Submitted:
11 October 2024
Posted:
14 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
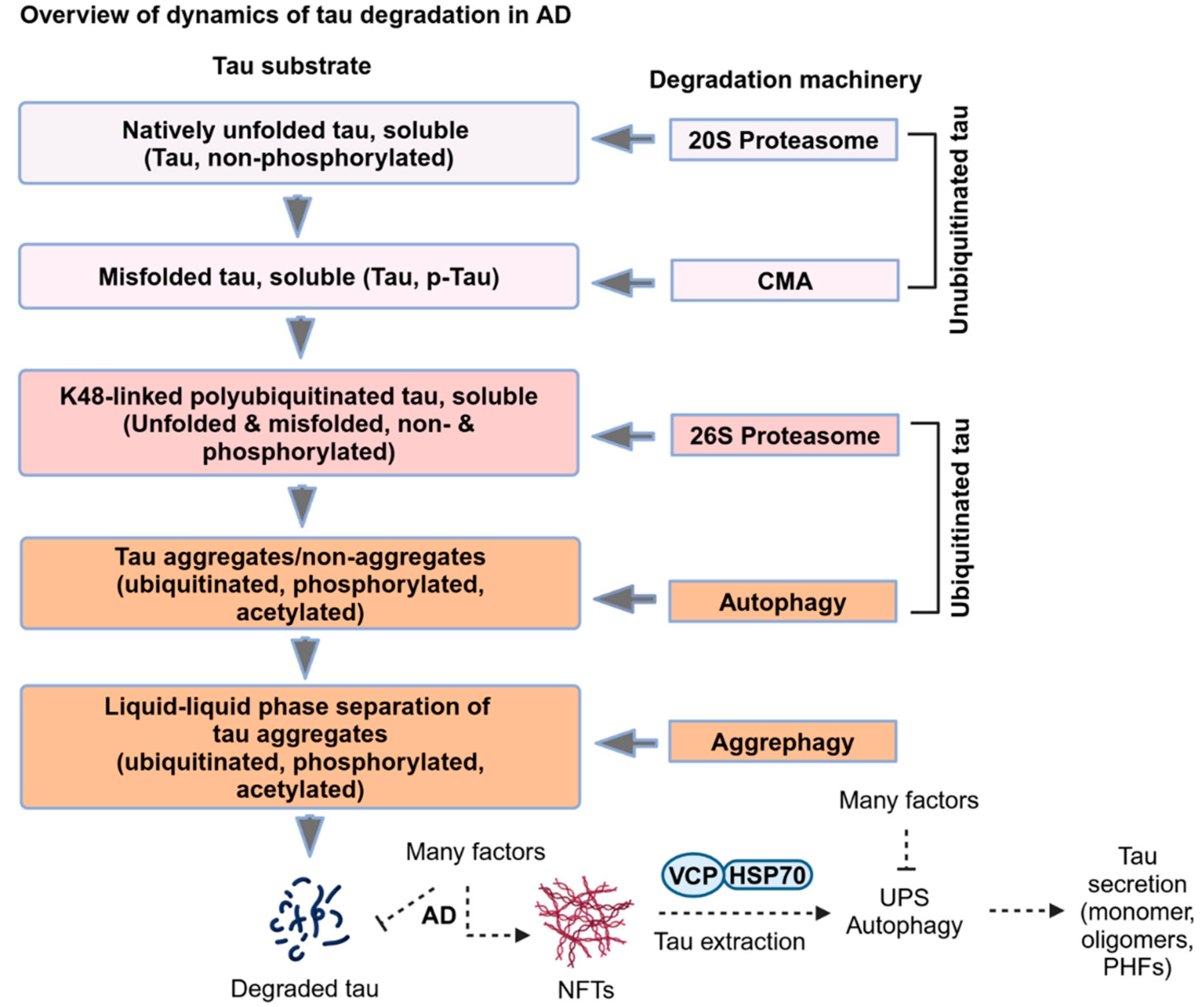
2. Dynamics of Tau Degradation in AD
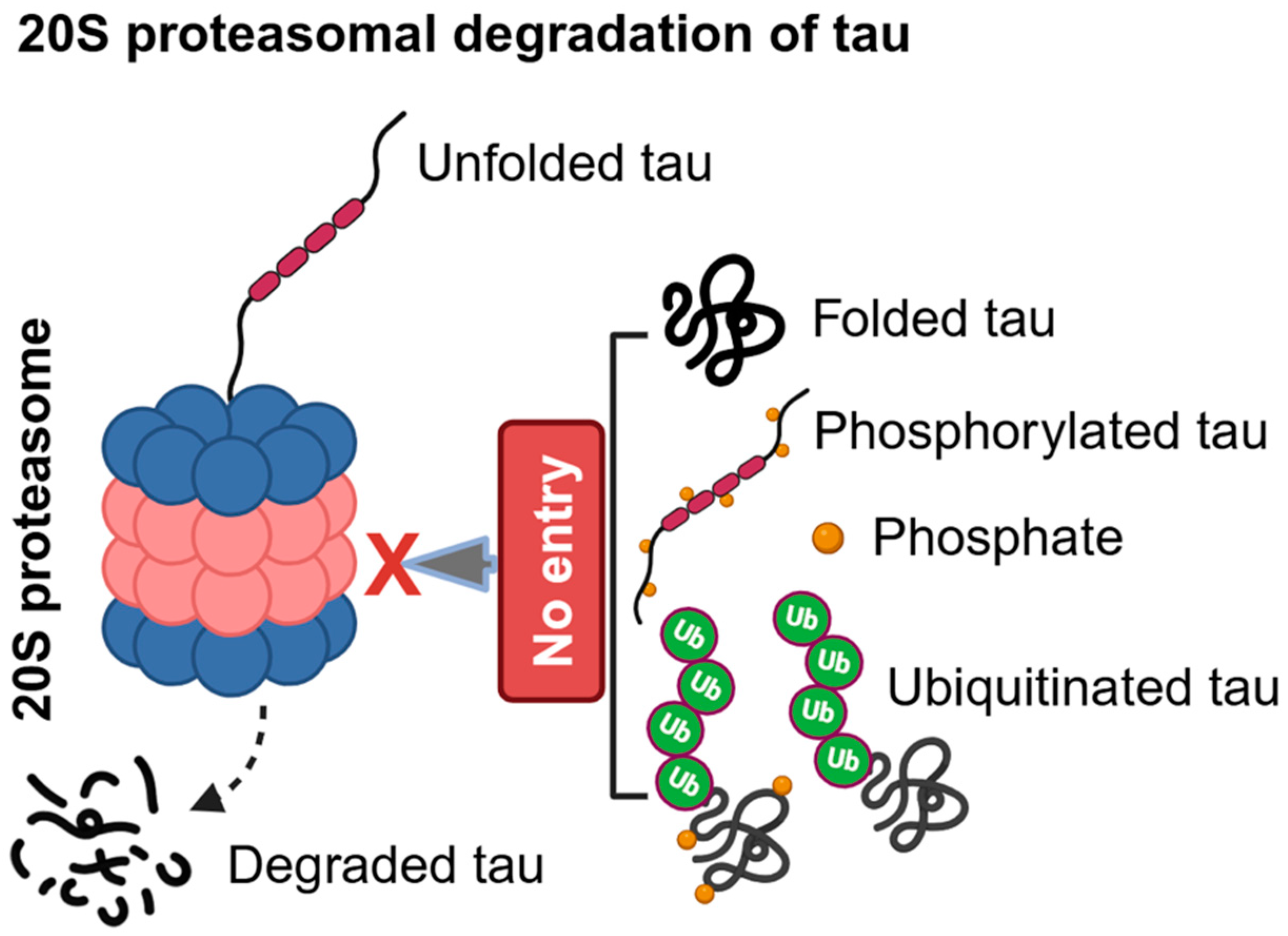
2.1. Ubiquitin-Independent Proteasomal Degradation of Tau in AD
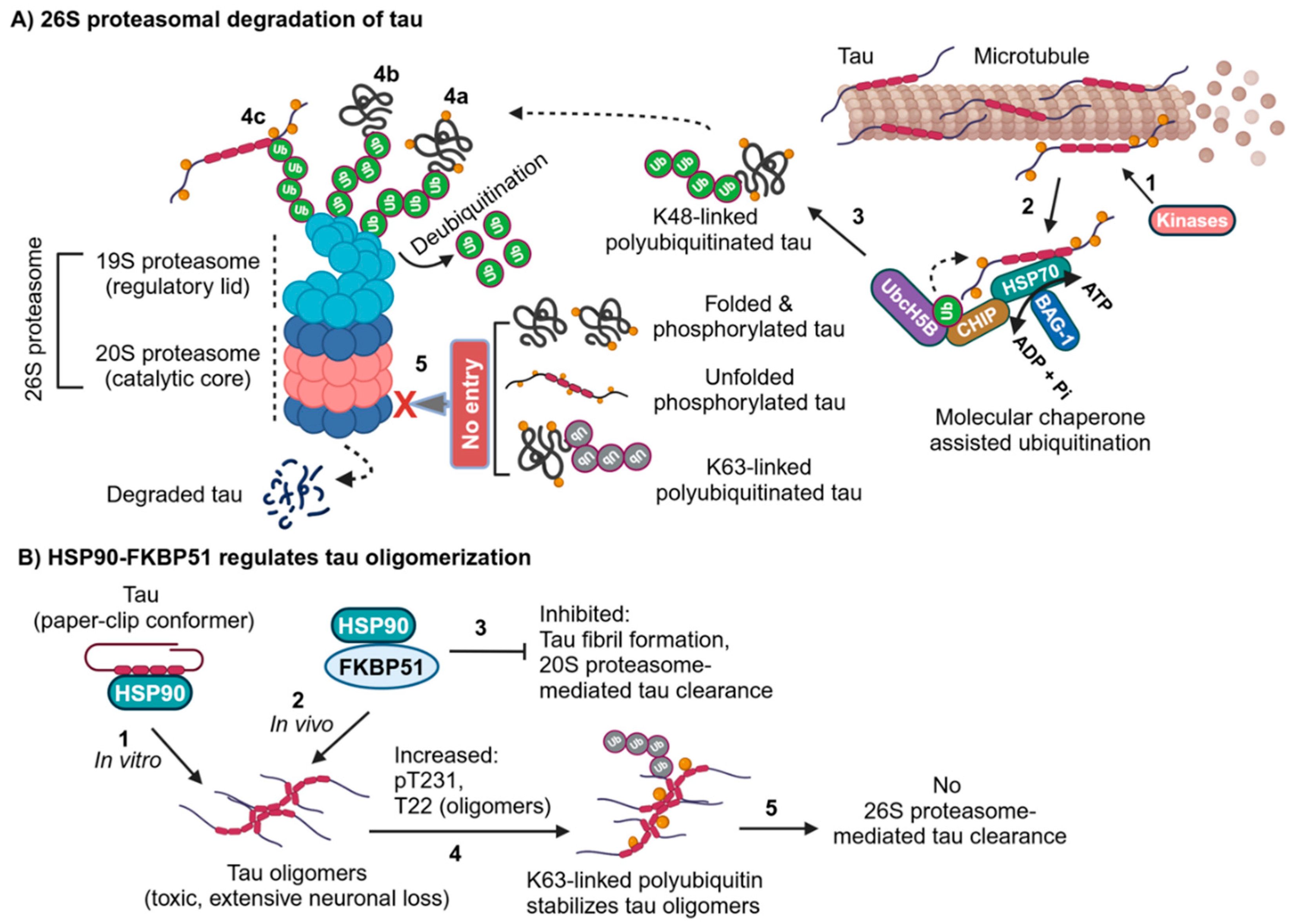
2.2. Molecular Chaperones Assisted Ubiquitin-Dependent Proteasomal Degradation of Tau in AD
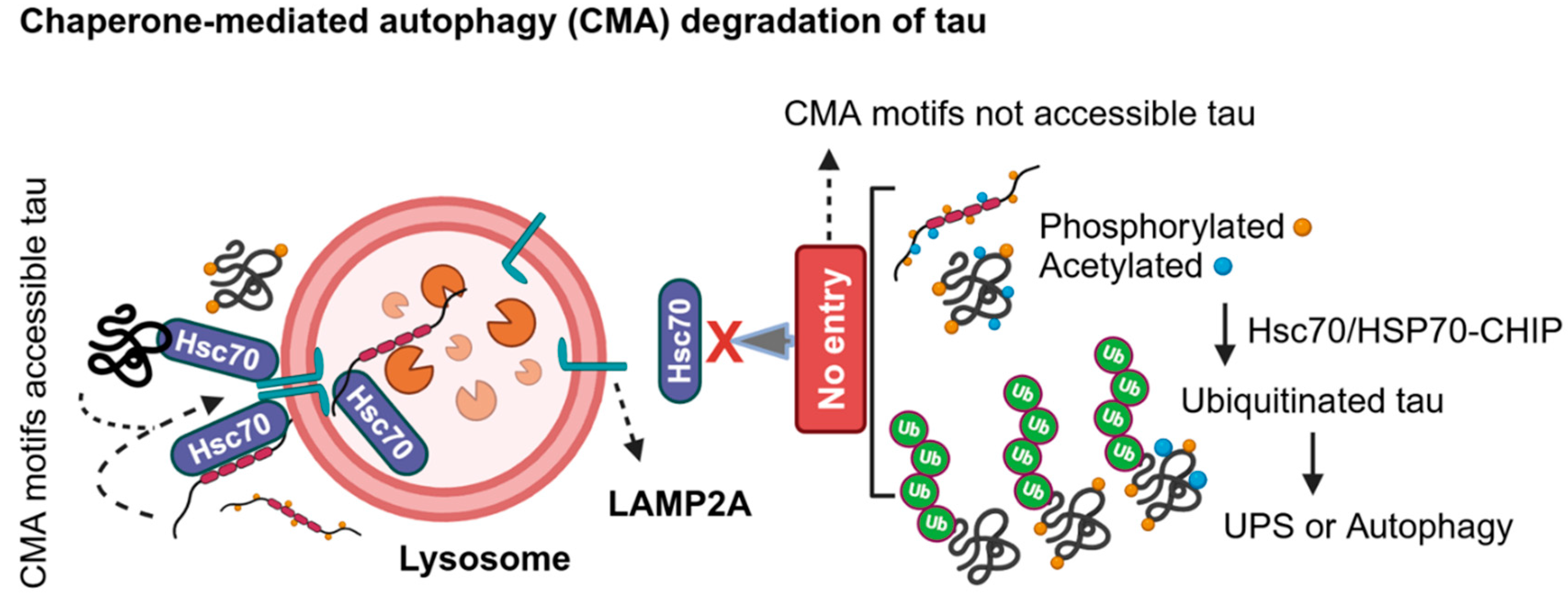
2.3. LC3-Independent Chaperone-Mediated Autophagy (CMA) Degradation of Tau in AD
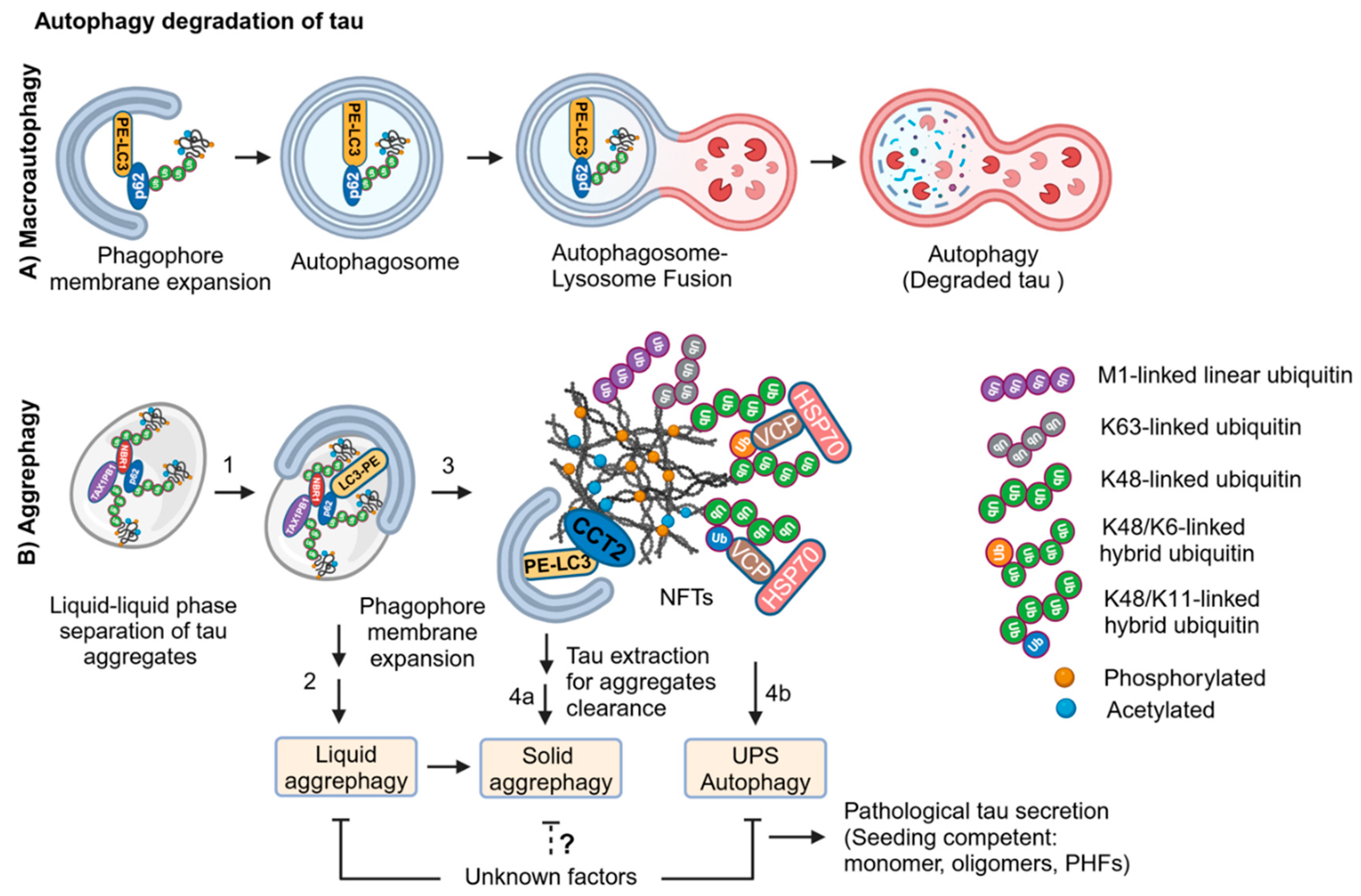
2.4. LC3-Dependent Autophagy Degradation of Tau in AD
2.5. Decoding of Hyperubiquitinated Tau Fibrils
2.6. Aggrephagy Degradation of Tau Aggregates
2.7. Tau Extraction from Hyperubiquitinated Neurofibrillary Tangles
3. Cellular Senescence in Alzheimer’s Disease (AD)
3.1. Cellular Senescence
3.2. Senescence in Neurons
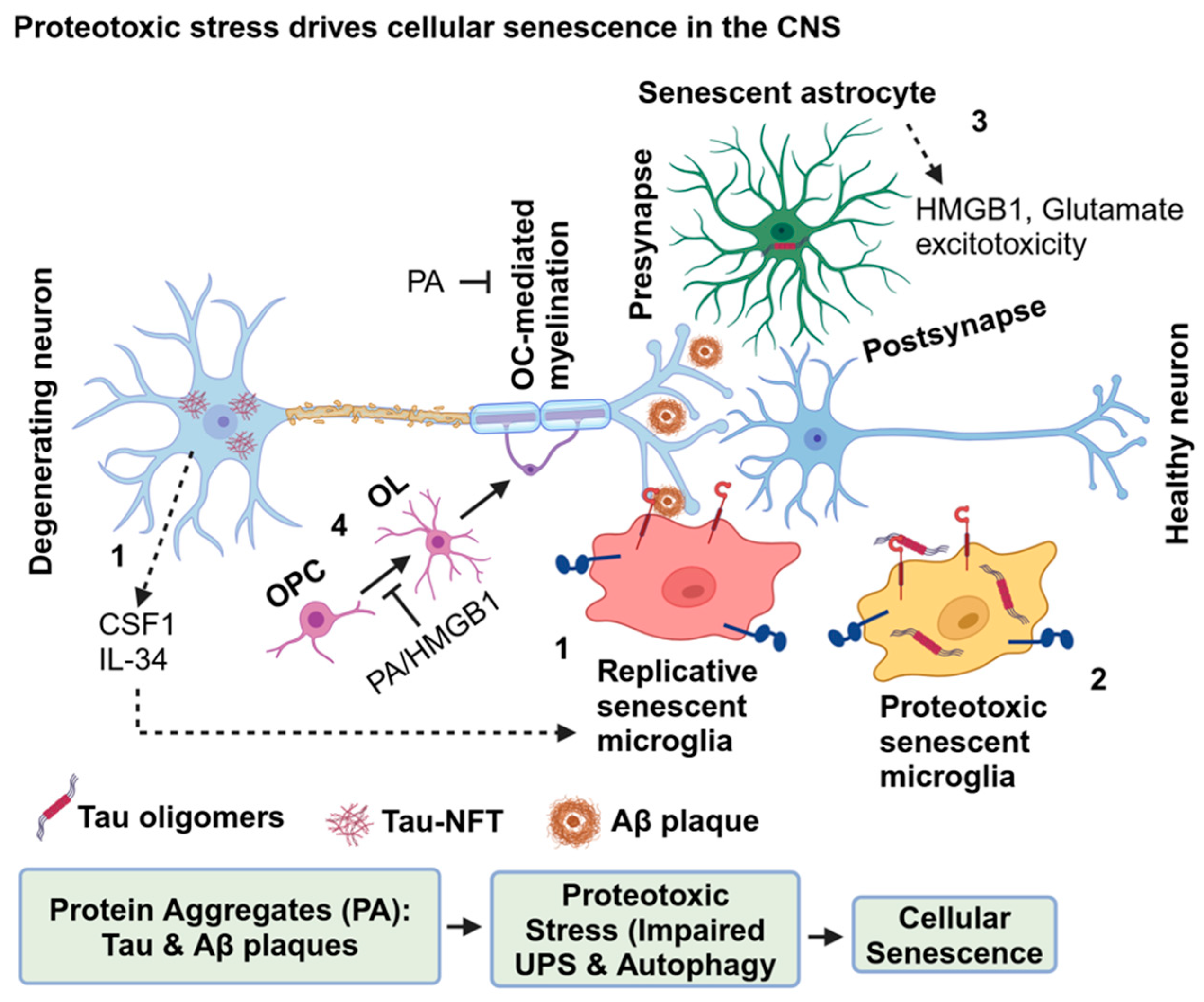
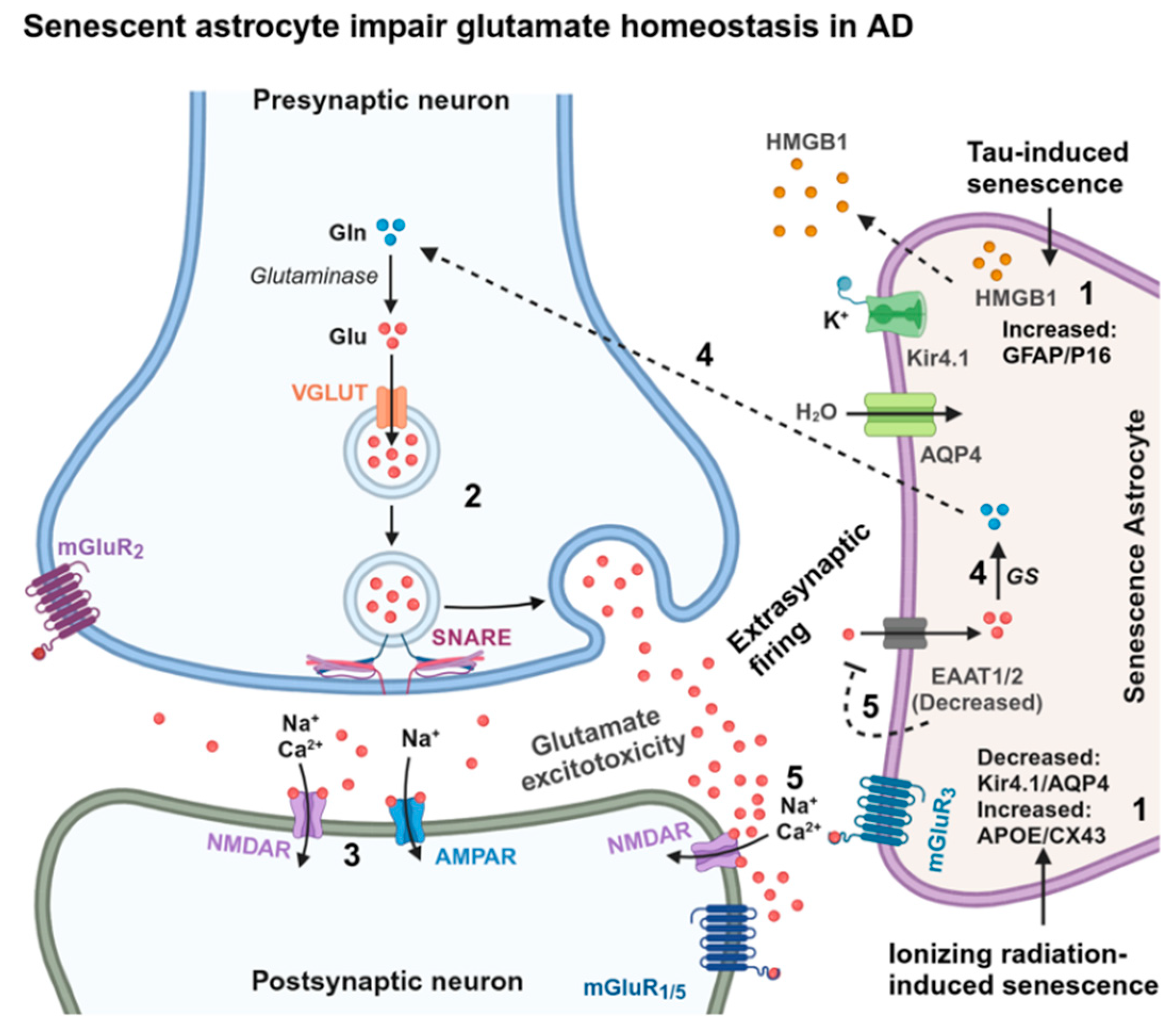
3.3. Senescence in Astrocytes
3.4. Senescence in Oligodendrocytes
3.5. Senescence in Microglia
3.6. Replicative Senescence
4. Proteotoxic Stress Drives Cellular Senescence
4.1. Proteasome in Senescence
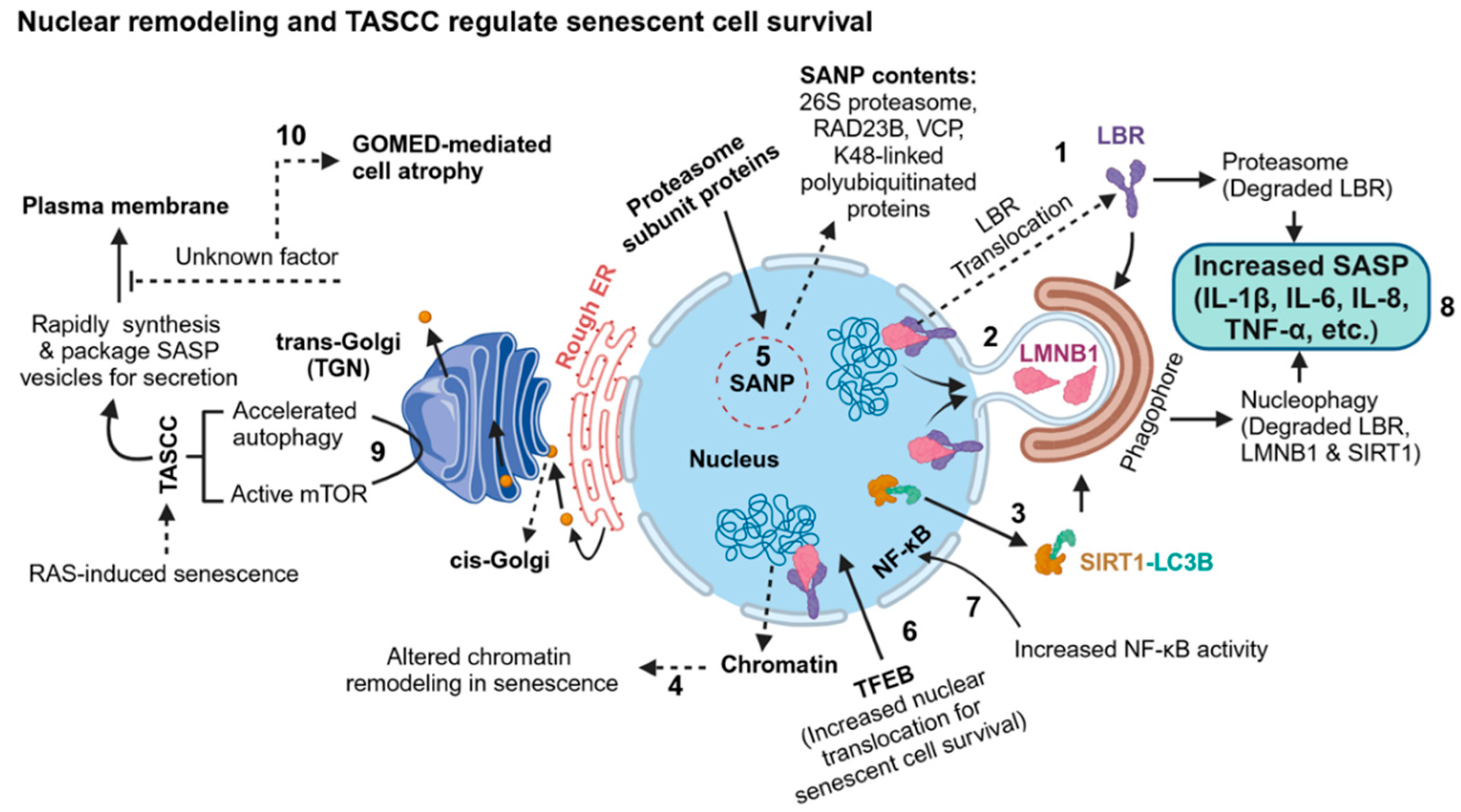
4.2. Autophagy in Senescence
5. Concluding Remarks
Funding
Acknowledgments
References
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2015, 17, 22–35. [Google Scholar] [CrossRef] [PubMed]
- LoPresti, P. Tau in Oligodendrocytes Takes Neurons in Sickness and in Health. Int. J. Mol. Sci. 2018, 19, 2408. [Google Scholar] [CrossRef] [PubMed]
- Poorkaj, P.; Bird, T.D.; Wijsman, E.; Nemens, E.; Garruto, R.M.; Anderson, L.; Andreadis, A.; Wiederholt, W.C.; Raskind, M.; Schellenberg, G.D. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 1998, 43, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, B.; Oblak, A.L.; Boeve, B.F.; Johnson, K.A.; Dickerson, B.C.; Goedert, M. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 2014, 41, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, K.-M.; Yang, L.; Dong, Q.; Yu, J.-T. Tauopathies: new perspectives and challenges. Mol. Neurodegener. 2022, 17, 1–29. [Google Scholar] [CrossRef]
- Lantero-Rodriguez, J.; Camporesi, E.; Montoliu-Gaya, L.; Gobom, J.; Piotrowska, D.; Olsson, M.; Burmann, I.M.; Becker, B.; Brinkmalm, A.; Burmann, B.M.; et al. Tau protein profiling in tauopathies: a human brain study. Mol. Neurodegener. 2024, 19, 1–25. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer's disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Masters, C.L.; Simms, G.; A Weinman, N.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. 1985, 82, 4245–4249. [Google Scholar] [CrossRef]
- Grundke-Iqbal, I.; Iqbal, K.; Quinlan, M.; Tung, Y.C.; Zaidi, M.S.; Wisniewski, H.M. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 1986, 261, 6084–6089. [Google Scholar] [CrossRef]
- Wood, J.G.; Mirra, S.S.; Pollock, N.J.; I Binder, L. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau). Proc. Natl. Acad. Sci. 1986, 83, 4040–4043. [Google Scholar] [CrossRef]
- Kosik, K.S.; Joachim, C.L.; Selkoe, D.J. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1986, 83, 4044–4048. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Lee, J.H.; Rubinsztein, D.C. Tau degradation: The ubiquitin–proteasome system versus the autophagy-lysosome system. Prog. Neurobiol. 2013, 105, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Chesser, A.S.; Pritchard, S.M.; Johnson, G.V.W. Tau Clearance Mechanisms and Their Possible Role in the Pathogenesis of Alzheimer Disease. Front. Neurol. 2013, 4, 122. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Bhaskar, K. Degradation and Transmission of Tau by Autophagic-Endolysosomal Networks and Potential Therapeutic Targets for Tauopathy. Front. Mol. Neurosci. 2020, 13, 586731. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef]
