Submitted:
25 October 2024
Posted:
28 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals and DC12 Administration
2.2. Density Gradient Centrifugation
2.3. SIRT5 Co-Immunoprecipitation
2.4. Immunoblotting
2.5. Fatty Acid Oxidation Flux Assays
2.6. Urinary Dicarboxylic Acids
2.7. Proteomics and Succinylomics
2.8. Recombinant Protein Expression and Purification
2.9. Enzyme Activity Assays
3. Results
3.1. Dicarboxylic Acid Feeding Recruits SIRT5 to Liver Peroxisomes
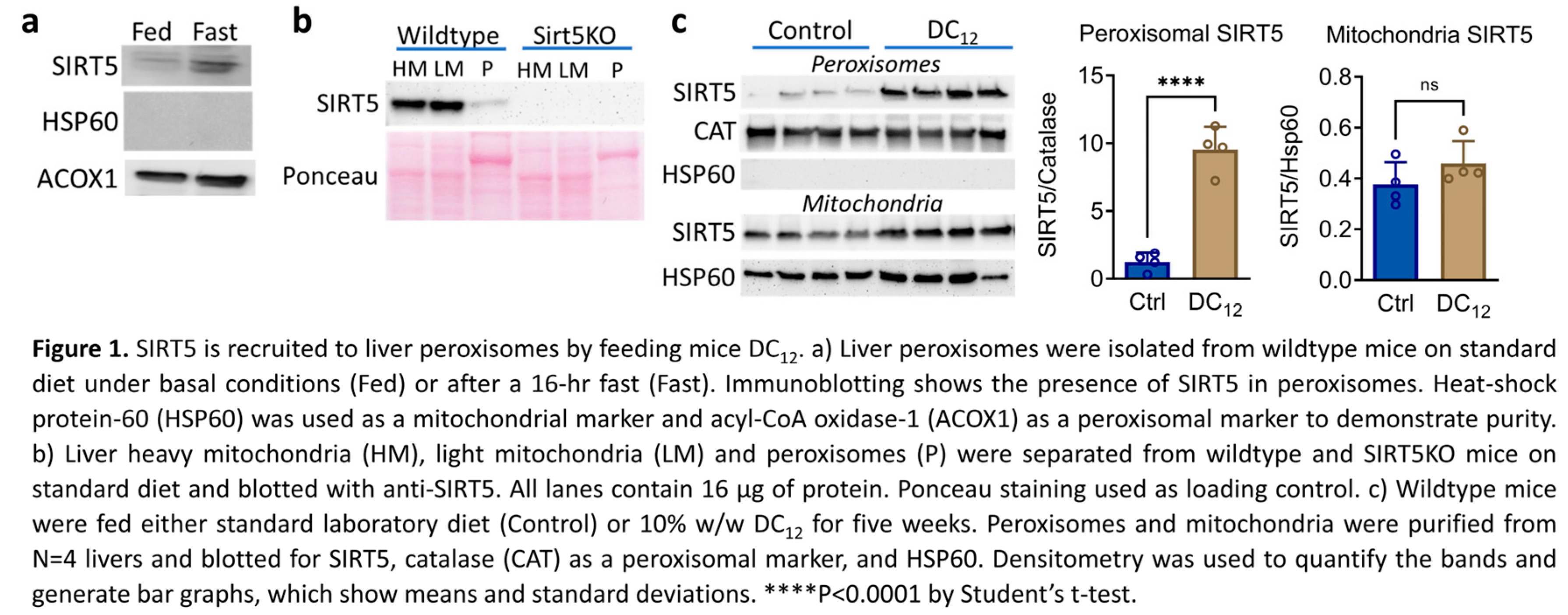
3.2. SIRT5 Knockout Mice Have Impaired DC12 Catabolism

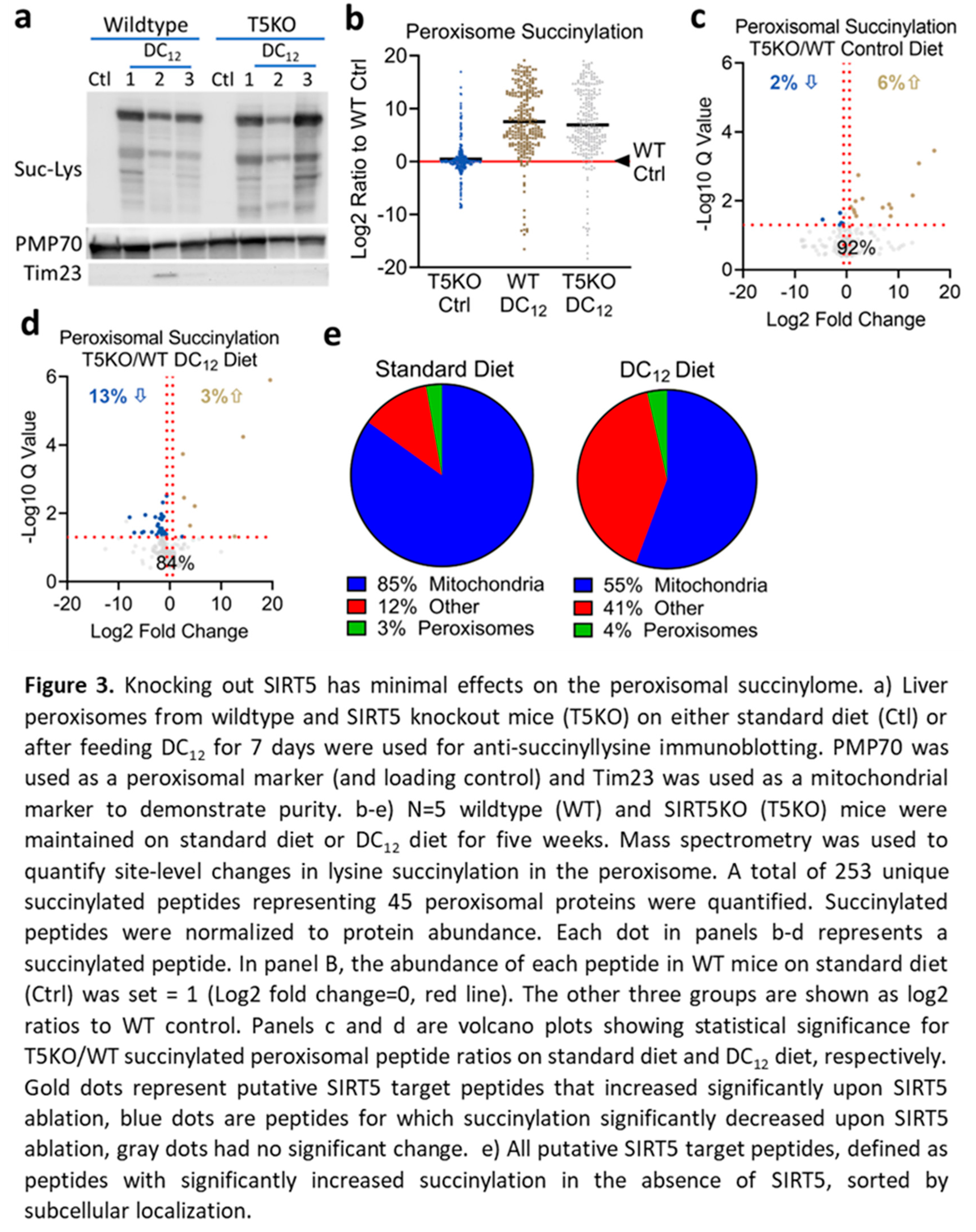
3.3. Knocking Out SIRT5 Has Minimal Effects on the Peroxisomal Succinylome
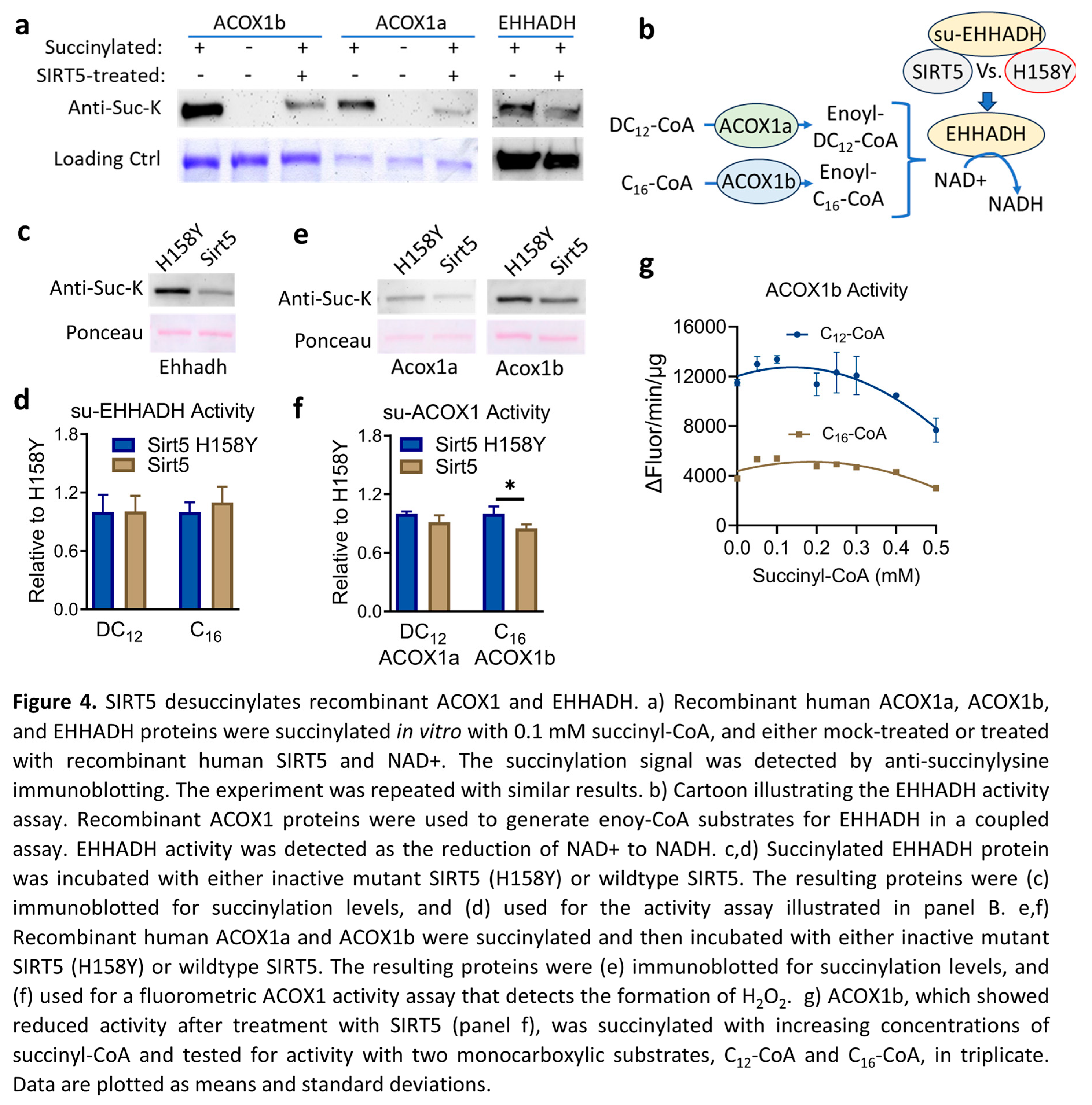
3.4. SIRT5 Desuccinylates Recombinant ACOX1 and EHHADH but the Impact on Enzyme Activity Is Minimal
4. Discussion
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wagner, G.R. and R.M. Payne, Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem 2013, 288, 29036–45. [Google Scholar] [CrossRef] [PubMed]
- Rardin, M.J. , et al., SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 2013, 18, 920–33. [Google Scholar] [CrossRef] [PubMed]
- Hirschey, M.D. and Y. Zhao, Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol Cell Proteomics 2015, 14, 2308–15. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y. , et al., SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell 2015, 59, 321–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F. , et al., SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep 2018, 19. [Google Scholar]
- Goetzman, E.S. , et al., Dietary dicarboxylic acids provide a non-storable alternative fat source that protects mice against obesity. J Clin Invest 2024, 134. [Google Scholar]
