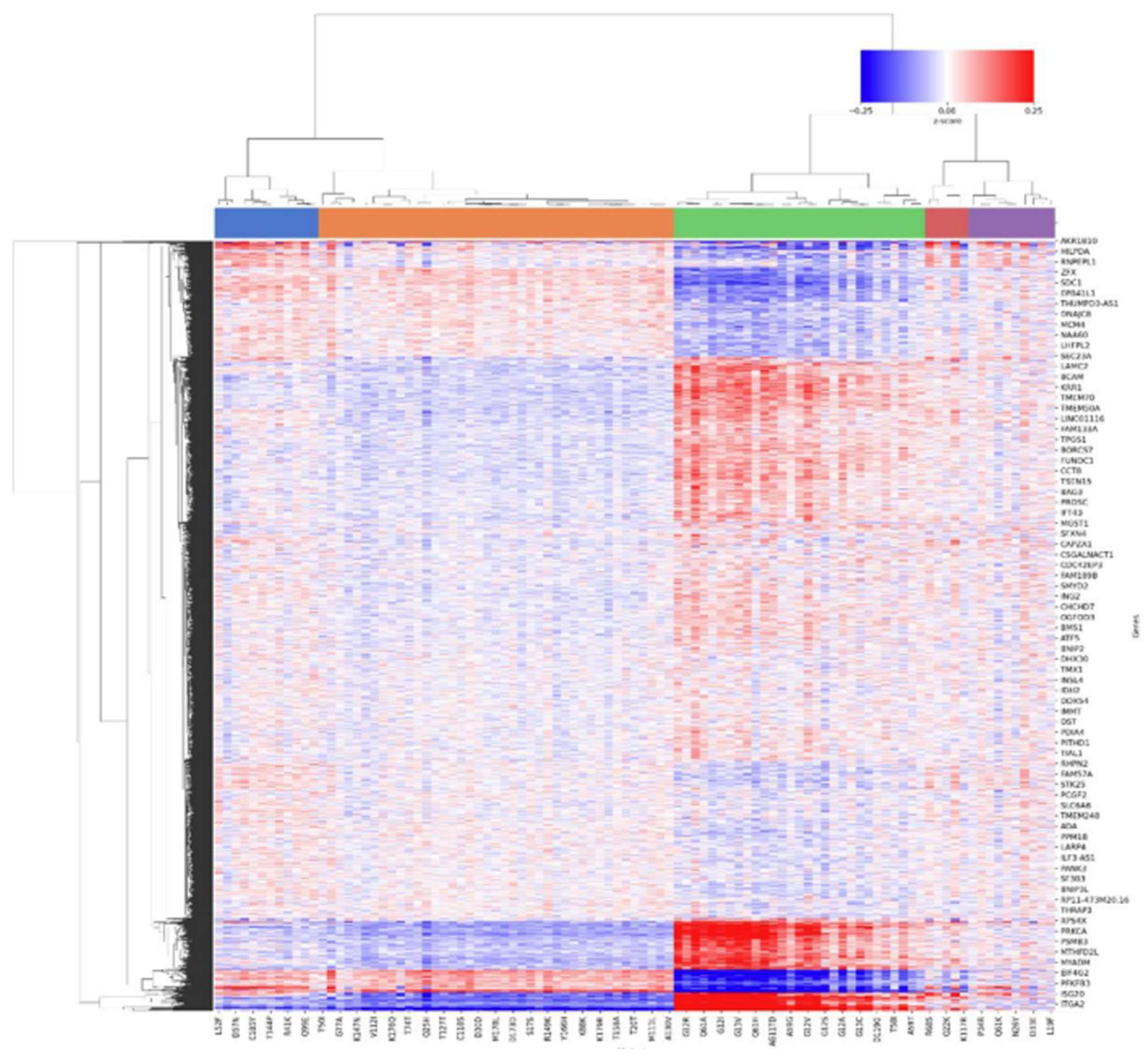
4. Conclusion
References
- AACR Project GENIE Consortium (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer discovery, 7(8), 818–831. [CrossRef]
- Adamson, B. , Norman, T. M., Jost, M., Cho, M. Y., Nuñez, J. K., Chen, Y., Villalta, J. E., Gilbert, L. A., Horlbeck, M. A., Hein, M. Y., Pak, R. A., Gray, A. N., Gross, C. A., Dixit, A., Parnas, O., Regev, A., & Weissman, J. S. (2016). A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell, 167(7), 1867–1882.e21. [CrossRef]
- Ahmed, R. , Baali, I., Erten, C., Hoxha, E., & Kazan, H. (2020). MEXCOwalk: mutual exclusion and coverage based random walk to identify cancer modules. Bioinformatics (Oxford, England), 36(3), 872–879. [CrossRef]
- Aibar, S. , González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H., Hulselmans, G., Rambow, F., Marine, J. C., Geurts, P., Aerts, J., van den Oord, J., Atak, Z. K., Wouters, J., & Aerts, S. (2017). SCENIC: single-cell regulatory network inference and clustering. Nature methods, 14(11), 1083–1086. [CrossRef]
- Ajiro, M. , Jia, R., Yang, Y., Zhu, J., & Zheng, Z. M. (2016). A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic acids research, 44(4), 1854–1870. [CrossRef]
- Babichev, S. , Lytvynenko, V., Korobchynskyi, M., & Sokur, I. (2019). Chapter 23 - Computational Epigenetics in Lung Cancer. In L. K. Wei (Ed.), Computational Epigenetics and Diseases (pp. 397–417). [CrossRef]
- Bader, G. D., & Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics, 4, 2. [CrossRef]
- Bailey, M. H. , Tokheim, C., Porta-Pardo, E., Sengupta, S., Bertrand, D., Weerasinghe, A., Colaprico, A., Wendl, M. C., Kim, J., Reardon, B., Ng, P. K., Jeong, K. J., Cao, S., Wang, Z., Gao, J., Gao, Q., Wang, F., Liu, E. M., Mularoni, L., Rubio-Perez, C., … Ding, L. (2018). Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell, 173(2), 371–385.e18. [CrossRef]
- Barel, G. , & Herwig, R. (2020). NetCore: a network propagation approach using node coreness. Nucleic acids research, 48(17), e98. [CrossRef]
- Barrett, T. , Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., Marshall, K. A., Phillippy, K. H., Sherman, P. M., Holko, M., Yefanov, A., Lee, H., Zhang, N., Robertson, C. L., Serova, N., Davis, S., & Soboleva, A. (2013). NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research, 41(Database issue), D991–D995. [CrossRef]
- Barry, T. , Mason, K., Roeder, K., & Katsevich, E. (2024). Robust differential expression testing for single-cell CRISPR screens at low multiplicity of infection. Genome biology, 25(1), 124. [CrossRef]
- Beisser, D. , Klau, G. W., Dandekar, T., Müller, T., & Dittrich, M. T. (2010). BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics (Oxford, England), 26(8), 1129–1130. [CrossRef]
- Belk, J. A. , Yao, W., Ly, N., Freitas, K. A., Chen, Y. T., Shi, Q., Valencia, A. M., Shifrut, E., Kale, N., Yost, K. E., Duffy, C. V., Daniel, B., Hwee, M. A., Miao, Z., Ashworth, A., Mackall, C. L., Marson, A., Carnevale, J., Vardhana, S. A., & Satpathy, A. T. (2022). Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer cell, 40(7), 768–786.e7. [CrossRef]
- Bell, J. L. , Wächter, K., Mühleck, B., Pazaitis, N., Köhn, M., Lederer, M., & Hüttelmaier, S. (2013). Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression?. Cellular and molecular life sciences : CMLS, 70(15), 2657–2675. [CrossRef]
- Berger, A. H. , Brooks, A. N., Wu, X., Shrestha, Y., Chouinard, C., Piccioni, F., Bagul, M., Kamburov, A., Imielinski, M., Hogstrom, L., Zhu, C., Yang, X., Pantel, S., Sakai, R., Watson, J., Kaplan, N., Campbell, J. D., Singh, S., Root, D. E., Narayan, R., … Boehm, J. S. (2016). High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer cell, 30(2), 214–228. [CrossRef]
- Beroukhim, R. , Mermel, C. H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J., Barretina, J., Boehm, J. S., Dobson, J., Urashima, M., Mc Henry, K. T., Pinchback, R. M., Ligon, A. H., Cho, Y. J., Haery, L., Greulich, H., Reich, M., Winckler, W., Lawrence, M. S., Weir, B. A., … Meyerson, M. (2010). The landscape of somatic copy-number alteration across human cancers. Nature, 463(7283), 899–905. [CrossRef]
- Bertoli, S. , Paubelle, E., Bérard, E., Saland, E., Thomas, X., Tavitian, S., Larcher, M. V., Vergez, F., Delabesse, E., Sarry, A., Huguet, F., Larrue, C., Bosc, C., Farge, T., Sarry, J. E., Michallet, M., & Récher, C. (2019). Ferritin heavy/light chain (FTH1/FTL) expression, serum ferritin levels, and their functional as well as prognostic roles in acute myeloid leukemia. European journal of haematology, 102(2), 131–142. [CrossRef]
