1. Introduction
Melatonin is a multifunctional effector which plays diverse physiological roles in plants, from seed germination to post-harvest fruit storage and seed longevity [
1]. However, the beneficial effects of this molecule as an anti-stress agent are of paramount importance due to its powerful antioxidant activity. Melatonin acts as a scavenger of reactive oxygen species (ROS) and reactive nitrogen species (RNS) such as O
2−, OH
−, NO and ONOO
− and also enhances the expression of superoxide generating enzymes (RbOHs), superoxide dismutase and the enzymes involved in detoxification of excess H
2O
2, particularly CAT, POD, and APX [
2]. At low concentrations, melatonin may act as a hormone-like signaling molecule involved in crosstalk with virtually all known plant hormones by modulating their signaling circuits and metabolism [
3].
The
Arabidopsis genome harbors several genes encoding four successive steps of melatonin biosynthesis. During the first two steps serotonin is derived from tryptophan by two enzymatic reactions carried out by TDC (tryptophan decarboxylase) and T5H (tryptamine 5-hydroxylase). The next step is committed SNAT (serotonin N-acyltransferase) responsible for the synthesis of
N-acetylserotonin, which is further catalyzed into melatonin by
O-methyltransferases COMT (caffeic acid O-methyltransferase) and ASMT (N-aceylserotonin O-methyltransferase) [
4]. Two melatonin metabolic genes
M2H (melatonin 2-hydroxylase) and
M3H (melatonin 3-hydroxylase) convert melatonin into 2-hydroxymelatonin and cyclic 3-hydroxymelatonin respectively [
5].
The signaling pathway of melatonin and its metabolites is proposed to act through the mitogen-activated protein kinase (MAPK) cascade, which is preceded by receptor-like kinases (RLKs), putative candidates for melatonin receptors in plants [
6]. Alternative signaling circuit includes putative MT receptor gene
CAND2 (also known as
PMTR1), which is associated with
GPA, encoding the α-subunit of the heterotrimeric G protein [
7].
Abscisic acid is conventionally considered a stress hormone since its level is greatly increased under a variety of abiotic stresses [
8]. ABA via binding to the PYR/PYL/RCAR (PYL) family of receptors, inhibits a class of A-type protein phosphatase 2Cs (PP2Cs). PP2C inactivation triggers phosphorylation of a collection of basic leucine zipper transcription factors (ABA-responsive element binding factors, ABFs) by a small set of class 3 sucrose nonfermenting-1-related protein kinase 2s (SNRK2s). Phosphorylated ABFs activate downstream target genes through the
cis-acting ABA response elements (ABREs). In addition to the canonic PYL-PP2C-SnRK2 core pathway, ABA responses in plants require a network of other signaling pathways that involve calcium, ROS, NO, phospholipids, and the various kinases [
9]. All these pathways can interact with each other through positive or negative feedback mechanisms producing a remarkable pleiotropy of effects in plant responses to abiotic stresses.
Melatonin-mediated action generally induces down-regulation of ABA biosynthesis genes and up-regulation of ABA catabolism genes resulting in a decrease of ABA level [
2,
3]. For example, in apple melatonin selectively down-regulated
MdNCED3, an ABA synthesis gene, and triggered its catabolic genes,
MdCYP707A1 and
MdCYP707A2, reducing ABA contents in drought-stressed plants [
10]. Under drought stress, exogeneous melatonin downregulated
NCED1 expression in maize and induced the expression of
ABA8ox1 (ABA-8- oxidase 1) and
ABA8ox3 responsible for ABA breakdown [
11]. Other examples include Chinese cabbage [
12], Chinese hickory
Carya cathayensis [
13], mango [
14], cucumber [
15] and ryegrass [
16]. A special mechanism by which melatonin antagonizes ABA action was recently described in
Arabidopsis. Melatonin offsets ABA action to delay leaf senescence via RBOHD-dependent H
2O
2 production that triggers [Ca
2+]cyt accumulation and subsequently inhibits K
+ efflux and delays cell death/leaf senescence in response to ABA [
17].
On the other hand, an increase in melatonin content can lead to up regulation of ABA-responsive genes testifying that crosstalk between MT and ABA is highly variable. Opposite responses were detected for
Elymus nutans [
18],
Brassica napus [
19] and
Raphanus sativus [
20]. Overexpression of
AtASMT caused massive melatonin accumulation and synergized with ABA to inhibit seed germination in
Arabidopsis [
21]. In case of barley, exogenously applied melatonin resulted in higher ABA concentration in the drought-primed plants than in the nonprimed plants when exposed to cold stress [
22]. Likewise, during arsenic stress, MT treatment of the susceptible rice cultivar Khitish up-regulated transcript levels of
NCED3, and down-regulated transcript levels of
ABA8ox1 elevating the endogenous ABA level. However, ABA concentration remained unaltered in tolerant rice cultivar Muktashri indicating that the response depends upon plant genotype [
23].
While much of the work conducted on melatonin-ABA interplay in plants has been targeted on influence of melatonin on ABA content and signal transduction, few studies have focused on the intersecting effects of ABA and melatonin on the expression of signaling and metabolic genes of these regulators. In rice, melatonin was shown to regulate ROS homeostasis under nitrogen limitation via the module OsbZIP79-OsABI5, composed of two ABA related
trans-factors. However, the module did not directly regulate melatonin biosynthesis [
24]. On the other hand, treatments with abscisic and methyl jasmonic acids simultaneously induced three isoforms of N-acetylserotonin methyltransferase in rice, the final enzyme in melatonin biosynthetic pathway [
25]
In this work, using exogenously applied ABA and melatonin as well as melatonin and ABA-related mutants, we showed that ABA is involved in downregulation of Arabidopsis ASMT gene under high light stress via the activity of the transcription factor ABI4 (ABSCISIC ACID INSENSITIVE 4). Additionally, we identified that MT-dependent regulation of ABA genes depends on CAND2 - GPA1 signaling circuit.