- Ranea-Robles, P. and S.M. Houten, The biochemistry and physiology of long-chain dicarboxylic acid metabolism. Biochem J 2023, 480, 607–627. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z. , et al., Compartmentation of Metabolism of the C12-, C9-, and C5-n-dicarboxylates in Rat Liver, Investigated by Mass Isotopomer Analysis: ANAPLEROSIS FROM DODECANEDIOATE. J Biol Chem 2015, 290, 18671–7. [Google Scholar] [CrossRef] [PubMed]
- Schluter, A. , et al, PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res 2010, 38, D800–5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. , et al., The fatty acid oxidation enzyme long-chain acyl-CoA dehydrogenase can be a source of mitochondrial hydrogen peroxide. Redox Biol 2019, 26, 101253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. and E. Goetzman, The enzyme activity of mitochondrial trifunctional protein is not altered by lysine acetylation or lysine succinylation. PLoS One 2021, 16, e0256619. [Google Scholar]
- Zhang, Y. , et al., SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PLoS One 2015, 10, e0122297. [Google Scholar]
- Hershberger, K.A. , et al., Ablation of Sirtuin5 in the postnatal mouse heart results in protein succinylation and normal survival in response to chronic pressure overload. J Biol Chem 2018, 293, 10630–10645. [Google Scholar] [CrossRef] [PubMed]
- Alleyn, M. , et al., The dawn of succinylation: a posttranslational modification. Am J Physiol Cell Physiol 2018, 314, C228–C232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. , et al., Lysine desuccinylase SIRT5 binds to cardiolipin and regulates the electron transport chain. J Biol Chem 2017, 292, 10239–10249. [Google Scholar] [CrossRef] [PubMed]
- Bharathi, S.S. , et al., Role of mitochondrial acyl-CoA dehydrogenases in the metabolism of dicarboxylic fatty acids. Biochem Biophys Res Commun 2020, 527, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, C. , et al., From sirtuin biology to human diseases: an update. J Biol Chem 2012, 287, 42444–52. [Google Scholar] [CrossRef] [PubMed]