- Puangmalai, N.; Sengupta, U.; Bhatt, N.; Gaikwad, S.; Montalbano, M.; Bhuyan, A.; Garcia, S.; McAllen, S.; Sonawane, M.; Jerez, C.; et al. Lysine 63-linked ubiquitination of tau oligomers contributes to the pathogenesis of Alzheimer’s disease. J. Biol. Chem. 2022, 298, 101766. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Sarmiento, J.; Troncoso, J.; Jackson, G.R.; Kayed, R. Identification of oligomers at early stages of tau aggregation in Alzheimer's disease. FASEB J. 2012, 26, 1946–1959. [Google Scholar] [CrossRef]
- Wesseling, H.; Mair, W.; Kumar, M.; Schlaffner, C.N.; Tang, S.; Beerepoot, P.; Fatou, B.; Guise, A.J.; Cheng, L.; Takeda, S.; et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer’s Disease. Cell 2020, 183, 1699–1713. [Google Scholar] [CrossRef]
- Li, L.; Jiang, Y.; Wang, J.-Z.; Liu, R.; Wang, X. Tau Ubiquitination in Alzheimer's Disease. Front. Neurol. 2022, 12, 786353. [Google Scholar] [CrossRef]
- Habib, J.A.; Lesenfants, J.; Vigneron, N.; Eynde, B.J.V.D. Functional Differences between Proteasome Subtypes. Cells 2022, 11, 421. [Google Scholar] [CrossRef]
- Nixon, R.A.; Rubinsztein, D.C. Mechanisms of autophagy–lysosome dysfunction in neurodegenerative diseases. Nat. Rev. Mol. Cell Biol. 2024, 1–21. [Google Scholar] [CrossRef] [PubMed]
- David, D.C.; Layfield, R.; Serpell, L.; Narain, Y.; Goedert, M.; Spillantini, M.G. Proteasomal degradation of tau protein. J. Neurochem. 2002, 83, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ukmar-Godec, T.; Fang, P.; de Opakua, A.I.; Henneberg, F.; Godec, A.; Pan, K.-T.; Cima-Omori, M.