2. Results
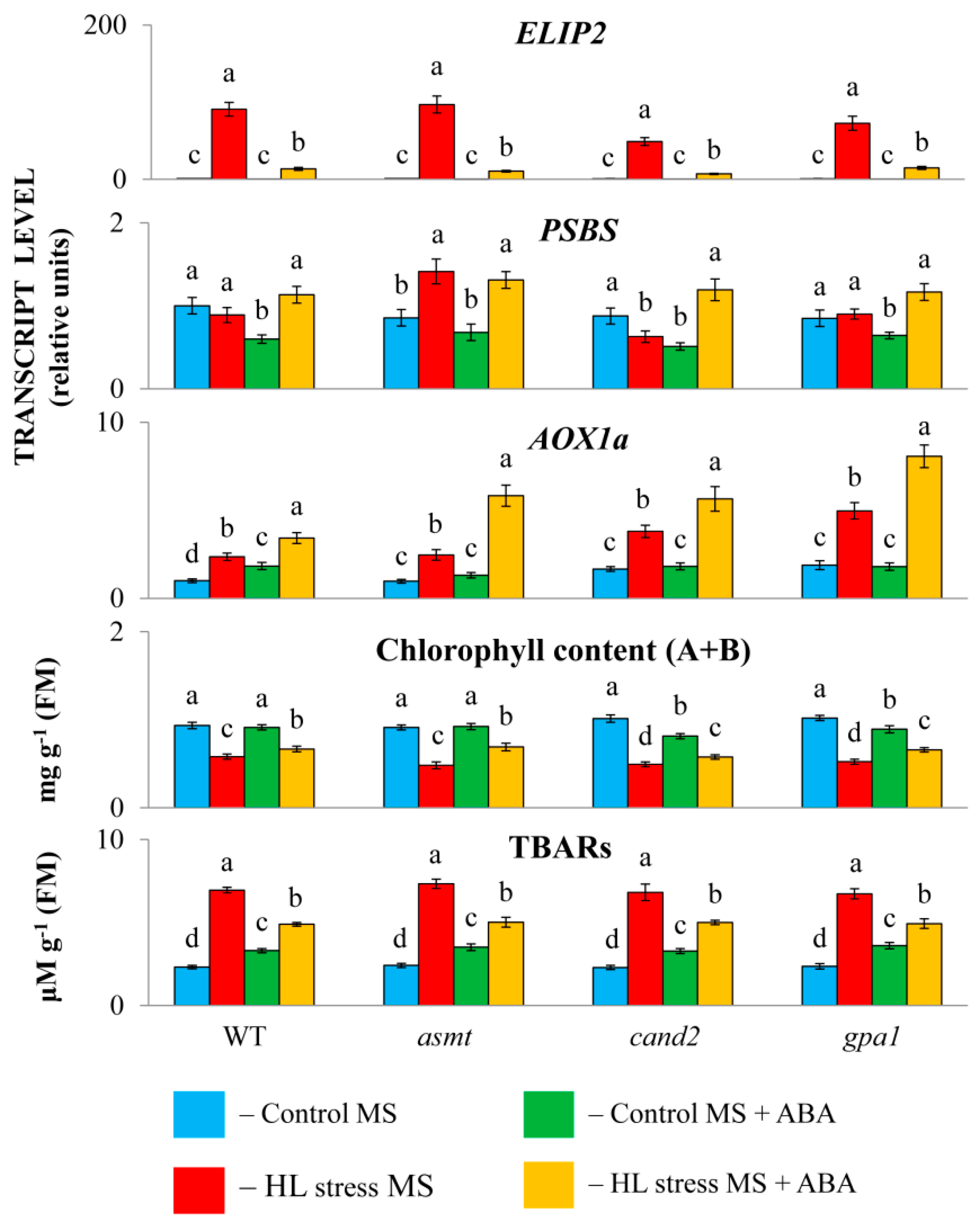
2.1. ABA Treatment Differentially Regulates Physiological Parameters and Stress-Induced Genes in Col0 and MT Mutants
ABA treatment under mild light (60 μmol m
-2 s
-1, 24 h) was perceived by wild type plants as a mild stress, as evidenced by the reduced transcript accumulation of
PsbS involved in quenching singlet excited chlorophylls and increased expression of stress marker gene
AOX1a which participates in the fine-tuning of mitochondrial membrane potential and the alleviation of ROS production (
Figure 1). Under HL stress (600 µmol m
-2 s
-1, 24 h), ABA additionally increased the accumulation of
AOX1a transcripts, and also reduced stress-induced levels of
ELIP2 transcripts (EARLY LIGHT-INDUCED PROTEIN 2), which is known to modulate chlorophyll synthesis to prevent photo-oxidative stress [
26]. MT mutants, when treated with ABA, did not differ significantly from the wild type in their responses under either control or stress conditions.
In parallel, the content of TBARs, a marker of membrane damage caused by lipid peroxidation, was significantly lifted by ABA treatment of plants under mild light, whereas it was decreased under combined application of HL and ABA compared to stress-induced levels. Hence, ABA treatment of control plants had a positive effect only under stress conditions. Exogenous ABA did not significantly change chlorophyll content in WT and asmt mutant under mild light, but promoted increased chlorophyll levels under excessive light compared to stress-repressed levels. On the other hand, MT signaling mutants, cand2 and gpa1, exhibited reduced chlorophyll levels under mild light when treated with ABA which indicates their elevated sensitivity to the hormone. However, as with the wild type, the chlorophyll content of these mutants increased after ABA treatment compared to their HL control values confirming plasticity of plant responses to ABA.