- Madsen, A.S. , et al., Investigating the Sensitivity of NAD+-dependent Sirtuin Deacylation Activities to NADH. J Biol Chem 2016, 291, 7128–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y., D. Cuebas, and H. Schulz, Channeling of 3-hydroxy-4-trans-decenoyl coenzyme A on the bifunctional beta-oxidation enzyme from rat liver peroxisomes and on the large subunit of the fatty acid oxidation complex from Escherichia coli. J Biol Chem 1986, 261, 15390–5. [Google Scholar] [CrossRef] [PubMed]
| Protein | Function | % Coverage |
|---|---|---|
| Uricase (UOX) | Oxidizes uric acid to 5-hydroxyisourate | 86 |
| Peroxisomal bifunctional enzyme type 2 (HSD17B4) | Steps 2 and 3 of β-oxidation | 74 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | Steps 2 and 3 of β-oxidation | 65 |
| Acyl-CoA dehydrogenase 11 (ACAD11) | Step 1 of very long-chain β-oxidation | 50 |
| Peroxisomal 2,4-dienoyl-CoA reductase (DECR2) | Reduces double bonds, β-oxidation | 49 |
| Peroxisomal acyl-coenzyme A oxidase 1 (ACOX1) | Step 1 of β-oxidation | 40 |
| Catalase (CAT) | Degrades H2O2 to H2O and O2 | 33 |
| ATP-binding cassette sub-family D member 3 (ABCD3) | Fatty acid transporter | 23 |
| Acyl-coenzyme A thioesterase 4 (ACOT4) | Cleaves CoA from succinyl-CoA | 21 |
| Non-specific lipid-transfer protein (SCP2) | Step 4 of β-oxidation | 21 |
| Peroxisomal sarcosine oxidase (PIPOX) | Metabolizes sarcosine & pipecolic acid | 16 |
| Lon protease homolog 2, peroxisomal (LONP2) | Degrades misfolded proteins | 14 |
| 3-ketoacyl-CoA thiolase B (ACAA1b) | Step 4 of β-oxidation | 13 |
| Very long-chain acyl-CoA synthetase (SLC27A2) | Activates fatty acids into acyl-CoAs | 12 |
| Peroxisomal coenzyme A diphosphatase (NUDT7) | Hydrolyzes CoA moiety of acyl-CoAs | 10 |
