Submitted:
14 October 2024
Posted:
15 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Literature Review
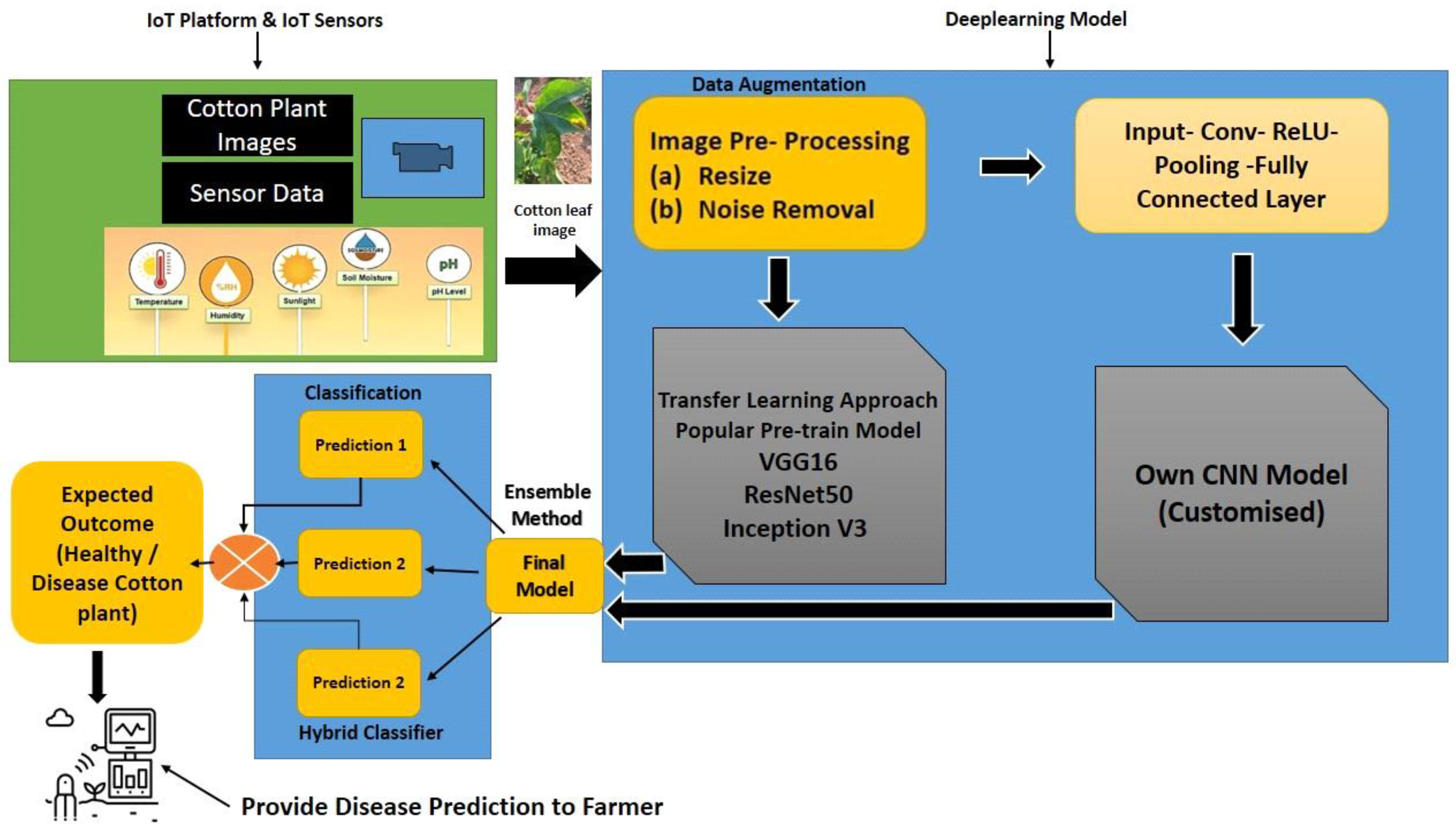
Proposed System
Data availability
Data Augmentation
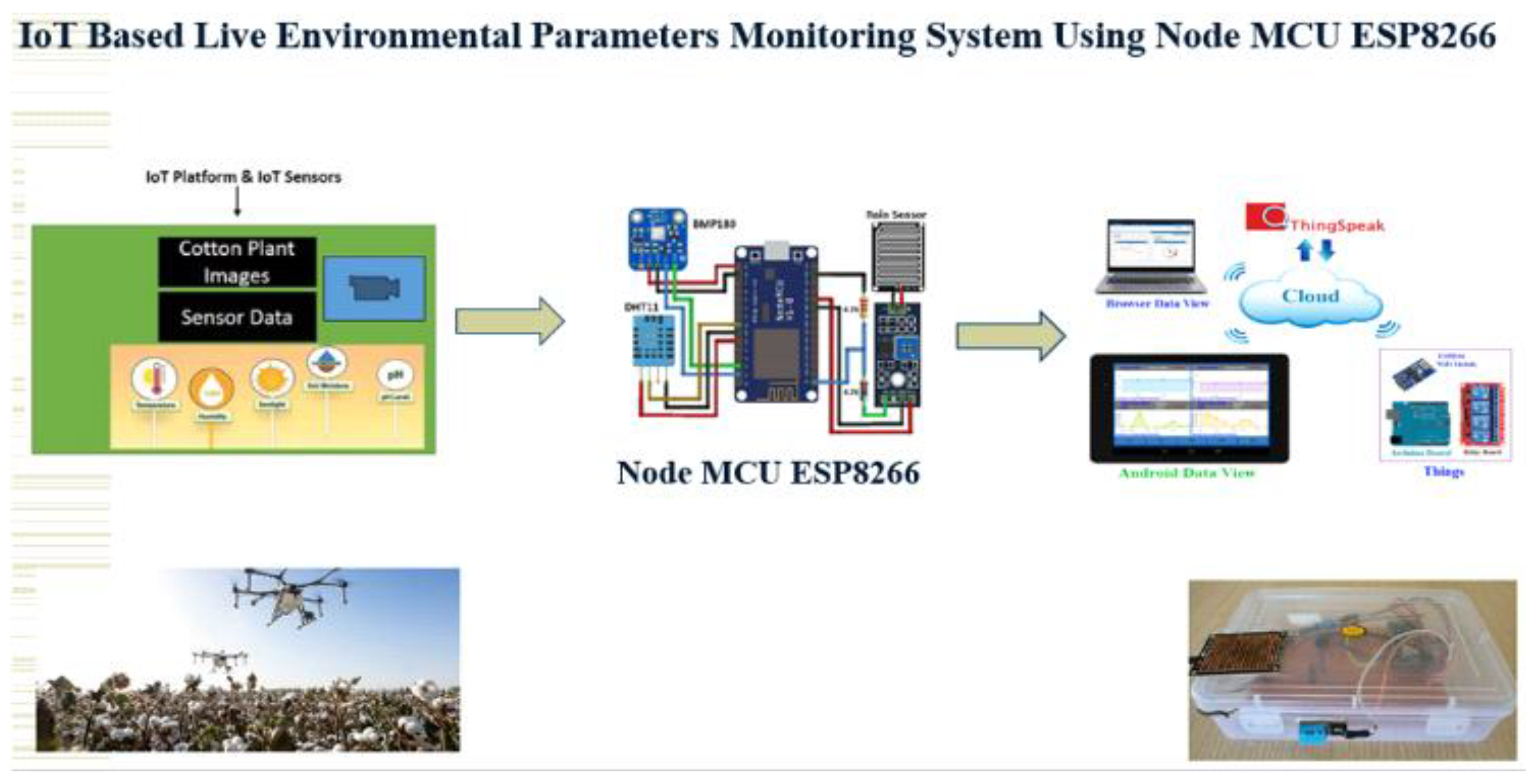
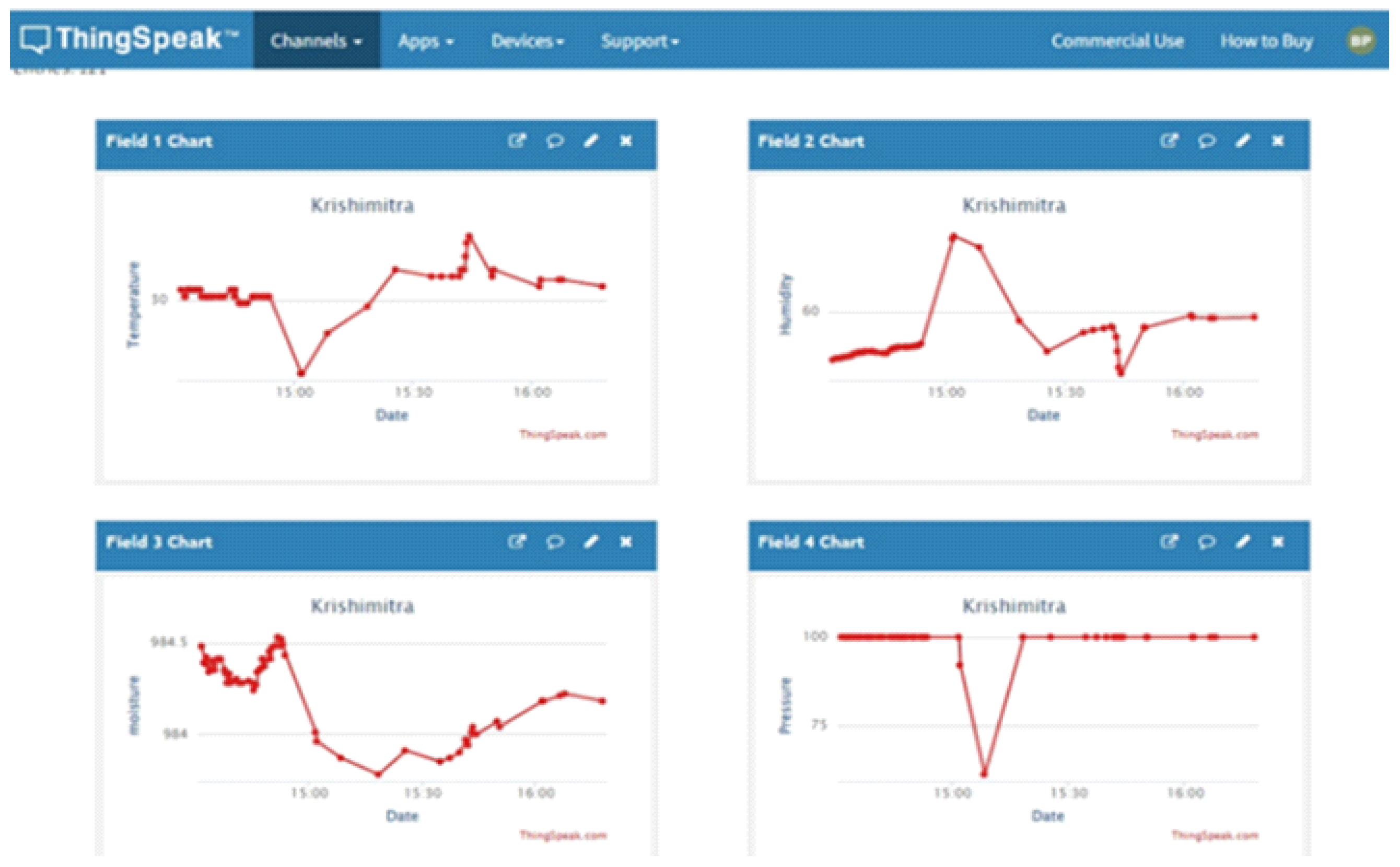
Role of IoT (Internet of Things) for Cotton Plant Diseases Detection
Components Details:
- climatic circumstances;
- improper use of fertilizers and insecticides;
- pest and disease attacks;
- Minimal revenue for small-scale farmers
- Low cost because of problems with quality
- Increased labor costs
- Depletion of soil
IoT Plays Major role for Cotton Farming
Classification using the Ensemble Learning Model
Result And Performance Analysis
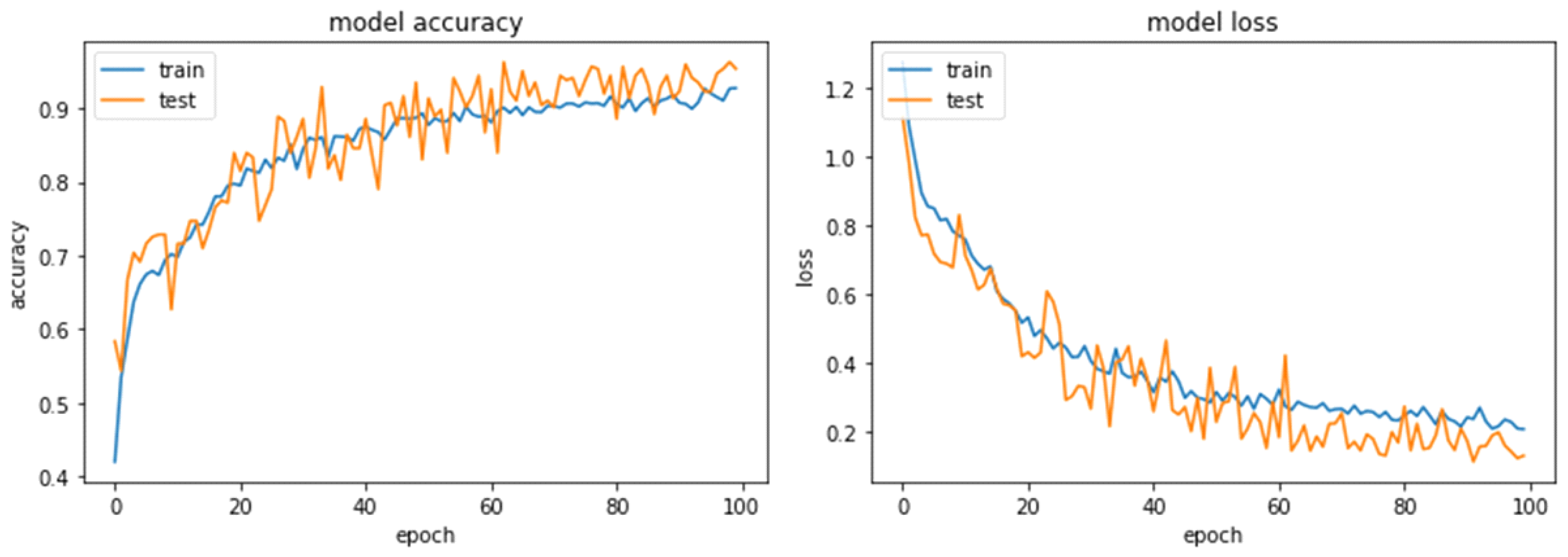
Conclusion
References
- Sharma, P. , et al. KrishiMitr (Farmer’s Friend): Using Machine Learning to Identify Diseases in Plants. Proceedings of the 2018 IEEE International Conference on Internet of Things and Intelligence System (IoTaIS); 2018.