2.2. Responses of MT-Related Genes to ABA Treatment of MT Mutants
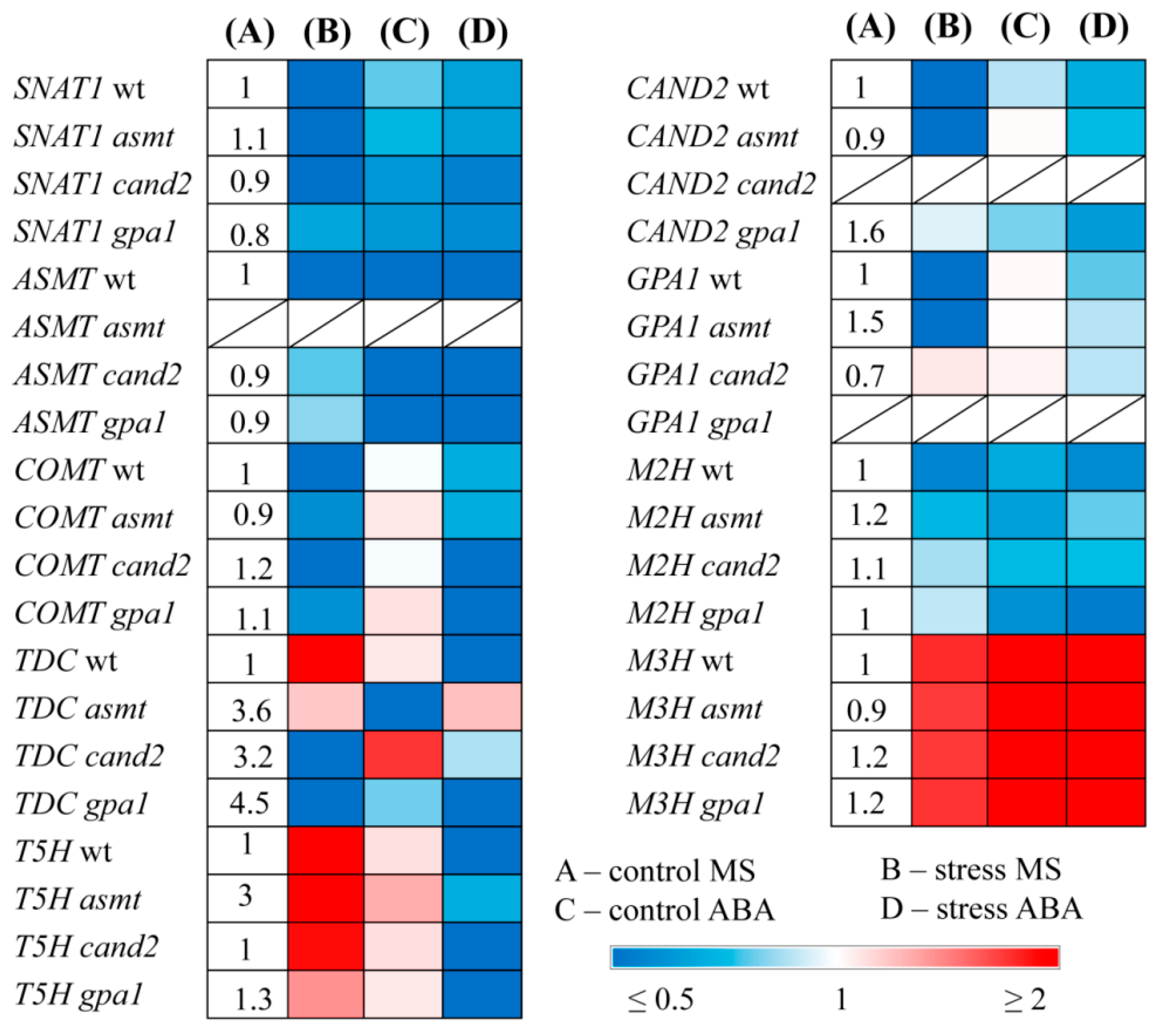
To elucidate the regulatory interactions between ABA and melatonin, we next examined the responses of MT related genes to ABA treatment in WT and MT mutants (
Figure 2). Under mild light, exogenous ABA drastically down-regulated
ASMT (4 fold), and to a lesser extent
SNAT1 both in WT and MT mutants and did not change transcript accumulation of
COMT and
T5H. TDK expression varied: it was not significantly affected in WT and
gpa1, but was reduced in
asmt and slightly increased in
cand2. It should be noted, however, that the steady state levels of
TDC transcripts in untreated MT mutants were 3 to 4 fold higher than in WT.
Under excessive irradiation, ABA maintained the expression of SNAT1 and COMT at a higher level as compared to untreated stressed plants, probably due to the stress mitigating effect of ABA. T5H was strongly inhibited in WT and all MT mutants except asmt, despite elevated steady state transcript level in this mutant. TDC transcript levels were decreased in WT and gpa1 and did not substantially differ from the control levels in asmt and cand2. However, ASMT expression was still downregulated by the hormone in WT and MT mutants under either control or stress conditions. These results indicate that ABA constantly suppresses ASMT in the context of differential responses of other MT biosynthesis genes.
The interactions between ABA and melatonin may also be mediated by the expression of MT catabolism genes, given that MT metabolites may have specific functions in plants. Melatonin is converted to 2-hydroxymelatonin (2-OM) and cyclic 3-hydroxymelatonin (3-OM) by members of the 2-oxoglutarate-dependent enzyme family M2H and M3H [
27]. The expression of
M2H, encoding melatonin 2-hydroxylase (M2H), was slightly reduced in WT and to a lesser extent in MT mutants upon exposure to HL or ABA and upon combined treatment with ABA and HL. In contrast, a second gene of plant melatonin metabolism, encoding melatonin 3-hydroxylase (
M3H), was upregulated 6- to 10-fold under HL. Moreover, its transcripts increased 20- to 40-fold in WT and MT mutants after ABA treatment under mild and high irradiation. These data support the idea that ABA may antagonistically regulate MT levels modulating the expression of MT catabolism genes.
We also tested whether the putative MT signaling genes CAND2 and GPA1, responded to ABA treatment. Exogenous ABA did not affect CAND2 and GPA1 expression under control radiation and contributed to their slightly reduced levels in high light conditions.