-S.; Chari, A.; Mandelkow, E.; Urlaub, H.; et al. Proteasomal degradation of the intrinsically disordered protein tau at single-residue resolution. Sci. Adv. 2020, 6, eaba3916. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Wei, W.; Yin, J.; Liu, G.-P.; Wang, Q.; Cao, F.-Y.; Wang, J.-Z. Proteasome inhibition increases tau accumulation independent of phosphorylation. Neurobiol. Aging 2009, 30, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.; Mali, S.M.; Sulkshane, P.; Xu, C.; Rozenberg, A.; Morag, R.; Sahoo, M.P.; Singh, S.K.; Ding, Z.; Wang, Y.; et al. The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag. Nat. Commun. 2021, 12, 1–21. [Google Scholar] [CrossRef]
- Ryder, B.D.; Wydorski, P.M.; Hou, Z.; Joachimiak, L.A. Chaperoning shape-shifting tau in disease. Trends Biochem. Sci. 2022, 47, 301–313. [Google Scholar] [CrossRef]
- Shimura, H.; Schwartz, D.; Gygi, S.P.; Kosik, K.S. CHIP-Hsc70 Complex Ubiquitinates Phosphorylated Tau and Enhances Cell Survival. J. Biol. Chem. 2004, 279, 4869–4876. [Google Scholar] [CrossRef]
- Petrucelli, L.; Dickson, D.; Kehoe, K.; Taylor, J.; Snyder, H.; Grover, A.; De Lucia, M.; McGowan, E.; Lewis, J.; Prihar, G.; et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004, 13, 703–714. [Google Scholar] [CrossRef]
- Takayama, S.; Bimston, D.N.; Matsuzawa, S.; Freeman, B.C.; Aime-Sempe, C.; Xie, Z.; Morimoto, R.I.; Reed, J.C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997, 16, 4887–4896. [Google Scholar] [CrossRef]
- Alberti, S.; Esser, C.; Höhfeld, J. BAG-1—a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperon- 2003, 8, 225–31. [Google Scholar] [CrossRef]
- Elliott, E.; Tsvetkov, P.; Ginzburg, I. BAG-1 Associates with Hsc70·Tau Complex and Regulates the Proteasomal Degradation of Tau Protein. J. Biol. Chem. 2007, 282, 37276–37284. [Google Scholar] [CrossRef] [PubMed]