- Blondel, V.D. , Guillaume, J., Lambiotte, R., & Lefebvre, E. (2008). Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment, 2008, P10008. [CrossRef]
- Bogdan, A. R. , Miyazawa, M., Hashimoto, K., & Tsuji, Y. (2016). Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends in biochemical sciences, 41(3), 274–286. [CrossRef]
- Borroto-Escuela, D. O. , Brito, I., Romero-Fernandez, W., Di Palma, M., Oflijan, J., Skieterska, K., Duchou, J., Van Craenenbroeck, K., Suárez-Boomgaard, D., Rivera, A., Guidolin, D., Agnati, L. F., & Fuxe, K. (2014). The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components. International journal of molecular sciences, 15(5), 8570–8590. [CrossRef]
- Butterworth, S. , Kordova, K., Chandrasekaran, S., Thomas, K. K., Torelli, F., Lockyer, E. J., Edwards, A., Goldstone, R., Koshy, A. A., & Treeck, M. (2023). High-throughput identification of Toxoplasma gondii effector proteins that target host cell transcription. Cell host & microbe, 31(10), 1748–1762.e8. [CrossRef]
- Canale, M. , Andrikou, K., Priano, I., Cravero, P., Pasini, L., Urbini, M., Delmonte, A., Crinò, L., Bronte, G., & Ulivi, P. (2022). The Role of TP53 Mutations in EGFR-Mutated Non-Small-Cell Lung Cancer: Clinical Significance and Implications for Therapy. Cancers, 14(5), 1143. [CrossRef]
- Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455(7216), 1061–1068. [CrossRef]
- Chen, P. H., Wu, J., Ding, C. C., Lin, C. C., Pan, S., Bossa, N., Xu, Y., Yang, W. H., Mathey-Prevot, B., & Chi, J. T. (2020). Kinome screen of ferroptosis reveals a novel role of ATM in regulating iron metabolism. Cell death and differentiation, 27(3), 1008–1022. [CrossRef]
- Chitra, U. , Park, T. Y., & Raphael, B. J. (2022). NetMix2: A Principled Network Propagation Algorithm for Identifying Altered Subnetworks. Journal of computational biology : a journal of computational molecular cell biology, 29(12), 1305–1323. [CrossRef]
- Cho, A. , Shim, J. E., Kim, E., Supek, F., Lehner, B., & Lee, I. (2016). MUFFINN: cancer gene discovery via network analysis of somatic mutation data. Genome biology, 17(1), 129. [CrossRef]
- Cho, H. , Berger, B., & Peng, J. (2015). Diffusion Component Analysis: Unraveling Functional Topology in Biological Networks. Research in computational molecular biology :... Annual International Conference, RECOMB... : proceedings. RECOMB (Conference : 2005- ), 9029, 62–64. [CrossRef]
- Chung, F. , Zhao, W. (2010). PageRank and Random Walks on Graphs. In: Katona, G.O.H., Schrijver, A., Szőnyi, T., Sági, G. (eds) Fete of Combinatorics and Computer Science. Bolyai Society Mathematical Studies, vol 20. Springer, Berlin, Heidelberg. [CrossRef]
- Ciriello, G., Cerami, E., Sander, C., & Schultz, N. (2012). Mutual exclusivity analysis identifies oncogenic network modules. Genome research, 22(2), 398–406. [CrossRef]
- Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., Christmas, R., Avila-Campilo, I., Creech, M., Gross, B., Hanspers, K., Isserlin, R., Kelley, R., Killcoyne, S., Lotia, S., Maere, S., Morris, J., Ono, K., Pavlovic, V., Pico, A. R., … Bader, G. D. (2007). Integration of biological networks and gene expression data using Cytoscape. Nature protocols, 2(10), 2366–2382. [CrossRef]
- Cowen, L. , Ideker, T., Raphael, B. J., & Sharan, R. (2017). Network propagation: a universal amplifier of genetic associations. Nature reviews. Genetics, 18(9), 551–562. [CrossRef]
- Dao, P. , Kim, Y. A., Wojtowicz, D., Madan, S., Sharan, R., & Przytycka, T. M. (2017). BeWith: A Between-Within method to discover relationships between cancer modules via integrated analysis of mutual exclusivity, co-occurrence and functional interactions. PLoS computational biology, 13(10), e1005695. [CrossRef]
- Datlinger, P. , Rendeiro, A. F., Boenke, T., Senekowitsch, M., Krausgruber, T., Barreca, D., & Bock, C. (2021). Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing. Nature methods, 18(6), 635–642. [CrossRef]
- Datlinger, P. , Rendeiro, A. F., Schmidl, C., Krausgruber, T., Traxler, P., Klughammer, J., Schuster, L. C., Kuchler, A., Alpar, D., & Bock, C. (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nature methods, 14(3), 297–301. [CrossRef]
- Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C. H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., Otim, O., Brown, C. T., Livi, C. B., Lee, P. Y., Revilla, R., Rust, A. G., Pan, Z.j, Schilstra, M. J., Clarke, P. J., Arnone, M. I., … Bolouri, H. (2002). A genomic regulatory network for development. Science (New York, N.Y.), 295(5560), 1669–1678. [CrossRef]
- Delgobo, M. , Gonçalves, R. M., Delazeri, M. A., Falchetti, M., Zandoná, A., Nascimento das Neves, R., Almeida, K., Fagundes, A. C., Gelain, D. P., Fracasso, J. I., Macêdo, G. B., Priori, L., Bassani, N., Bishop, A. J. R., Forcelini, C. M., Moreira, J. C. F., & Zanotto-Filho, A. (2021). Thioredoxin reductase-1 levels are associated with NRF2 pathway activation and tumor recurrence in non-small cell lung cancer. Free radical biology & medicine, 177, 58–71. [CrossRef]
- Demirel, H. C. , Arici, M. K., & Tuncbag, N. (2022). Computational approaches leveraging integrated connections of multi-omic data toward clinical applications. Molecular omics, 18(1), 7–18. [CrossRef]
- DeNicola, G. M. , Karreth, F. A., Humpton, T. J., Gopinathan, A., Wei, C., Frese, K., Mangal, D., Yu, K. H., Yeo, C. J., Calhoun, E. S., Scrimieri, F., Winter, J. M., Hruban, R. H., Iacobuzio-Donahue, C., Kern, S. E., Blair, I. A., & Tuveson, D. A. (2011). Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature, 475(7354), 106–109. [CrossRef]
- De Vusser, K., Winckelmans, E., Martens, D., Lerut, E., Kuypers, D., Nawrot, T., & Naesens, M. (2020). Intrarenal arteriosclerosis and telomere attrition associate with dysregulation of the cholesterol pathway. Aging, 12(9), 7830–7847. [CrossRef]
- Dey, P. , Li, J., Zhang, J., Chaurasiya, S., Strom, A., Wang, H., Liao, W. T., Cavallaro, F., Denz, P., Bernard, V., Yen, E. Y., Genovese, G., Gulhati, P., Liu, J., Chakravarti, D., Deng, P., Zhang, T., Carbone, F., Chang, Q., Ying, H., … DePinho, R. A. (2020). Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer discovery, 10(4), 608–625. [CrossRef]
- Ding, C. , Fan, X., & Wu, G. (2017). Peroxiredoxin 1 - an antioxidant enzyme in cancer. Journal of cellular and molecular medicine, 21(1), 193–202. [CrossRef]
- Ding, L., Getz, G., Wheeler, D. A., Mardis, E. R., McLellan, M. D., Cibulskis, K., Sougnez, C., Greulich, H., Muzny, D. M., Morgan, M. B., Fulton, L., Fulton, R. S., Zhang, Q., Wendl, M. C., Lawrence, M. S., Larson, D. E., Chen, K., Dooling, D. J., Sabo, A., Hawes, A. C., … Wilson, R. K. (2008). Somatic mutations affect key pathways in lung adenocarcinoma. Nature, 455(7216), 1069–1075. [CrossRef]
- Dittrich, M. T. , Klau, G. W., Rosenwald, A., Dandekar, T., & Müller, T. (2008). Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics (Oxford, England), 24(13), i223–i231. [CrossRef]
- Dixit, A. , Parnas, O., Li, B., Chen, J., Fulco, C. P., Jerby-Arnon, L., Marjanovic, N. D., Dionne, D., Burks, T., Raychowdhury, R., Adamson, B., Norman, T. M., Lander, E. S., Weissman, J. S., Friedman, N., & Regev, A. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell, 167(7), 1853–1866.e17. [CrossRef]
- Doncheva, N. T. , Morris, J. H., Holze, H., Kirsch, R., Nastou, K. C., Cuesta-Astroz, Y., Rattei, T., Szklarczyk, D., von Mering, C., & Jensen, L. J. (2023). Cytoscape stringApp 2.0: Analysis and Visualization of Heterogeneous Biological Networks. Journal of proteome research, 22(2), 637–646. [CrossRef]
- Escala-Garcia, M. , Abraham, J., Andrulis, I. L., Anton-Culver, H., Arndt, V., Ashworth, A., Auer, P. L., Auvinen, P., Beckmann, M. W., Beesley, J., Behrens, S., Benitez, J., Bermisheva, M., Blomqvist, C., Blot, W., Bogdanova, N. V., Bojesen, S. E., Bolla, M. K., Børresen-Dale, A. L., Brauch, H., … Schmidt, M. K. (2020). A network analysis to identify mediators of germline-driven differences in breast cancer prognosis. Nature communications, 11(1), 312. [CrossRef]
- Galindez, G. , Sadegh, S., Baumbach, J., Kacprowski, T., & List, M. (2022). Network-based approaches for modeling disease regulation and progression. Computational and structural biotechnology journal, 21, 780–795. [CrossRef]
- Gligorijević, N. , Dobrijević, Z., Šunderić, M., Robajac, D., Četić, D., Penezić, A., Miljuš, G., & Nedić, O. (2022). The Insulin-like Growth Factor System and Colorectal Cancer. Life (Basel, Switzerland), 12(8), 1274. [CrossRef]
- Gortzak-Uzan, L. , Ignatchenko, A., Evangelou, A. I., Agochiya, M., Brown, K. A., St Onge, P., Kireeva, I., Schmitt-Ulms, G., Brown, T. J., Murphy, J., Rosen, B., Shaw, P., Jurisica, I., & Kislinger, T. (2008). A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. Journal of proteome research, 7(1), 339–351. [CrossRef]