2.3. ABA Mutations Affect Melatonin Content and the Expression of MT Related Genes
To further delineate possible links between melatonin and ABA, we assessed the effect of modulated endogenous ABA status on the responses of MT biosynthesis and signaling genes to stress and MT treatment The study included ABA biosynthesis mutants
aba2 and
aba3 with the lowered level of endogenous ABA [
28], and mutants with impaired
ABI3, ABI4 and
ABI5 genes encoding three well-characterized transcription factors. These regulators of ABA signaling are members of the B3-, APETALA2- (AP2), and basic leucine zipper- (bZIP) domain families, respectively, which have been shown to regulate overlapping subsets of ABA-inducible genes [
29].
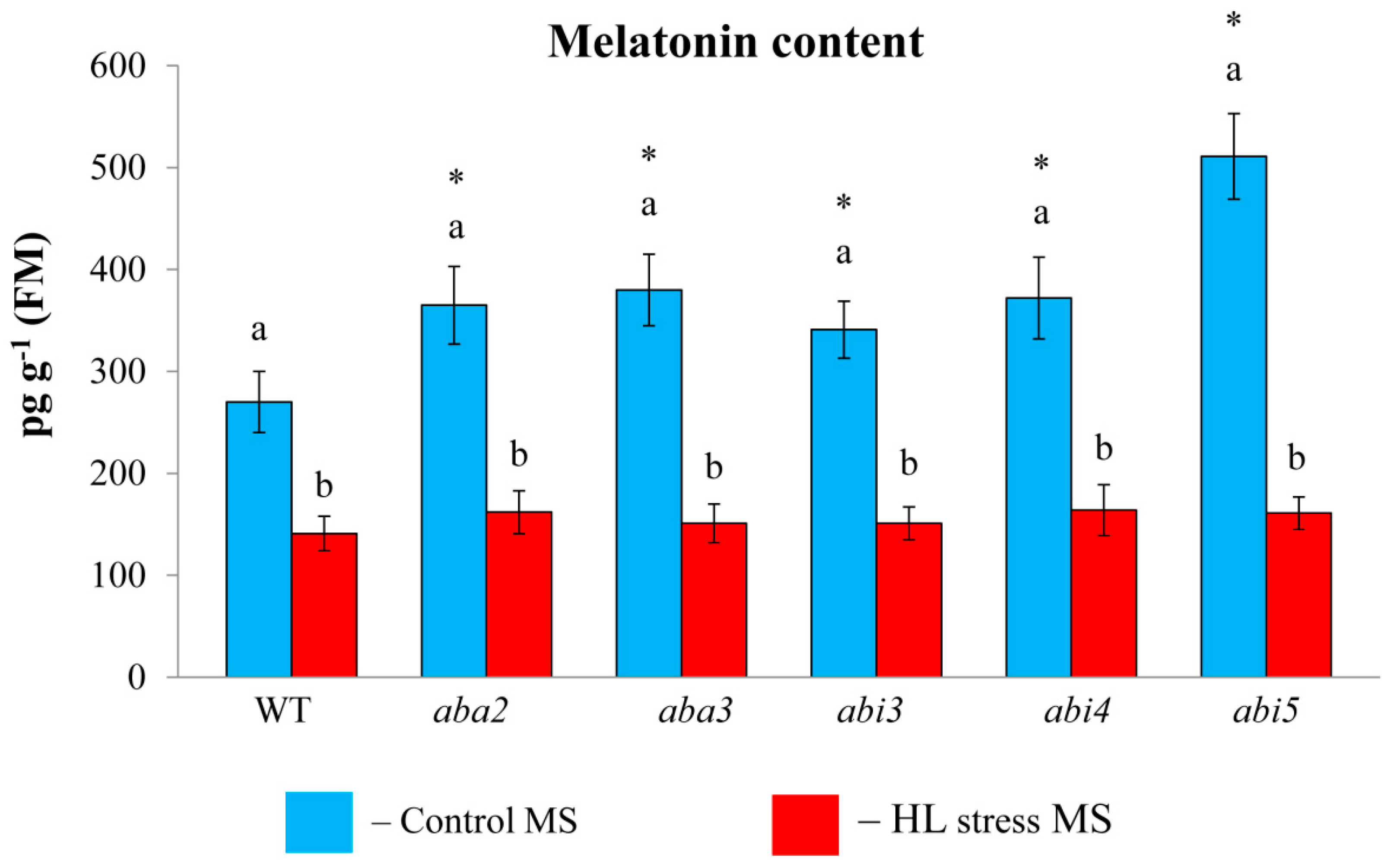
Considering that the effect of MT treatment may depend on the level of endogenous MT, we first assessed melatonin content in ABA mutants under control conditions and under photooxidative stress (
Figure 3). All the mutants displayed elevated steady state MT levels (2.5 fold) as compared to WT, especially in
abi5, consistent with the increased steady state levels of
ASMT transcripts in this mutant. Following HL exposure, melatonin content decreased in the mutants with altered ABA status.