- Weickert, S.; Wawrzyniuk, M.; John, L.H.; Rüdiger, S.G.D.; Drescher, M. The mechanism of Hsp90-induced oligomerizaton of Tau. Sci. Adv. 2020, 6, eaax6999. [Google Scholar] [CrossRef] [PubMed]
- Blair, L.J.; Nordhues, B.A.; Hill, S.E.; Scaglione, K.M.; O’leary, J.C.; Fontaine, S.N.; Breydo, L.; Zhang, B.; Li, P.; Wang, L.; et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Investig. 2013, 123, 4158–4169. [Google Scholar] [CrossRef] [PubMed]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Investig. 2007, 117, 648–658. [Google Scholar] [CrossRef]
- Dickey, C.A.; Yue, M.; Lin, W.-L.; Dickson, D.W.; Dunmore, J.H.; Lee, W.C.; Zehr, C.; West, G.; Cao, S.; Clark, A.M.K.; et al. Deletion of the Ubiquitin Ligase CHIP Leads to the Accumulation, But Not the Aggregation, of Both Endogenous Phospho- and Caspase-3-Cleaved Tau Species. J. Neurosci. 2006, 26, 6985–6996. [Google Scholar] [CrossRef]
- Cuervo, A.; Dice, J. Regulation of Lamp2a Levels in the Lysosomal Membrane. Traffic 2000, 1, 570–583. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Dice, J.F. A Receptor for the Selective Uptake and Degradation of Proteins by Lysosomes. Science 1996, 273, 501–503. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Bourdenx, M.; Martín-Segura, A.; Scrivo, A.; Rodriguez-Navarro, J.A.; Kaushik, S.; Tasset, I.; Diaz, A.; Storm, N.J.; Xin, Q.; Juste, Y.R.; et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 2021, 184, 2696–2714. [Google Scholar] [CrossRef]
- Massey, A.C.; Kaushik, S.; Sovak, G.; Kiffin, R.; Cuervo, A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. 2006, 103, 5805–5810. [Google Scholar] [CrossRef]
- Wang, Y.; Martinez-Vicente, M.; Krüger, U.; Kaushik, S.; Wong, E.; Mandelkow, E.-M.; Cuervo, A.M.; Mandelkow, E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 2009, 18, 4153–4170. [Google Scholar] [CrossRef] [PubMed]