- Griffiths, J. I. , Chen, J., Cosgrove, P. A., O'Dea, A., Sharma, P., Ma, C., Trivedi, M., Kalinsky, K., Wisinski, K. B., O'Regan, R., Makhoul, I., Spring, L. M., Bardia, A., Adler, F. R., Cohen, A. L., Chang, J. T., Khan, Q. J., & Bild, A. H. (2021). Serial single-cell genomics reveals convergent subclonal evolution of resistance as early-stage breast cancer patients progress on endocrine plus CDK4/6 therapy. Nature cancer, 2(6), 658–671. [CrossRef]
- Hagberg, A.A., Schult, D.A. and Swart, P.J. (2008) Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux, G., Vaught, T. and Millman, J., Eds., Proceedings of 7th Python in Science Conference (SciPy2008), 11-15. Available in https://www.osti.gov/biblio/960616.
- Hecker, D. , Lauber, M., Behjati Ardakani, F., Ashrafiyan, S., Manz, Q., Kersting, J., Hoffmann, M., Schulz, M. H., & List, M. (2023). Computational tools for inferring transcription factor activity. Proteomics, 23(23-24), e2200462. [CrossRef]
- Hein, M. Y. , & Weissman, J. S. (2022). Functional single-cell genomics of human cytomegalovirus infection. Nature biotechnology, 40(3), 391–401. [CrossRef]
- Holland, C. H. , Tanevski, J., Perales-Patón, J., Gleixner, J., Kumar, M. P., Mereu, E., Joughin, B. A., Stegle, O., Lauffenburger, D. A., Heyn, H., Szalai, B., & Saez-Rodriguez, J. (2020). Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome biology, 21(1), 36. [CrossRef]
- Horn, H. , Lawrence, M. S., Chouinard, C. R., Shrestha, Y., Hu, J. X., Worstell, E., Shea, E., Ilic, N., Kim, E., Kamburov, A., Kashani, A., Hahn, W. C., Campbell, J. D., Boehm, J. S., Getz, G., & Lage, K. (2018). NetSig: network-based discovery from cancer genomes. Nature methods, 15(1), 61–66. [CrossRef]
- Hotelling, H. (1992). The Generalization of Student’s Ratio. In: Kotz, S., Johnson, N.L. (eds) Breakthroughs in Statistics. Springer Series in Statistics. Springer, New York, NY. [CrossRef]
- Huang, X. , Zhang, H., Guo, X., Zhu, Z., Cai, H., & Kong, X. (2018). Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. Journal of hematology & oncology, 11(1), 88. [CrossRef]
- Ideker, T., Ozier, O., Schwikowski, B., & Siegel, A. F. (2002). Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics (Oxford, England), 18 Suppl 1, S233–S240. [CrossRef]
- Jacob, L. , Neuvial, P., & Dudoit, S. (2012). More power via graph-structured tests for differential expression of gene networks. Annals of Applied Statistics, 6(2), 561–600. [CrossRef]
- Jaitin, D. A. , Weiner, A., Yofe, I., Lara-Astiaso, D., Keren-Shaul, H., David, E., Salame, T. M., Tanay, A., van Oudenaarden, A., & Amit, I. (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell, 167(7), 1883–1896.e15. [CrossRef]
- Jeay, S. , Gaulis, S., Ferretti, S., Bitter, H., Ito, M., Valat, T., Murakami, M., Ruetz, S., Guthy, D. A., Rynn, C., Jensen, M. R., Wiesmann, M., Kallen, J., Furet, P., Gessier, F., Holzer, P., Masuya, K., Würthner, J., Halilovic, E., Hofmann, F., … Graus Porta, D. (2015). A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097. eLife, 4, e06498. [CrossRef]
- Jensen, T. I., Mikkelsen, N. S., Gao, Z., Foßelteder, J., Pabst, G., Axelgaard, E., Laustsen, A., König, S., Reinisch, A., & Bak, R. O. (2021). Targeted regulation of transcription in primary cells using CRISPRa and CRISPRi. Genome research, 31(11), 2120–2130. [CrossRef]
- Jin, X. , Simmons, S. K., Guo, A., Shetty, A. S., Ko, M., Nguyen, L., Jokhi, V., Robinson, E., Oyler, P., Curry, N., Deangeli, G., Lodato, S., Levin, J. Z., Regev, A., Zhang, F., & Arlotta, P. (2020). In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science (New York, N.Y.), 370(6520), eaaz6063. [CrossRef]
- Jones, S., Zhang, X., Parsons, D. W., Lin, J. C., Leary, R. J., Angenendt, P., Mankoo, P., Carter, H., Kamiyama, H., Jimeno, A., Hong, S. M., Fu, B., Lin, M. T., Calhoun, E. S., Kamiyama, M., Walter, K., Nikolskaya, T., Nikolsky, Y., Hartigan, J., Smith, D. R., … Kinzler, K. W. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, N.Y.), 321(5897), 1801–1806. [CrossRef]
- Kanehisa, M. , Furumichi, M., Sato, Y., Kawashima, M., & Ishiguro-Watanabe, M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic acids research, 51(D1), D587–D592. [CrossRef]
- Keat Wei, L. , & Au, A. (2017). Chapter 12 - Computational Epigenetics. In T. O. Tollefsbol (Ed.), Handbook of Epigenetics (Second Edition) (Second Edition, pp. 167–190). [CrossRef]
- Kim, E. M. , Park, J. K., Hwang, S. G., Kim, W. J., Liu, Z. G., Kang, S. W., & Um, H. D. (2014). Nuclear and cytoplasmic p53 suppress cell invasion by inhibiting respiratory complex-I activity via Bcl-2 family proteins. Oncotarget, 5(18), 8452–8465. [CrossRef]