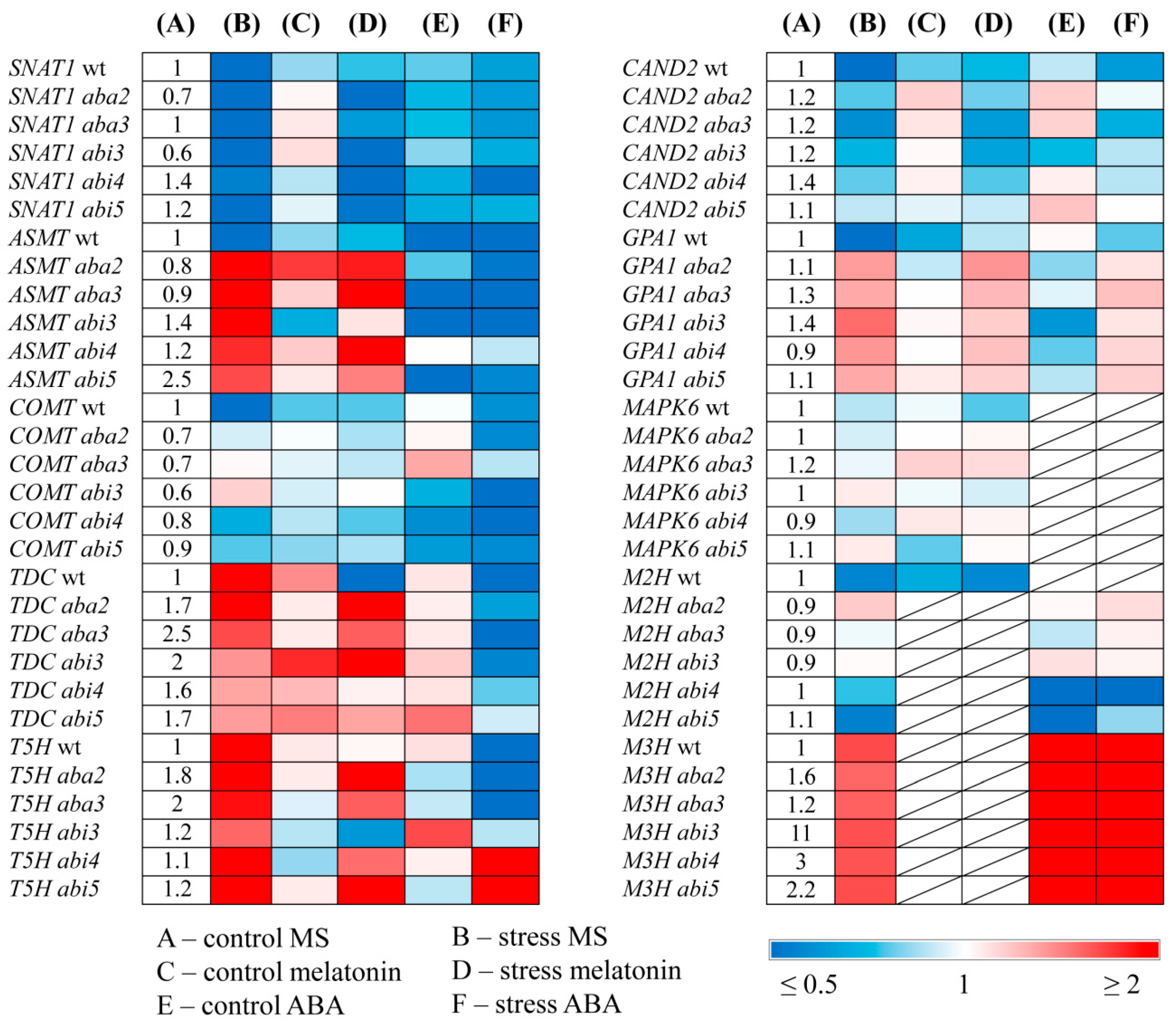
The expression of MT synthesis genes
SNAT and
COMT was downregulated under HL in the mutants, though less than in WT or did not change (
Figure 4). However, in contrast to WT,
ASMT transcript levels increased, especially in
aba3 and
aba2 (5 and 6 fold respectively) and remained elevated after combined-treatment with HL and melatonin. Thus, defects of ABA synthesis or signaling contributed to the induction of
ASMT. These results imply that
ASMT expression negatively correlated with ABA content and signaling.
T5H and
TDC were also activated by stress and stress
+MT, in ABA mutants, although in WT stress-induced elevation of both genes was mitigated by MT.
HL mildly reduced the expression of CAND2, and did not reliably affect GPA1 in ABA mutants, in contrast to WT in which both genes were dramatically downregulated (3-4 fold). Under combined MT+HL treatment no further changes of CAND2 and GPA1 expression occurred in tested ABA mutants while in WT plants the transcript levels of both genes increased as compared to those in untreated stressed plants. Hence, the expression patterns of CAND2 and GPA1 under stress conditions depend on ABA status with ABA deficiency promoting increased expression of MT signaling genes. The results also suggest possible involvement of additional factors in ABA dependent regulation of GPA1 expression since the expression pattern of GPA1 in ABA mutants did not strictly follow that of CAND2.
Given that melatonin signaling pathway may act through the mitogen-activated protein kinase (MAPK) cascade, [
6] we also analyzed expression profiles of
MAPK6. However, no statistically significant changes occurred following HL or HL+MT exposure in either WT or ABA mutants
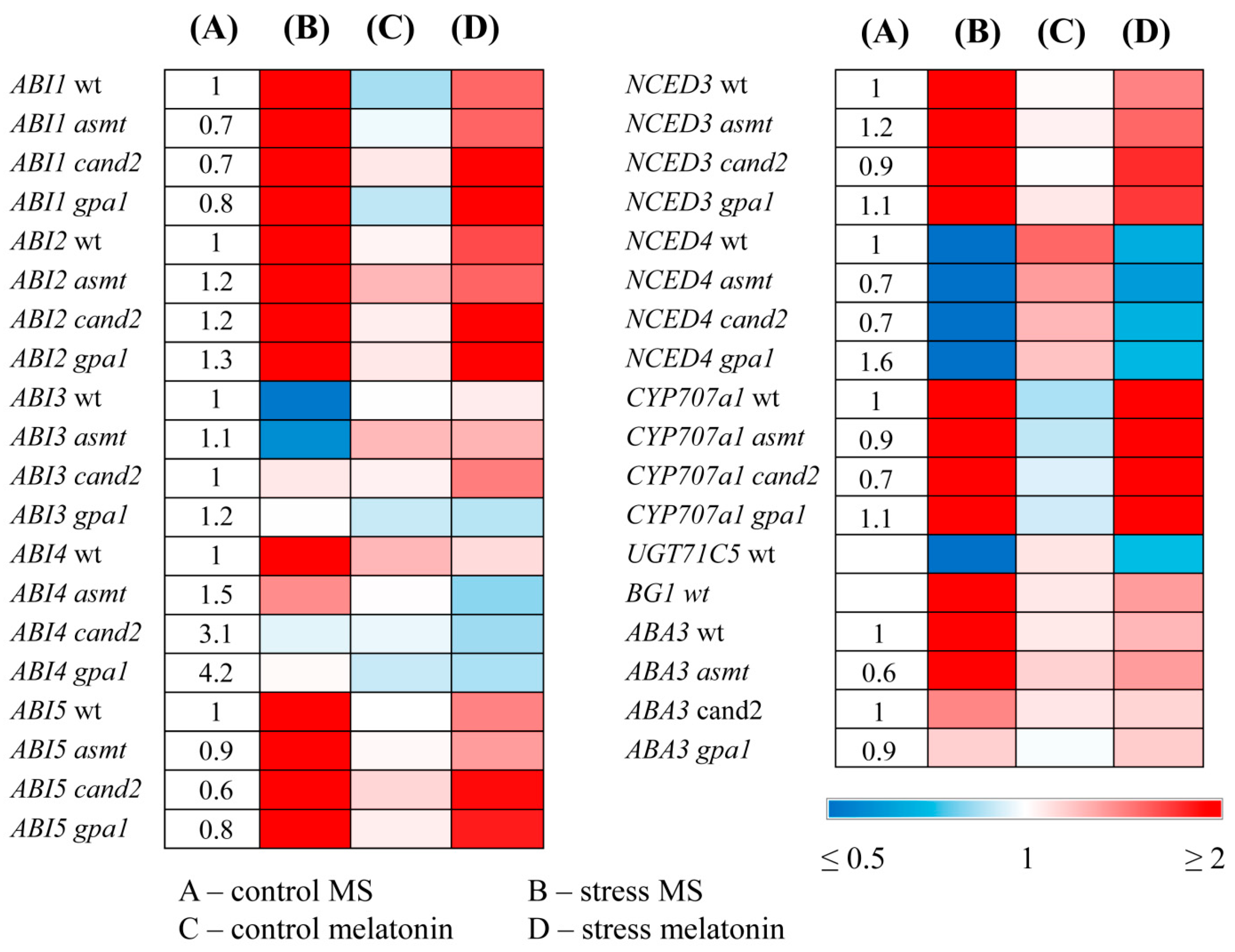
2.4. MT Treatment Alters the Expression of ABA Related Genes in MT Mutants
Next, we studied whether melatonin regulated the expression of ABA related genes in MT mutants. In wild type plants, key ABA synthesis and signaling genes
NCED3, ABA3, ABI1, ABI2, ABI4, ABI5 were upregulated and
ABI3 was downregulated by excess irradiation, whereas melatonin attenuated these responses (
Figure 5). Such alleviation may indicate a more stable status of the plant exposed to melatonin, when stress-induced response of ABA genes is less pronounced. At the same time, this reaction may be the result of competitive relations between these regulators for a common set of transcription targets under stress, leading to partial repression of ABA genes by melatonin. Of note,
NCED4, which plays a major role in degradation of beta-carotene, a substrate for ABA synthesis [
30] was strongly downregulated by HL stress in accord with stress protective role of ABA and to a lesser extent following MT+HL treatment alleviating the effects of stress.