- Caballero, B.; Bourdenx, M.; Luengo, E.; Diaz, A.; Sohn, P.D.; Chen, X.; Wang, C.; Juste, Y.R.; Wegmann, S.; Patel, B.; et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Min, S.-W.; Cho, S.-H.; Zhou, Y.; Schroeder, S.; Haroutunian, V.; Seeley, W.W.; Huang, E.J.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Cohen, T.J.; Guo, J.L.; Hurtado, D.E.; Kwong, L.K.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M.Y. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef]
- Cohen, T.J.; Guo, J.L.; Hurtado, D.E.; Kwong, L.K.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M.Y. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef]
- Kim, M.-S.; Mun, Y.-S.; Lee, S.-E.; Cho, W.-Y.; Han, S.-H.; Kim, D.-H.; Yoon, S.-Y. Tau acetylation at K280 regulates tau phosphorylation. Int. J. Neurosci. 2022, 133, 1394–1398. [Google Scholar] [CrossRef]
- Abreha, M.H.; Dammer, E.B.; Ping, L.; Zhang, T.; Duong, D.M.; Gearing, M.; Lah, J.J.; Levey, A.I.; Seyfried, N.T. Quantitative Analysis of the Brain Ubiquitylome in Alzheimer’s Disease. Proteomics 2018, 18, e1800108. [Google Scholar] [CrossRef]
- Nakayama, Y.; Sakamoto, S.; Tsuji, K.; Ayaki, T.; Tokunaga, F.; Ito, H. Identification of linear polyubiquitin chain immunoreactivity in tau pathology of Alzheimer’s disease. Neurosci. Lett. 2019, 703, 53–57. [Google Scholar] [CrossRef]
- Cripps, D.; Thomas, S.N.; Jeng, Y.; Yang, F.; Davies, P.; Yang, A.J. Alzheimer Disease-specific Conformation of Hyperphosphorylated Paired Helical Filament-Tau Is Polyubiquitinated through Lys-48, Lys-11, and Lys-6 Ubiquitin Conjugation. J. Biol. Chem. 2006, 281, 10825–10838. [Google Scholar] [CrossRef]
- Tan, J.M.M.; Wong, E.S.P.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.-P.; Ho, M.W.L.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef]