- Adhao, A.S.; Pawar, V.R. Automatic Cotton Leaf Disease Diagnosis and Controlling Using Raspberry Pi and IoT. Intelligent Communication and Computational; 2019.
- Prashar, K.; Talwar, R.; Kant, C. CNN based on Overlapping Pooling Method and Multi-layered Learning with SVM & KNN for American Cotton Leaf Disease Recognition. Proceedings of IEEE; 2019.
- Alves, A.N.; Souza, W.S.R.; Borges, D.L. Cotton pest’s classification in field-based images using deep residual networks. Comput. Electron. Agric. 2020, 174, 105488. [Google Scholar] [CrossRef]
- Afroze, A.S.; Beham, M.P.; Tamilselvi, R.; Maraikkayar, S.M.A.; Rajakumar, K. Cotton Leaf Disease Detection Using Texture and Gradient Features. Int. J. Eng. Adv. Technol. 2019, 9, 700–703. [Google Scholar] [CrossRef]
- Nerkar, B.; Talbar, S. Improving Leaf Disease Detection and Localization Accuracy Using Bio-Inspired Machine Learning. Applied Computer Vision and Image Processing. 2020.
- Patki, S.S.; Sable, G.S. Cotton Leaf Disease Detection & Classification using Multi SVM. Int. J. Adv. Res. Comput. Commun. Eng. 2017, 5, 165–168. [Google Scholar]
- Bhong, V.S.; Pawar, B.V. Study and Analysis of Cotton Leaf Disease Detection Using Image Processing. Int. J. Adv. Res. Sci. Eng. Technol. 2017, 3, 1447–1454. [Google Scholar]
- Dubey, Y.K.; Mushrif, M.M.; Tiple, S. Superpixel based roughness measure for cotton leaf diseases detection and classification. Proceedings of the 2018 4th International Conference on Recent Advances in Information Technology (RAIT); Dhanbad, India; 2018.
- Bodhe, K.D.; Taiwade, H.V.; Yadav, V.P.; Aote, N.V. Implementation of Prototype for Detection & Diagnosis of Cotton Leaf Diseases using Rule Based System for Farmers. Proceedings of the 2018 3rd International Conference on Communication and Electronics Systems (ICCES); Coimbatore, India; 2018.
- Jenifa, R.; Ramalakshmi, R.; Ramachandran, V. Cotton Leaf Disease Classification using Deep Convolution Neural Network for Sustainable Cotton Production. Proceedings of the IEEE International Conference on Clean Energy and Energy Efficient Electronics Circuit for Sustainable Development (INCCES); Krishnankoil, India; 2019.
- Jenifa, R.; Ramalakshmi, R.; Ramachandran, V. Classification of Cotton Leaf Disease Using Multi-Support Vector Machine. Proceedings of the 2019 IEEE International Conference on Intelligent Techniques in Control, Optimization and Signal Processing (INCOS); Tamilnadu, India; 2019.
- Prashar, K.; Talwar, R.; Kant, C. CNN based on Overlapping Pooling Method and Multi-layered Learning with SVM & KNN for American Cotton Leaf Disease Recognition. Proceedings of the International Conference on Automation, Computational and Technology Management (ICACTM); London, UK; 2019.