The catabolism of ABA from ABA to phaseic acid is catalyzed by a cytochrome P450 monooxygenase (P450) encoded by
CYP707As [
31]. ABA can also be glucosylated by a UDP-glucosyltransferase encoded by
UGT71C5 to ABA-glucose ester (ABA-GE), which is an inactive form of ABA [
32]. ABA-GE, in its turn, can rapidly be transformed to active ABA via
AtBG1 and
AtBG2 encoded β-glucosidases when the environment conditions change. MT treatment under HL did not substantially affect
CYP707A transcript levels compared to their increased values under HL.
UGT71C5 expression was significantly downregulated while that of
AtBG1 dramatically upregulated under HL which may contribute to increased ABA levels, and these alterations were attenuated by MT. Therefore, melatonin treatment primarily affects genes involved in the metabolism of reversible forms of ABA.
In the melatonin-deficient asmt mutant, the response of ABA genes to stress and melatonin was similar to that of the wild-type, although less pronounced for some genes. However, in cand2 and gpa1 mutants ABI4 was not regulated by either stress or melatonin Moreover, MT insensitive cand2 and gpa1 exhibited elevated steady state levels of ABI4 transcripts (3- and 4-fold, respectively, as compared to WT) suggesting that ABI4 is constitutively derepressed in the cand2 and gpa1 background and further response to stress treatment is not possible. Steady state levels of ABI4 transcripts in MT deficient asmt were also significantly increased (1.5-fold). These results imply that the transcription factor ABI4 may play a role in the interactions between ABA and melatonin.
2.5. ASMT as a Potential Target Gene for ABI4 under HL Stress
Finally, we tested whether MT metabolism and signaling genes were transcriptionally altered in ABA mutants (
Figure 4). Steady state levels of
TDC and
T5H transcripts were significantly elevated and that of
COMT, SNAT and
GPA1 reduced in
aba2 and
aba3 mutants relative to WT.
ASMT and
TDC transcript abundance was increased in
abi5.and
abi3 while the expression of
CAND2 was not significantly altered.
Under ABA treatment or ABA treatment coupled with HL, the expression of SNAT and COMT slightly decreased in WT and ABA mutants as compared to untreated samples. ABA+HL application also down regulated TDC and T5H in WT, aba2, aba3 and abi3 as compared to their stress induced levels, but had no impact in abi4 and abi5. These data suggest that ABA may act through ABI4 and ABI5 transcription factors in stress dependent regulation of TDC and T5H. It should be noted, however, that TDC and T5H have also been identified as participants in IAA biosynthesis. Therefore, ABA-dependent expression of these genes may be related to ABA/auxin interactions, given that the expression of downstream SNAT and COMT in the melatonin biosynthetic pathway followed that of WT in all ABA mutants tested upon ABA+HL.