| Protein | Lysine Residue | Log2 FC KO/WT |
|---|---|---|
| Increased Succinylation (q<0.05) | ||
| Hydroxysteroid dehydrogenase like 2 (HSDL2) | K481 | 16.9 |
| Enoyl-CoA isomerase (ECH1) | K320 | 14.0 |
| Enoyl-CoA isomerase (ECH1) | K76 | 12.8 |
| Acyl-coenzyme A diphosphatase (NUDT19) | K351 | 8.6 |
| Hydroxysteroid dehydrogenase like 2 (HSDL2) | K390 | 8.5 |
| Alpha-methylacyl-CoA racemase (AMACR) | K204 | 8.2 |
| Hydroxysteroid dehydrogenase like 2 (HSDL2) | K295 | 7.0 |
| Prostaglandin reductase-3 (ZADH2) | K214 | 3.1 |
| Dehydrogenase/reductase SDR family member 4 (DHRS4) | K106 | 2.3 |
| Catalase (CAT) | K522 | 1.9 |
| Peroxisomal 2,4-dienoyl-CoA reductase (DECR2) | K230 | 1.7 |
| ATP-binding cassette sub-family D member 3 (ABCD3) | K6 | 1.5 |
| Alanine--glyoxylate aminotransferase (AGXT) | K322 | 1.4 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | K355 | 0.8 |
| Decreased Succinylation (q<0.05) | ||
| Dehydrogenase/reductase SDR family member 4 (DHRS4) | K99 | -0.7 |
| Catalase (CAT) | K98 | -1.0 |
| Peroxisomal trans-2-enoyl-CoA reductase (PECR) | K83 | -1.3 |
| Enoyl-CoA isomerase (ECH1) | K230 | -1.3 |
| 2-Hydroxyacid oxidase 1 (HAO1) | K249 | -4.6 |
| Protein | Lysine Residue | Log2 FC KO/WT |
|---|---|---|
| Increased Succinylation (q<0.05) | ||
| Peroxisomal ATPase (PEX1) | K1205 | 14.3 |
| Peroxisomal bifunctional enzyme type 2 (HSD17B4) | K50/K57 | 12.6 |
| Alanine--glyoxylate aminotransferase (AGXT) | K411/K314 | 4.9 |
| Hydroxysteroid dehydrogenase like 2 (HSDL2) | K295 | 4.0 |
| Catalase (CAT) | K477 | 2.8 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | K348/K355 | 2.6 |
| Acyl-coenzyme A diphosphatase (NUDT19) | K351 | 2.4 |
| Decreased Succinylation (q<0.05) | ||
| Uricase (UOX) | K27 | -0.6 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | K579 | -0.7 |