- Kumari, U.; Prasad, S.J.; Mounika, G. Leaf Disease Detection: Feature Extraction with K-means clustering and Classification with ANN. Proceedings of the 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC); Erode, India; 2019.
- Plant Disease Handbook: Cotton. Texas A&M AgriLife Extension Service. Available from:https://plantdiseasehandbook.tamu.edu/industry-specialty/fiber-oil-specialty/cotton/.
- Sarangdhar, A.; Pawar, V.R. Machine learning regression technique for cotton leaf disease detection and controlling using IoT. Proceedings of the 2017 International Conference on Electronics, Communication and Aerospace Technology (ICECA); Coimbatore, India; 2017.
- Dubey, Y.K.; Mushrif, M.M.; Tiple, S. Superpixel based roughness measure for cotton leaf diseases detection and classification. Proceedings of the 2018 4th International Conference on Recent Advances in Information Technology (RAIT); Dhanbad, India; 2018.
- Bodhe, K.D.; Taiwade, H.V.; Yadav, V.P.; Aote, N.V. Implementation of Prototype for Detection & Diagnosis of Cotton Leaf Diseases using Rule Based System for Farmers. Proceedings of the 2018 3rd International Conference on Communication and Electronics Systems (ICCES); Coimbatore, India; 2018.
- Bhimte, N.R.; Thool, V.R. Diseases Detection of Cotton Leaf Spot Using Image Processing and SVM Classifier. Proceedings of the 2018 Second International Conference on Intelligent Computing and Control Systems (ICICCS); Madurai, India; 2018.
- Shah, N.; Jain, S. Detection of Disease in Cotton Leaf using Artificial Neural Network. Proceedings of the 2019 Amity International Conference on Artificial Intelligence (AICAI); Dubai, UAE; 2019.
- Ahmed, N.; Amin, I.; Mansoor, S. Cotton Leaf Curl Disease (Geminiviridae). Encyclopedia of Virology (Fourth Edition). 2021.
- Mushtaq, R.; Shahzad, K.; Bashir, A. Isolation of biotic stress resistance genes from cotton (Gossypium arboreum) and their analysis in model plant tobacco (Nicotiana tabacum) for resistance against cotton leaf curl disease complex. J. Virol. Methods 2019, 276, 113760. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Iqbal, Z.; Amin, I. Real-time quantitative PCR assay for the quantification of virus and satellites causing leaf curl disease in cotton in Pakistan. J. Virol. Methods 2017, 248, 54–60. [Google Scholar] [CrossRef]
- Akmal, M.; Baig, M.S.; Khan, J.A. Suppression of cotton leaf curl disease symptoms in Gossypium hirsutum through over expression of host-encoded miRNAs. J. Biotechnol. 2017, 263, 21–29. [Google Scholar] [CrossRef]
- Briddon, R.W.; Akbar, F.; Mansoor, S. Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2015, 186, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.; Mansoor, S.; Amin, I. Diversity of alphasatellites associated with cotton leaf curl disease in Pakistan. Virol. Rep. 2016, 6, 41–52. [Google Scholar] [CrossRef]
- Zhang, J.H.; Kong, F.; Zhai, Z.F. Automatic image segmentation method for cotton leaves with disease under natural environment. J. Integr. Agric. 2018, 17, 1800–1814. [Google Scholar] [CrossRef]
- Khattab, A.; Habib, S.E.; Ismail, H.; Zayan, S.; Fahmy, Y.; Khairy, M.M. An IoT-based cognitive monitoring system for early plant disease forecast. Comput. Electron. Agric. 2019, 166, 105028. [Google Scholar] [CrossRef]
- He, D.F.; Zhao, X.; Zhang, R. Genetic variation in LBL1 contributes to depth of leaf blades lobes between cotton subspecies, Gossypium barbadense and Gossypium hirsutum. J. Integr. Agric. 2018, 17, 2394–2404. [Google Scholar] [CrossRef]
- Sarwar, M.W.; Riaz, A.; Mubin, M. Homology modeling and docking analysis of ßC1 protein encoded by Cotton leaf curl Multan betasatellite with different plant flavonoids. Heliyon 2019, 5, e01303. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Suassuna, N.D.; Bettiol, W. Management of Ramularia leaf spot on cotton using integrated control with genotypes, a fungicide and Trichoderma asperellum. Crop. Prot. 2017, 94, 28–32. [Google Scholar] [CrossRef]
- Ashry, N.A.; Ghonaim, M.M.; Mogazy, A.M. Physiological and molecular genetic studies on two elicitors for improving the tolerance of six Egyptian soybean cultivars to cotton leaf worm. Plant Physiol. Biochem. 2018, 130, 224–234. [Google Scholar] [CrossRef]
- Celik, Y.; Ulker, E. An Improved Marriage in Honey Bees Optimization Algorithm for Single Objective Unconstrained Optimization. Sci. World J. 2013, 2013, 370172. [Google Scholar] [CrossRef]
- Mareli, M.; Twala, B. An adaptive Cuckoo search algorithm for optimisation. Appl. Comput. Inform. 2018, 14, 107–115. [Google Scholar] [CrossRef]
- Chuang, K.S.; Tzeng, H.L.; Chen, S.; Wu, J.; Chen, T.J. Fuzzy c-means clustering with spatial information for image segmentation. Comput. Med. Imaging Graph. 2006, 30, 9–15. [Google Scholar] [CrossRef]
- Ershad, S.F. Texture Classification Approach Based on Combination of Edge & Co-occurrence and Local Binary Pattern. Proceedings of the Int'l Conf. IP, Comput Vision, and Pattern Recognition; 2011.