- French, M.E.; Koehler, C.F.; Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discov. 2021, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, D.; Sato, Y.; Ito, H.; Tokunaga, F. Linear Ubiquitin Code: Its Writer, Erasers, Decoders, Inhibitors, and Implications in Disorders. Int. J. Mol. Sci. 2020, 21, 3381. [Google Scholar] [CrossRef] [PubMed]
- Jahan, A.S.; Elbæk, C.R.; Damgaard, R.B. Met1-linked ubiquitin signalling in health and disease: inflammation, immunity, cancer, and beyond. Cell Death Differ. 2021, 28, 473–492. [Google Scholar] [CrossRef] [PubMed]
- Furthmann, N.; Bader, V.; Angersbach, L.; Blusch, A.; Goel, S.; Sánchez-Vicente, A.; Krause, L.J.; Chaban, S.A.; Grover, P.; Trinkaus, V.A.; et al. NEMO reshapes the α-Synuclein aggregate interface and acts as an autophagy adapter by co-condensation with p62. Nat. Commun. 2023, 14, 1–24. [Google Scholar] [CrossRef]
- Goel, S.; Oliva, R.; Jeganathan, S.; Bader, V.; Krause, L.J.; Kriegler, S.; Stender, I.D.; Christine, C.W.; Nakamura, K.; Hoffmann, J.-E.; et al. Linear ubiquitination induces NEMO phase separation to activate NF-κB signaling. Life Sci. Alliance 2023, 6, e202201607. [Google Scholar] [CrossRef]
- Wu, C.-J.; Conze, D.B.; Li, T.; Srinivasula, S.M.; Ashwell, J.D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 2006, 8, 398–406. [Google Scholar] [CrossRef]
- Kanayama, A.; Seth, R.B.; Sun, L.; Ea, C.-K.; Hong, M.; Shaito, A.; Chiu, Y.-H.; Deng, L.; Chen, Z.J. TAB2 and TAB3 Activate the NF-κB Pathway through Binding to Polyubiquitin Chains. Mol. Cell 2004, 15, 535–548. [Google Scholar] [CrossRef]
- Iha, H.; Peloponese, J.-M.; Verstrepen, L.; Zapart, G.; Ikeda, F.; Smith, C.D.; Starost, M.F.; Yedavalli, V.; Heyninck, K.; Dikic, I.; et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 2008, 27, 629–641. [Google Scholar] [CrossRef]