| Acyl-CoA dehydrogenase-11 (ACAD11) | K422 | -0.9 |
| Uricase (UOX) | K118 | -1.0 |
| Uricase (UOX) | K80 | -1.1 |
| Bile acid-CoA: amino acid N-acyltransferase (BAAT) | K346 | -1.2 |
| Peroxisomal sarcosine oxidase (PIPOX) | K104 | -1.3 |
| ATP-binding cassette sub-family D member 3 (ABCD3) | K260 | -1.4 |
| Bile acid-CoA: amino acid N-acyltransferase (BAAT) | K406 | -1.4 |
| Non-specific lipid-transfer protein (SCP2) | K26 | -1.5 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | K527 | -1.6 |
| 2-Hydroxyacid oxidase 1 (HAO1) | K353 | -1.6 |
| Peroxisomal bifunctional enzyme type 2 (HSD17B4) | K81 | -1.6 |
| Peroxisomal bifunctional enzyme type 1 (EHHADH) | K459 | -1.6 |
| Peroxisomal bifunctional enzyme type 2 (HSD17B4) | K57 | -1.8 |
| Dehydrogenase/reductase SDR family member 4 (DHRS4) | K99 | -1.8 |
| Peroxisomal bifunctional enzyme type 2 (HSD17B4) | K65/K68 | -1.8 |
| Hydroxysteroid dehydrogenase like 2 (HSDL2) | K87 | -1.8 |
| Acyl-CoA dehydrogenase-11 (ACAD11) | K90 | -1.9 |
| Peroxisomal 2,4-dienoyl-CoA reductase (DECR2) | K70/K71 | -1.9 |
| Peroxisomal trans-2-enoyl-CoA reductase (PECR) | K32 | -1.9 |
| Peroxisomal sarcosine oxidase (PIPOX) | K153 | -2.1 |
| Peroxisomal sarcosine oxidase (PIPOX) | K98 | -2.1 |
| Alpha-methylacyl-CoA racemase (AMACR) | K57 | -2.3 |
| Bifunctional epoxide hydrolase 2 (EPHX2) | K482 | -2.4 |
| 2-hydroxyacyl-CoA lyase 1 (HACL1) | K21 | -2.5 |
| Enoyl-CoA isomerase (ECH1) | K230 | -2.8 |
| Peroxisomal coenzyme A diphosphatase (NUDT7) | K20 | -3.5 |
| 2-hydroxyacyl-CoA lyase 1 (HACL1) | K238 | -4.8 |
| 2-Hydroxyacid oxidase 1 (HAO1) | K309 | -5.1 |
| Acyl-CoA oxidase-2 (ACOX2) | K678 | -5.4 |
| Peroxisomal trans-2-enoyl-CoA reductase (PECR) | K17 | -6.9 |
| Acyl-CoA oxidase-2 (ACOX2) | K303 | -7.8 |
| NAD-capped RNA hydrolase (NUDT12) | K282 | -8.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