ABA also negatively regulated ASMT transcript accumulation, under both mild and high light in WT plants and in aba2, aba3, abi3, and abi5. However, ASMT did not respond to ABA treatment in the abi4 mutant. These results suggest that ASMT can be a potential target gene for ABI4 thus supporting the notion that the transcription factor ABI4 is a key player in the interactions between ABA and melatonin.
`Furthermore, the
ABI4-disrupted mutant showed a 2-3-fold decrease in
M2H transcript accumulation following ABA treatment and a 2-fold increase in transcript levels compared to the dramatically ABA-induced
M3H expression in WT and other ABA mutants. These results confirm that ABI4 is an important factor in the regulation of the interactions between melatonin and ABA, However, the exact biological meaning of ABA-dependent shifts in the expression of melatonin catabolism genes remains to be clarified, taking into account that, on the one hand, 2-OHM triggers the production of ROS [
33], and on the other hand, that 3-OHM is a powerful antioxidant exhibiting even higher antioxidant activity, than melatonin [
5].
2.6. In Silico Promotor Analysis of ASMT Gene
A role for the ABI4 transcription factor in the regulation of MT synthesis implies that promoter regions of MT related genes contain
cis-regulatory elements that are potential binding sites for ABI4. ABI4 was shown to bind a number of DNA sequences including Coupling Element 1 (CE1) CACCG [
34], two ABE motifs GC(C/G)GCTT(T) [
35], B ELEMENT CGTGAT [
36] and S-box and S-box similar sequence CACYKSCA [
37]. A motif CCAC has also been suggested to bind ABI4 particularly when adjacent to, or overlapping with, a G box motif [
38]. In addition, DRE and ABRE core sequences have the potential to be bound by ABI4 [
35].
Detailed
in silico analysis of
ASMT promoter region within 1000 bp from ATG start codon (basal part), showed one potential ABI4-binding element CE1 (nt 467 to 472) (
Figure 6). The binding affinity of CE1 (CACCG) is considered higher than that observed with other DNA-binding sites (Finkelstein et al., 2011). We also identified one ABRE-like element (nt 742 to749) within basal part. Five more elements, one CE1 (nt 1953 to 1958) and four CCAC motifs (nt 1005 to 1010; 1261 to 1266; 1954 to 1959; and 2113 to 2118) were found in distal region of the
ASMT promoter as its maximum length can reach 2,483 bp according to the AGRIS database (
https://agris knowledgebase.org/AtcisDB/getpromseq.html?id=At4g35160). We therefore hypothesize that
ASMT could be a direct transcriptional target of ABI4.
It should be noted that sequence recognition by ABI4 may be more flexible than known canonical sequences. According to the results of [
29] many of the direct ABI4 transcriptional targets do not contain the previously characterized ABI4 binding motifs. Nevertheless, some of them were able to interact with ABI4 in electrophoretic mobility shift assays. Moreover, ABI4-regulated promoters could be synergistically induced by specific combinations of ABI4 and some bZIP factors, even when no canonical ABI4 binding sites were present. Given a relatively small number of direct transcriptional targets for ABI4 within proximal part, we can also hypothesize that
ASMT promoter can be regulated indirectly via
cis-regulatory elements recognized by ABI4-inducible transcription factors.
Functional characterization of the
ASMT promoter was carried out by testing the ability to drive expression of the reporter β-glucuronidase (
GUS) gene. To this end three 5′-deletion fragments (−1000 bp, −1847 bp, and −2483 bp) upstream of the
ASMT translation start codon were fused to the open reading frame of the
GUS reporter gene. ABA treatment (50 µM ABA for 6 h) of
ASMT pro::GUS fusions in the wild type background resulted in a considerable decrease in accumulation of
ASMT pro::GUS transcripts as compared to untreated leaf discs (
Figure S1). The signal also decreased in plants containing1000 bp
ASMT promoter fragment in
abi4 background. At the same time, plants containing the 1847 bp and 2483 bp fragments in
abi4 background did not differ from untreated plants when exposed to ABA suggesting that ABI4 was required for ABA induced decrease in
ASMT promoter activity. Given that distal region of the
ASMT promoter includes one CE1 and four CCAC motifs it was concluded that ABI4 can directly play role in the ability of the ASMT promoter to respond to ABA treatment.
3. Discussion
Overall, we found that melatonin and ABA are able to mutually influence the expression of the genes involved in their metabolism and signaling. In particular, the
ASMT which encodes enzyme of the final step of melatonin biosynthesis was suppressed by melatonin treatment under both mild and HL in the wild type. However, the expression remained virtually unchanged in the mutants with the disrupted genes for ABA synthesis and signaling under mild light and was even upregulated under combined MT+HL treatment. Therefore, deficiencies in ABA content or perception contribute to increased expression of
ASMT gene, which has been proposed to be stress-regulated [
39].
In parallel, the ASMT was downregulated by ABA treatment in the wild type plants and to a lesser extent in aba2, aba3, abi3 and abi5 mutants with the compromised ABA status. But the ASMT did not respond to ABA in abi4 with inactivated gene for transcription factor ABI4. We therefore hypothesize that application of ABA induces repression in ASMT promoter activity in WT via interaction with the ABI4 transcriptional factor, and this repression is absent in the abi4 mutant. These results are consistent with the notion that ASMT is a potential target gene for ABI4 which is probably involved in melatonin biosynthesis as a negative regulator.
It should be noted that ABI4 has been suggested to act as both a transcription activator and a repressor, which is determined by its interaction with other proteins, protein modification or binding to discrete DNA binding sites leading to either activation or repression of target genes [
35]
. ABI4 binds to a number of GC-enriched DNA sequences providing regulation of target genes involved in a large functional spectrum of activities. At least 2 Coupling Element1 (CE1) sequences (CACCG) are present in
ASMT promotor region and one ABRE-like element (TACGTGTA), in addition to 7 CCAC motifs which also correlated with
ABI4 signaling [
38].