- Kim, P. M. , & Tidor, B. (2003). Limitations of quantitative gene regulation models: a case study. Genome research, 13(11), 2391–2395. [CrossRef]
- Kolberg, L. , Raudvere, U., Kuzmin, I., Adler, P., Vilo, J., & Peterson, H. (2023). g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic acids research, 51(W1), W207–W212. [CrossRef]
- Lebrigand, K., Magnone, V., Barbry, P., & Waldmann, R. (2020). High throughput error corrected Nanopore single cell transcriptome sequencing. Nature communications, 11(1), 4025. [CrossRef]
- Leiserson, M. D. , Vandin, F., Wu, H. T., Dobson, J. R., Eldridge, J. V., Thomas, J. L., Papoutsaki, A., Kim, Y., Niu, B., McLellan, M., Lawrence, M. S., Gonzalez-Perez, A., Tamborero, D., Cheng, Y., Ryslik, G. A., Lopez-Bigas, N., Getz, G., Ding, L., & Raphael, B. J. (2015). Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature genetics, 47(2), 106–114. [CrossRef]
- Levi, H. , Elkon, R., & Shamir, R. (2021). DOMINO: a network-based active module identification algorithm with reduced rate of false calls. Molecular systems biology, 17(1), e9593. [CrossRef]
- Littman, R. , Cheng, M., Wang, N., Peng, C., & Yang, X. (2023). SCING: Inference of robust, interpretable gene regulatory networks from single cell and spatial transcriptomics. iScience, 26(7), 107124. [CrossRef]
- Liu, C. , Ma, Y., Zhao, J., Nussinov, R., Zhang, Y.-C., Cheng, F., & Zhang, Z.-K. (2020). Computational network biology: Data, models, and applications. Physics Reports, 846, 1–66. [CrossRef]
- Lotfollahi, M. , Klimovskaia Susmelj, A., De Donno, C., Hetzel, L., Ji, Y., Ibarra, I. L., Srivatsan, S. R., Naghipourfar, M., Daza, R. M., Martin, B., Shendure, J., McFaline-Figueroa, J. L., Boyeau, P., Wolf, F. A., Yakubova, N., Günnemann, S., Trapnell, C., Lopez-Paz, D., & Theis, F. J. (2023). Predicting cellular responses to complex perturbations in high-throughput screens. Molecular systems biology, 19(6), e11517. [CrossRef]
- Love, M. I. , Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 15(12), 550. [CrossRef]
- Lu, B., Chen, X. B., Ying, M. D., He, Q. J., Cao, J., & Yang, B. (2018). The Role of Ferroptosis in Cancer Development and Treatment Response. Frontiers in pharmacology, 8, 992. [CrossRef]
- Luck, K. , Kim, D. K., Lambourne, L., Spirohn, K., Begg, B. E., Bian, W., Brignall, R., Cafarelli, T., Campos-Laborie, F. J., Charloteaux, B., Choi, D., Coté, A. G., Daley, M., Deimling, S., Desbuleux, A., Dricot, A., Gebbia, M., Hardy, M. F., Kishore, N., Knapp, J. J., … Calderwood, M. A. (2020). A reference map of the human binary protein interactome. Nature, 580(7803), 402–408. [CrossRef]
- Margolin, A. A. , Nemenman, I., Basso, K., Wiggins, C., Stolovitzky, G., Dalla Favera, R., & Califano, A. (2006). ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC bioinformatics, 7 Suppl 1(Suppl 1), S7. [CrossRef]
- Martínez-Jiménez, F. , Muiños, F., Sentís, I., Deu-Pons, J., Reyes-Salazar, I., Arnedo-Pac, C., Mularoni, L., Pich, O., Bonet, J., Kranas, H., Gonzalez-Perez, A., & Lopez-Bigas, N. (2020). A compendium of mutational cancer driver genes. Nature reviews. Cancer, 20(10), 555–572. [CrossRef]
- McInnes, L, Healy, J. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction, ArXiv e-prints 1802.03426,. [CrossRef]
- Meyers, S. , Demeyer, S., & Cools, J. (2023). CRISPR screening in hematology research: from bulk to single-cell level. Journal of hematology & oncology, 16(1), 107. [CrossRef]
- Miyake, N. , Chikumi, H., Takata, M., Nakamoto, M., Igishi, T., & Shimizu, E. (2012). Rapamycin induces p53-independent apoptosis through the mitochondrial pathway in non-small cell lung cancer cells. Oncology reports, 28(3), 848–854. [CrossRef]
- Muzellec, B. , Teleńczuk, M., Cabeli, V., & Andreux, M. (2023). PyDESeq2: a python package for bulk RNA-seq differential expression analysis. Bioinformatics (Oxford, England), 39(9), btad547. [CrossRef]
- Otto, J. E. , Ursu, O., Wu, A. P., Winter, E. B., Cuoco, M. S., Ma, S., Qian, K., Michel, B. C., Buenrostro, J. D., Berger, B., Regev, A., & Kadoch, C. (2023). Structural and functional properties of mSWI/SNF chromatin remodeling complexes revealed through single-cell perturbation screens. Molecular cell, 83(8), 1350–1367.e7. [CrossRef]