- ChandraPrabha, K.; Bharathi, C. Texture Analysis using GLCM & GLRLM Feature Extraction Methods. Int. J. Res. Appl. Sci. Eng. Technol. 2019, 7, 2059–2064. [Google Scholar]
- Wang, J.H.; Liu, T.W.; Luo, X.; Wang, L. An LSTM Approach to Short Text Sentiment Classification with Word Embeddings. Proceedings of the 2018 Conference on Computational Linguistics and Speech Processing; 2018.
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep learning for image-based cassava disease detection. Front. Plant Sci. 2017, 8, 1852. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, L.G.; Araujo, R.M.; Nachtigall, G.R. Classification of apple tree disorders using convolutional neural networks. Proceedings of the 2016 IEEE 28th International Conference on Tools with Artificial Intelligence (ICTAI); 2016. p. 472-6.
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Cotton Disease Management. Government of India. Available from: https://farmer.gov.in/imagedefault/ipm/cotton.pdf.
- Ahmed, S. , et al. Cotton Production and Bacterial Blight Management in Pakistan: A Review. Pak. J. Phytopathol. 2017, 29, 251–270. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef]
- Prajapati, B.S.; Shah, J.; Dabhi, V.K. A survey on detection and classification of cotton leaf diseases. In: 2016 IEEE International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT). 2016.
- Revathi, P.; Hemlata, M. Advance computing enrichment evaluation of cotton leaf spot disease detection using image edge detection. In: 2012 IEEE International Conference on Computing, Communication and Networking Technologies (ICCCNT). 2012.
- Rothe, P.R.; Kshirsagar, P.R. Cotton leaf disease identification using pattern recognition techniques. In: 2015 International Conference on Pervasive Computing (ICPC). 2015.
- Shakeel, W.; Rehman, A.; Razaque, A. , et al. Early detection of Cercospora cotton plant disease by using machine learning technique. In: 2020 IEEE 30th International Conference on Computer Theory and Applications (ICCTA). Alexandria, Egypt; 2020:12-14.
- Warne, P.; Ganorkar, S. Detection of diseases on cotton leaves using K-mean clustering method. Int. Res. J. Eng. Technol. 2015, 2, 425–431. [Google Scholar]
- Rothe, P.R.; Kshirsagar, P.R. SVM-based classifier system for recognition of cotton leaf diseases. Int. J. Emerg. Technol. Comput. Appl. Sci. 2014, 8, 427–432. [Google Scholar]
- Rothe, P.R.; Kshirsagar, P.R. A study and implementation of active contour model for feature extraction: With diseased cotton leaf as example. Int. J. Curr. Eng. Technol. 2014, 4, 812–816. [Google Scholar]
- Patil, S.; Zambre, R. Classification of cotton leaf spot disease using support vector machine. Int. J. Eng. Res. 2014, 3, 1511–1514. [Google Scholar]
- Chaudhari, V.; Patil, C.Y. Disease detection of cotton leaves using advanced image processing. Int. J. Adv. Comput. Res. 2014, 4, 653. [Google Scholar]
- Revathi, P.; Hemalatha, M. Classification of cotton leaf spot diseases using image processing edge detection techniques. In: Emerging Trends in Science, Engineering and Technology (INCOSET), 2012 International Conference. 2012:169-173.