Furthermore, in this study, we found that ASMT pro::GUS fusions did not respond to ABA treatment in abi4 mutant background, although ABA decreased ASMT promoter activity in WT background. These results imply that a functional ABI4 is necessary for ABA dependent regulation of ASMT expression. Consistent with the fact that ABI4 is involved in ABA-melatonin crosstalk, MT-insensitive cand2 and gpa1 exhibited increased steady state levels of ABI4 transcripts, suggesting that ABI4 is constitutively derepressed in genotypes defective in MT signaling.
Although MT may act directly as an anti-stress agent, the complexity of MT-dependent gene expression suggests that MT mediates many important functions through a melatonin receptor(s) and downstream signal transduction pathways in plants. To date, studies of the melatonin- CAND2-GPA1-signaling pathway have documented its involvement in osmotic, salt, drought and HL stress tolerance, in addition to biotic stresses. Here we have showed that an intricate network of ABA related genes under HL, at least in part, depends on CAND2. This is particularly relevant for ABA signaling, since, in contrast to WT, the expression levels of ABA signaling genes in cand2 and gpa1 mutants did not respond significantly to MT treatment.
The identification of molecular links between MT and ABA synthesis and signaling provides a basis for understanding how disruptions of these two regulators can feed back to influence each other. According to results of this study, inactivating components of MT signaling circuit results in compensatory effects in ABA signaling. On the other hand, deficiencies in ABA synthesis and signaling entail increased expression of MT synthesis genes and elevated steady state MT levels. The existence of such compensatory mechanisms gives rise to a certain parallelism in the action of these two regulatory molecules and can have a broad impact beyond the immediate activity of the inactivated components.
In this context, ABA-dependent modulations in the expression of melatonin catabolism are of special interest. ABA treatment promoted downregulation of M2M, which may result in decreased levels of 2-OM with its ROS-inducing mode of action and consequently decreased ROS production. In parallel, M3H upregulation, particularly in the abi4 mutant, may trigger 3-OM overproduction and enhance antioxidant activity. Therefore, the ABA-induced decrease in MT content, which can be regarded as a manifestation of the antagonistic relationship between ABA and MT, may actually lead to an increase in antioxidant capacity and stress resistance
The functional redundancy in the effects of MT and ABA allows the plant to trigger alternative pathways in response to environmental challenges or developmental demands. At the same time, they provoke antagonism between ABA and MT under both control and stress conditions. As we have already outlined, examples of antagonistic relations between MT and ABA were shown for a number of plant species [
11,
12,
13,
14,
15,
16,
17]. On the other hand, synergism in MT and ABA effects was also demonstrated in several studies [
18,
19,
20,
21,
22]. Moreover, dual role of melatonin as a signaling molecule in ABA-melatonin interplay was revealed during seed formation and seed germination of
Arabidopsis. Knock-out mutants for melatonin receptor
PMTR1 contained higher ABA concentrations in developing seeds, but accumulated lower ABA levels in dry and imbibed seeds than the wild-type Col-0 [
40]. Hence, multiplicity of possible functions defined by ABA-melatonin crosstalk imply multivariant responses depending on physiological state and genetic background.
The interactions between melatonin and ABA are likely to be integrated with other hormone signals. Mutual impact of ABA, gibberellic acid (GA) and MT has been shown for cucumber. Exogenous melatonin increased the expression of ABA catabolism genes (monooxygenase, CYP707A1 and CYP707A2) and GA biosynthesis genes (GA20ox and GA3ox), and also suppressed the ABA biosynthesis gene 9-cis-epoxycarotenoid dioxygenase (NCED2). This led to a decrease in ABA levels and an increase in GA
3 and GA
4 content, and accelerated seed germination [
15]. A similar picture was observed during the germination of cotton seeds (
Gossypium hirsutum L.) under salt stress conditions [
41].
In ryegrass (
Lolium perenne L.), exogenous melatonin reduced the ABA content by suppressing the expression of biosynthesis (
LpZEP and
LpNCED1), downregulated ABA signaling genes (
LpABI3 and
LpABI5), but activated cytokinin (CK) synthesis genes (
LpIPT2 and
LpOG1), thereby increasing the content of endogenous CK and activating CK signaling circuits [
42]. In our prior research we showed that melatonin may act synergistically with CK in
Arabidopsis via melatonin-mediated activation or repression of the CK synthesis and signaling genes [
16]. Conversely, CK was involved in expression regulation of the genes for melatonin metabolism and signal transduction, with the melatonin biosynthesis gene
ASMT being a key component in the crosstalk between the CK and MT metabolic pathways. In this study we revealed that
ASMT is a potential target for ABI4 in ABA-melatonin intersection. Therefore, fluctuation in melatonin activity induced by various hormones may, at least in part, be mediated by ASMT. These data are consistent with RNA sequencing and qRT-PCR data obtained by [
43], which showed that in the walnut,
Juglans regia, members of the
ASMT gene family participate in regulatory networks of phytohormones such as ABA, salicylic acid and CK, regulating flower development and stress tolerance. In addition,
in silico analysis of the promoter regions revealed 11 hormone-response motifs including the ABA response element ABRE, auxin response element AuxRR-core, jasmonic acid (JA) response elements CGTCA and TGACG gibberellin response element P-box and salicylic acid response element TCA.
Hormone-like behavior of MT while interacting with classical hormones, as well as participation of PMTR1/CAND2 receptor in such reactions, makes it possible to classify melatonin as a new phytohormone especially when it acts in physiologically effective concentrations using high-affinity binding sites. At the same time, MT is believed to emerge as the first-line antioxidant capable of operating in cardinally higher concentrations [
44,
45]. The evolution in modes of action and the acquisition of secondary hormone-like properties have required the incorporation of melatonin into highly interconnected web involved in hormone metabolism and signaling [
46]. This contributed to the generation of joint pathways that ensured the inclusion of melatonin in the plant hormone system. The interplay between ABA and melatonin, resulting from a considerable overlap in effects, is an example of such integration, with MT-dependent regulation of ABA genes via CAND2/GPA1 signaling circuit and ABI4/ASMT acting as elements of this bond.