- Oughtred, R. , Stark, C., Breitkreutz, B. J., Rust, J., Boucher, L., Chang, C., Kolas, N., O'Donnell, L., Leung, G., McAdam, R., Zhang, F., Dolma, S., Willems, A., Coulombe-Huntington, J., Chatr-Aryamontri, A., Dolinski, K., & Tyers, M. (2019). The BioGRID interaction database: 2019 update. Nucleic acids research, 47(D1), D529–D541. [CrossRef]
- Ozturk, K., & Carter, H. (2022). Predicting functional consequences of mutations using molecular interaction network features. Human genetics, 141(6), 1195–1210. [CrossRef]
- Ozturk, K. , Panwala, R., Sheen, J., Ford, K., Payne, N., Zhang, D. E., Hutter, S., Haferlach, T., Ideker, T., Mali, P., & Carter, H. (2023). Interface-guided phenotyping of coding variants in the transcription factor RUNX1 with SEUSS. bioRxiv : the preprint server for biology, 2023.08.03.551876. [CrossRef]
- Pacalin, N. M. , Steinhart, Z., Shi, Q., Belk, J. A., Dorovskyi, D., Kraft, K., Parker, K. R., Shy, B. R., Marson, A., & Chang, H. Y. (2024). Bidirectional epigenetic editing reveals hierarchies in gene regulation. Nature biotechnology, 10.1038/s41587-024-02213-3. Advance online publication. [CrossRef]
- Papalexi, E. , Mimitou, E. P., Butler, A. W., Foster, S., Bracken, B., Mauck, W. M., 3rd, Wessels, H. H., Hao, Y., Yeung, B. Z., Smibert, P., & Satija, R. (2021). Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nature genetics, 53(3), 322–331. [CrossRef]
- Parsons, D. W., Jones, S., Zhang, X., Lin, J. C., Leary, R. J., Angenendt, P., Mankoo, P., Carter, H., Siu, I. M., Gallia, G. L., Olivi, A., McLendon, R., Rasheed, B. A., Keir, S., Nikolskaya, T., Nikolsky, Y., Busam, D. A., Tekleab, H., Diaz, L. A., Jr, Hartigan, J., … Kinzler, K. W. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science (New York, N.Y.), 321(5897), 1807–1812. [CrossRef]
- Pataky T., C. (2012). One-dimensional statistical parametric mapping in Python. Computer methods in biomechanics and biomedical engineering, 15(3), 295–301. [CrossRef]
- Picart-Armada, S., Barrett, S. J., Willé, D. R., Perera-Lluna, A., Gutteridge, A., & Dessailly, B. H. (2019). Benchmarking network propagation methods for disease gene identification. PLoS computational biology, 15(9), e1007276. [CrossRef]
- Pratapa, A. , Jalihal, A. P., Law, J. N., Bharadwaj, A., & Murali, T. M. (2020). Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nature methods, 17(2), 147–154. [CrossRef]
- Rath, S. , Sharma, R., Gupta, R., Ast, T., Chan, C., Durham, T. J., Goodman, R. P., Grabarek, Z., Haas, M. E., Hung, W. H. W., Joshi, P. R., Jourdain, A. A., Kim, S. H., Kotrys, A. V., Lam, S. S., McCoy, J. G., Meisel, J. D., Miranda, M., Panda, A., Patgiri, A., … Mootha, V. K. (2021). MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic acids research, 49(D1), D1541–D1547. [CrossRef]
- Razick, S. , Magklaras, G., & Donaldson, I. M. (2008). iRefIndex: a consolidated protein interaction database with provenance. BMC bioinformatics, 9, 405. [CrossRef]
- Replogle, J. M. , Saunders, R. A., Pogson, A. N., Hussmann, J. A., Lenail, A., Guna, A., Mascibroda, L., Wagner, E. J., Adelman, K., Lithwick-Yanai, G., Iremadze, N., Oberstrass, F., Lipson, D., Bonnar, J. L., Jost, M., Norman, T. M., & Weissman, J. S. (2022). Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell, 185(14), 2559–2575.e28. [CrossRef]
- Reyna, M. A. , Leiserson, M. D. M., & Raphael, B. J. (2018). Hierarchical HotNet: identifying hierarchies of altered subnetworks. Bioinformatics (Oxford, England), 34(17), i972–i980. [CrossRef]
- Ricoult, S. J. , Yecies, J. L., Ben-Sahra, I., & Manning, B. D. (2016). Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene, 35(10), 1250–1260. [CrossRef]
- Robinson, M. D. , McCarthy, D. J., & Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England), 26(1), 139–140. [CrossRef]
- Safari-Alighiarloo, N., Taghizadeh, M., Rezaei-Tavirani, M., Goliaei, B., & Peyvandi, A. A. (2014). Protein-protein interaction networks (PPi) and complex diseases. Gastroenterology and hepatology from bed to bench, 7(1), 17–31. Available in https://pubmed.ncbi.nlm.nih.gov/25436094/.
- Saint-Antoine, M. , & Singh, A. (2023). Benchmarking Gene Regulatory Network Inference Methods on Simulated and Experimental Data. bioRxiv : the preprint server for biology, 2023.05.12.540581. [CrossRef]
- Saki, K., Mansouri, V., Abdi, S., Fathi, M., Razzaghi, Z., & Haghazali, M. (2021). Assessment of common and differentially expressed proteins between diabetes mellitus and fatty liver disease: a network analysis. Gastroenterology and hepatology from bed to bench, 14(Suppl1), S94–S101. Available in https://pubmed.ncbi.nlm.nih.gov/35154608/.