- Adsule, B.; Bhattad, J. Leaves classification using SVM and NN for disease detection. Int. J. Innov. Res. Comput. Commun. Eng. 2015, 3, 5488–5495. [Google Scholar]
- Abhang, D.; Autade, P.; Bhokare, S.; Dawange, P. A survey on, analysis and detection of disease on cotton leaves. Int. J. Adv. Res. Innov. Ideas Educ. 2015, 4, 236–243. [Google Scholar]
- Arjunagi, S. , et al. A survey on detection and computing the amount of plant diseases using image processing. IAETSD J. Adv. Res. Appl. Sci. 2017, 4, 344–354. [Google Scholar]
- Sartin, M.; Da Silva, A.; Kappas, C. Image segmentation with artificial neural network for nutrient deficiency in cotton crop. J. Comput. Sci. 2014, 10, 1084–1093. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. A review of the use of convolutional neural networks in agriculture. J. Agric. Sci. 2018, 156, 312–322. [Google Scholar] [CrossRef]
- Gawas, M.; Patil, H.; Govekar, S.S. An integrative approach for secure data sharing in vehicular edge computing using blockchain. Peer-to-Peer Netw. Appl. 2021, 14, 2840–2857. [Google Scholar] [CrossRef]
- Hebbar, N.; Patil, H.Y.; Agarwal, K. Web powered CT scan diagnosis for brain hemorrhage using deep learning. In: 2020 IEEE 4th Conference on Information & Communication Technology (CICT). 2020:1-5. [CrossRef]
- Patil, P.; Patil, H. X-ray imaging based pneumonia classification using deep learning and adaptive clip limit based CLAHE algorithm. In: 2020 IEEE 4th Conference on Information & Communication Technology (CICT). 2020:1-4. [CrossRef]
- Barua, S.; Patil, H.; Desai, P.D.; Arun, M. Deep learning-based smart colored fabric defect detection system. In Applied Computer Vision and Image Processing. Advances in Intelligent Systems and Computing; Iyer, B., Rajurkar, A., Gudivada, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1155, pp. 333–342. [Google Scholar] [CrossRef]
- Patil, P.; Ranganathan, M.; Patil, H. Ship image classification using deep learning method. In Applied Computer Vision and Image Processing. Advances in Intelligent Systems and Computing; Iyer, B., Rajurkar, A., Gudivada, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1155, pp. 343–351. [Google Scholar] [CrossRef]
- Patil, B.V.; Patil, P.S. Computational method for cotton plant disease detection of crop management using deep learning and internet of things platforms. In: Evolutionary Computing and Mobile Sustainable Networks: Proceedings of ICECMSN 2020. Springer; 2021.
- Patil, H.Y.; Chandra, A. Deep learning based kinship verification on KinFaceW-I dataset. In: TENCON 2019 - 2019 IEEE Region 10 Conference (TENCON). 2019:2529-2532. [CrossRef]
- iKisan. Cotton disease management. Available from: https://www.ikisan.com/ap-cotton-disease-management.htm. Accessed June 21, 2024.


| Sr. No. | Category of Diseases | Examples |
|---|---|---|
| 1 | Bacterial diseases | Bacterial Blight Crown Gall |
| 2 | Fungal diseases | Anthracnose Leaf Spot Alternaria |
| 3 | Viral diseases | Leaf Curl Leaf Crumple Leaf Roll |
| 4 | Diseases(Due to insects) | White flies Leaf insects |
| Sr. No. | Author [Citation] | Dataset Used | Adopted Methodology | Features | Challenges |
|---|---|---|---|---|---|
| 1 | Parul Sharma et al. [1] | PlantVillage database, field-based dataset, Internet image |
Convolutional neural network (CNN) | High accuracy (>93%) was obtained with very noisy images | Required more disease coverage |
| 2 | Adhao and Pawar [2] | Images are Captured using digital camera | Support Vector Machine (SVM) | Cost-effective and independent | Lower overall accuracy for disease detection (83.26%) Highly prone to noise |
| 3 | Prashar et al. [3] | 40 infected images capture through digital camera. | KNN and SVM | Accurately localize the disease region | Higher time complexity |
| 4 | Alves et al. [4] | Live field-based images & Internet image. | deep residual networks | Achieved the highest accuracy higher F1-Score |
Lower learning rate |
| 5 | Afroze et al. [5] | Images of normal and diseased leaves are gathered from the Internet and used in the field. | Local Binary Pattern (LBP) is used to extract the gradient feature and texture of the K-Nearest neighbor classifier. | Reduced computing time and increased classification accuracy | Reduced specificity and sensitivity |