- Schnitzler, G. R. , Kang, H., Fang, S., Angom, R. S., Lee-Kim, V. S., Ma, X. R., Zhou, R., Zeng, T., Guo, K., Taylor, M. S., Vellarikkal, S. K., Barry, A. E., Sias-Garcia, O., Bloemendal, A., Munson, G., Guckelberger, P., Nguyen, T. H., Bergman, D. T., Hinshaw, S., Cheng, N., … Engreitz, J. M. (2024). Convergence of coronary artery disease genes onto endothelial cell programs. Nature, 626(8000), 799–807. [CrossRef]
- Schraivogel, D. , Gschwind, A. R., Milbank, J. H., Leonce, D. R., Jakob, P., Mathur, L., Korbel, J. O., Merten, C. A., Velten, L., & Steinmetz, L. M. (2020). Targeted Perturb-seq enables genome-scale genetic screens in single cells. Nature methods, 17(6), 629–635. [CrossRef]
- Sedgwick, P. (2014). Spearman's rank correlation coefficient. BMJ (Clinical research ed.), 349, g7327. [CrossRef]
- Shahamatdar, S. , He, M. X., Reyna, M. A., Gusev, A., AlDubayan, S. H., Van Allen, E. M., & Ramachandran, S. (2020). Germline Features Associated with Immune Infiltration in Solid Tumors. Cell reports, 30(9), 2900–2908.e4. [CrossRef]
- Shannon, P. , Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research, 13(11), 2498–2504. [CrossRef]
- Shrestha, R. , Hodzic, E., Sauerwald, T., Dao, P., Wang, K., Yeung, J., Anderson, S., Vandin, F., Haffari, G., Collins, C. C., & Sahinalp, S. C. (2017). HIT'nDRIVE: patient-specific multidriver gene prioritization for precision oncology. Genome research, 27(9), 1573–1588. [CrossRef]
- Sillars-Hardebol, A. H. , Carvalho, B., Beliën, J. A., de Wit, M., Delis-van Diemen, P. M., Tijssen, M., van de Wiel, M. A., Pontén, F., Fijneman, R. J., & Meijer, G. A. (2012). BCL2L1 has a functional role in colorectal cancer and its protein expression is associated with chromosome 20q gain. The Journal of pathology, 226(3), 442–450. [CrossRef]
- Soranzo, N. , Bianconi, G., & Altafini, C. (2007). Comparing association network algorithms for reverse engineering of large-scale gene regulatory networks: synthetic versus real data. Bioinformatics (Oxford, England), 23(13), 1640–1647. [CrossRef]
- Spisak, S. , Chen, D., Likasitwatanakul, P., Doan, P., Li, Z., Bala, P., Vizkeleti, L., Tisza, V., De Silva, P., Giannakis, M., Wolpin, B., Qi, J., & Sethi, N. S. (2024). Identifying regulators of aberrant stem cell and differentiation activity in colorectal cancer using a dual endogenous reporter system. Nature communications, 15(1), 2230. [CrossRef]
- Sunshine, S. , Puschnik, A. S., Replogle, J. M., Laurie, M. T., Liu, J., Zha, B. S., Nuñez, J. K., Byrum, J. R., McMorrow, A. H., Frieman, M. B., Winkler, J., Qiu, X., Rosenberg, O. S., Leonetti, M. D., Ye, C. J., Weissman, J. S., DeRisi, J. L., & Hein, M. Y. (2023). Systematic functional interrogation of SARS-CoV-2 host factors using Perturb-seq. Nature communications, 14(1), 6245. [CrossRef]
- Swaney, D. L. , Ramms, D. J., Wang, Z., Park, J., Goto, Y., Soucheray, M., Bhola, N., Kim, K., Zheng, F., Zeng, Y., McGregor, M., Herrington, K. A., O'Keefe, R., Jin, N., VanLandingham, N. K., Foussard, H., Von Dollen, J., Bouhaddou, M., Jimenez-Morales, D., Obernier, K., … Krogan, N. J. (2021). A protein network map of head and neck cancer reveals PIK3CA mutant drug sensitivity. Science (New York, N.Y.), 374(6563), eabf2911. [CrossRef]
- Szklarczyk, D. , Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N. T., Morris, J. H., Bork, P., Jensen, L. J., & Mering, C. V. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research, 47(D1), D607–D613. [CrossRef]
- Tian, R. , Gachechiladze, M. A., Ludwig, C. H., Laurie, M. T., Hong, J. Y., Nathaniel, D., Prabhu, A. V., Fernandopulle, M. S., Patel, R., Abshari, M., Ward, M. E., & Kampmann, M. (2019). CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron, 104(2), 239–255.e12. [CrossRef]
- Traag, V. A. , Waltman, L., & van Eck, N. J. (2019). From Louvain to Leiden: guaranteeing well-connected communities. Scientific reports, 9(1), 5233. [CrossRef]
- Ursu, O. , Neal, J. T., Shea, E., Thakore, P. I., Jerby-Arnon, L., Nguyen, L., Dionne, D., Diaz, C., Bauman, J., Mosaad, M. M., Fagre, C., Lo, A., McSharry, M., Giacomelli, A. O., Ly, S. H., Rozenblatt-Rosen, O., Hahn, W. C., Aguirre, A. J., Berger, A. H., Regev, A., … Boehm, J. S. (2022). Massively parallel phenotyping of coding variants in cancer with Perturb-seq. Nature biotechnology, 40(6), 896–905. [CrossRef]
- Vandin, F., Clay, P., Upfal, E., & Raphael, B. J. (2012). Discovery of mutated subnetworks associated with clinical data in cancer. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 55–66. [CrossRef]
- Vanunu, O., Magger, O., Ruppin, E., Shlomi, T., & Sharan, R. (2010). Associating genes and protein complexes with disease via network propagation. PLoS computational biology, 6(1), e1000641. [CrossRef]
- Vaske, C. J. , Benz, S. C., Sanborn, J. Z., Earl, D., Szeto, C., Zhu, J., Haussler, D., & Stuart, J. M. (2010). Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics (Oxford, England), 26(12), i237–i245. [CrossRef]
- Vastrik, I. , D'Eustachio, P., Schmidt, E., Gopinath, G., Croft, D., de Bono, B., Gillespie, M., Jassal, B., Lewis, S., Matthews, L., Wu, G., Birney, E., & Stein, L. (2007). Reactome: a knowledge base of biologic pathways and processes. Genome biology, 8(3), R39. [CrossRef]
- Vogelstein, B., & Kinzler, K. W. (2004). Cancer genes and the pathways they control. Nature medicine, 10(8), 789–799. [CrossRef]
- Walhout A., J. (2006). Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome research, 16(12), 1445–1454. [CrossRef]
- Wessels, H. H. , Méndez-Mancilla, A., Hao, Y., Papalexi, E., Mauck, W. M., 3rd, Lu, L., Morris, J. A., Mimitou, E. P., Smibert, P., Sanjana, N. E., & Satija, R. (2023). Efficient combinatorial targeting of RNA transcripts in single cells with Cas13 RNA Perturb-seq. Nature methods, 20(1), 86–94. [CrossRef]
- Wolf, F. A. , Angerer, P., & Theis, F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome biology, 19(1), 15. [CrossRef]
- Wood, L. D., Parsons, D. W., Jones, S., Lin, J., Sjöblom, T., Leary, R. J., Shen, D., Boca, S. M., Barber, T., Ptak, J., Silliman, N., Szabo, S., Dezso, Z., Ustyanksky, V., Nikolskaya, T., Nikolsky, Y., Karchin, R., Wilson, P. A., Kaminker, J. S., Zhang, Z., … Vogelstein, B. (2007). The genomic landscapes of human breast and colorectal cancers. Science (New York, N.Y.), 318(5853), 1108–1113. [CrossRef]
- Wu, D. , Poddar, A., Ninou, E., Hwang, E., Cole, M. A., Liu, S. J., Horlbeck, M. A., Chen, J., Replogle, J. M., Carosso, G. A., Eng, N. W. L., Chang, J., Shen, Y., Weissman, J. S., & Lim, D. A. (2022). Dual genome-wide coding and lncRNA screens in neural induction of induced pluripotent stem cells. Cell genomics, 2(11), 100177. [CrossRef]
- Wu, G., Feng, X., & Stein, L. (2010). A human functional protein interaction network and its application to cancer data analysis. Genome biology, 11(5), R53. [CrossRef]
- Yachie-Kinoshita, A. , & Kaizu, K. (2019). Cell Modeling and Simulation. In S. Ranganathan, M. Gribskov, K. Nakai, & C. Schönbach (Eds.), Encyclopedia of Bioinformatics and Computational Biology (pp. 864–873). [CrossRef]
- Yang, L., Chen, R., Goodison, S., & Sun, Y. (2021). An efficient and effective method to identify significantly perturbed subnetworks in cancer. Nature computational science, 1(1), 79–88. [CrossRef]
- Yang, L. , Chen, R., Melendy, T., Goodison, S., & Sun, Y. (2023). Identifying Significantly Perturbed Subnetworks in Cancer Using Multiple Protein-Protein Interaction Networks. Cancers, 15(16), 4090. [CrossRef]
- Yao, D. , Binan, L., Bezney, J., Simonton, B., Freedman, J., Frangieh, C. J., Dey, K., Geiger-Schuller, K., Eraslan, B., Gusev, A., Regev, A., & Cleary, B. (2023). Scalable genetic screening for regulatory circuits using compressed Perturb-seq. Nature biotechnology, 10.1038/s41587-023-01964-9. Advance online publication. [CrossRef]
- Yehia, A. M. , Elsakka, E. G. E., Abulsoud, A. I., Abdelmaksoud, N. M., Elshafei, A., Elkhawaga, S. Y., Ismail, A., Mokhtar, M. M., El-Mahdy, H. A., Hegazy, M., Elballal, M. S., Mohammed, O. A., El-Husseiny, H. M., Midan, H. M., El-Dakroury, W. A., Zewail, M. B., Abdel Mageed, S. S., Moustafa, Y. M., Mostafa, R. M., Elkady, M. A., … Doghish, A. S. (2023). Decoding the role of miRNAs in multiple myeloma pathogenesis: A focus on signaling pathways. Pathology, research and practice, 248, 154715. [CrossRef]
- Yepes, S. , Tucker, M. A., Koka, H., Xiao, Y., Jones, K., Vogt, A., Burdette, L., Luo, W., Zhu, B., Hutchinson, A., Yeager, M., Hicks, B., Freedman, N. D., Chanock, S. J., Goldstein, A. M., & Yang, X. R. (2020). Using whole-exome sequencing and protein interaction networks to prioritize candidate genes for germline cutaneous melanoma susceptibility. Scientific reports, 10(1), 17198. [CrossRef]
- Yu, X. , Abbas-Aghababazadeh, F., Chen, Y. A., & Fridley, B. L. (2021). Statistical and Bioinformatics Analysis of Data from Bulk and Single-Cell RNA Sequencing Experiments. Methods in molecular biology (Clifton, N.J.), 2194, 143–175. [CrossRef]
- Zeng, Y. , Shi, Y., Xu, L., Zeng, Y., Cui, X., Wang, Y., Yang, N., Zhou, F., & Zhou, Y. (2021). Prognostic Value and Related Regulatory Networks of MRPL15 in Non-Small-Cell Lung Cancer. Frontiers in oncology, 11, 656172. [CrossRef]
- Zhao, Q. , Lin, X., & Wang, G. (2022). Targeting SREBP-1-Mediated Lipogenesis as Potential Strategies for Cancer. Frontiers in oncology, 12, 952371. [CrossRef]
- Zheng, G. X. , Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., Ziraldo, S. B., Wheeler, T. D., McDermott, G. P., Zhu, J., Gregory, M. T., Shuga, J., Montesclaros, L., Underwood, J. G., Masquelier, D. A., Nishimura, S. Y., Schnall-Levin, M., Wyatt, P. W., Hindson, C. M., Bharadwaj, R., … Bielas, J. H. (2017). Massively parallel digital transcriptional profiling of single cells. Nature communications, 8, 14049. [CrossRef]
- Zheng, X. , Wu, B., Liu, Y., Simmons, S. K., Kim, K., Clarke, G. S., Ashiq, A., Park, J., Wang, Z., Tong, L., Wang, Q., Xu, X., Levin, J. Z., & Jin, X. (2023). Massively parallel in vivo Perturb-seq reveals cell type-specific transcriptional networks in cortical development. bioRxiv : the preprint server for biology, 2023.09.18.558077. [CrossRef]
- Zhou, Y. H., Xia, K., & Wright, F. A. (2011). A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics Oxford, 27(19), 2672–2678. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