| 6 | Nerkar and Talbar [6] | Images are Captured using digital camera | bio-inspired ML-based classification algorithm | Improved classification accuracy Increases the performance of system based on disease localization. |
Higher computational complexity in terms of cost and time |
| 7 | Patki et al. [7] | 103 infected images capture through digital camera | Multi SVM (Multi Support Vector Machine) classifier | Higher detection accuracy | Higher average recognition time |
| 8 | Bhong et al. [8] | images capture through digital camera | K-means technique for clustering | straightforward and precise | The identification procedure has a slower recognition rate. |
| 9 | Dubey et al. [9] | images capture through digital camera | Support Vector Machine (SVM) | Higher classification accuracy | Higher testing time |
| 10 | Bodhe et al. [10] |
images capture through digital camera | Rule Based System | Higher detection and diagnosis Higher accuracy in classification |
Very time-consuming and expensive |
| 11 | Jenifa et al. [11] | 600 images capture through digital camera | Deep Convolution Neural Network | Improved classification rate | Highly prone to noise Higher misclassification |
| 12 | Jenifa et al. [12] | Real time images capture by camera | Multi-Support Vector Machine | less prone to over fits | suffers from multiple local minima |
| 13 | Dong et al. [13] | Own database formed for 5 diseases. | CBR and fuzzy logic | get a high success percentage for diagnostics | Inadequate database size and manual selection reduce the effectiveness of the diagnosis. |
| 14 | Kumari et al. [14] | plant village data base | K-means clustering and ANN | Improved average classification accuracy (92.5 %.) |
Samples are misclassified |
| 15 | Sarangdhar and Pawar [15] | Images capture through Nikon digital camera | Support Vector Machine | Higher overall classification accuracy | Highly prone to noise |
| Soil Type | Soil Moisture Level |
|---|---|
| Fine (Clay) | Below 60% |
| Medium (Black & loamy) | Below 70% |
| Coarse (Sandy) | Below 80% |
| Temperature | Humidity | Soil Moisture | Rail fall | Symptoms | Disease type |
|---|---|---|---|---|---|
| 240-280 C | High | 80-90% | YES | Yellow to brown colored cotyledon leaves turn brown and drop off | Fussarium wilt |
| 350-430 C | High | 50-60% | YES | Sudden and complete wilting of the plant | Root rot |
| 290-330 C | High | 80-90% | YES | Small reddish-colored spots and cotyledons. Water soaked small | Boll spotting |
| 250-300 C | Very high | 60-70% | 25.4 to 76.2 mm | Angular leaf spots on leaf on steam and leaf- lesions on young leaves | Angular leaf spots |
| Temperature | Humidity | Soil Moisture | Rail fall | Symptoms | Disease type |
| 100-300 C | Very high | 80-90% | YES | Brown rounded or irregular spots on leaf cracked centres cause canker on steam | Alternaria leaf spot |
| 250-300 C | Very high | 60-70% | YES | Irregular translucent spots on leaf- leaves become yellowish brown and finally fall off | Bacterial blight |
| 350-430 C | High | 50-60% | NO | Purple dark brown or blakish borders and white centers | Cercospora |
| Parameters | Proposed Own CNN model |
|---|---|
| Training Accuracy | 0.92 |
| Training Loss | 0.209 |
| Testing Accuracy | 0.95 |
| Testing Loss | 0.132 |
| Training Time | 29s 466ms/step |
| Testing Time | 29s 467ms/step |
| Epoch | 100 |
| Performance (%) |
VGG 16 |
ResNet 50 |
Inception V3 |
Proposed (Stack ensemble) |
|---|---|---|---|---|
| Accuracy | 0.9844 | 0.9881 | 0.9930 | 0.9966 |
| Precision | 0.9380 | 0.9525 | 0.9728 | 0.9866 |
| F1-score | 0.9382 | 0.9527 | 0.9726 | 0.9869 |
| MCC | 0.9290 | 0.9457 | 0.9683 | 0.9849 |
| Specificity | 0.9911 | 0.9932 | 0.9960 | 0.99811 |
| Sensitivity | 0.9380 | 0.9529 | 0.9723 | 0.9871 |
| RMSE | 0.0704 | 0.060 | 0.0453 | 0.0346 |
| MSE | 0.0049 | 0.0036 | 0.0020 | 0.0422 |
| MAE | 0.1889 | 0.1373 | 0.0766 | 0.0422 |
| PPV | 0.9380 | 0.9525 | 0.9728 | 0.98662 |
| NPV | 0.9911 | 0.9932 | 0.9960 | 0.99810 |
| FPR | 0.0088 | 0.0067 | 0.0039 | 0.99818 |
| FNR | 0.0614 | 0.0470 | 0.0276 | 0.01280 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