- Rahmanto, A.S.; Blum, C.J.; Scalera, C.; Heidelberger, J.B.; Mesitov, M.; Horn-Ghetko, D.; Gräf, J.F.; Mikicic, I.; Hobrecht, R.; Orekhova, A.; et al. K6-linked ubiquitylation marks formaldehyde-induced RNA-protein crosslinks for resolution. Mol. Cell 2023, 83, 4272–4289. [Google Scholar] [CrossRef]
- Zhao, S.; Cordes, J.; Caban, K.M.; Götz, M.J.; Mackens-Kiani, T.; Veltri, A.J.; Sinha, N.K.; Weickert, P.; Kaya, S.; Hewitt, G.; et al. RNF14-dependent atypical ubiquitylation promotes translation-coupled resolution of RNA-protein crosslinks. Mol. Cell 2023, 83, 4290–4303. [Google Scholar] [CrossRef]
- Tracz, M.; Bialek, W. Beyond K48 and K63: non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.-J.; Rape, M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.G.; Doerner, K.; Castellanos, E.R.; Haakonsen, D.L.; Werner, A.; Wang, N.; Yang, X.W.; Martinez-Martin, N.; Matsumoto, M.L.; Dixit, V.M.; et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Johansen, T. Aggrephagy: Selective Disposal of Protein Aggregates by Macroautophagy. Int. J. Cell Biol. 2012, 2012, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bauer, B.; Martens, S.; Ferrari, L. Aggrephagy at a glance. J. Cell Sci. 2023, 136. [Google Scholar] [CrossRef]
- Zhang, Z.; Klionsky, D.J. CCT2, a newly identified aggrephagy receptor in mammals, specifically mediates the autophagic clearance of solid protein aggregates. Autophagy 2022, 18, 1483–1485. [Google Scholar] [CrossRef]
- Ma, X.; Lu, C.; Chen, Y.; Li, S.; Ma, N.; Tao, X.; Li, Y.; Wang, J.; Zhou, M.; Yan, Y.-B.; et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell 2022, 185, 1325–1345. [Google Scholar] [CrossRef]
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018, 28, 405–415. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Danieli, A.; Martens, S. p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Turco, E.; Savova, A.; Gere, F.; Ferrari, L.; Romanov, J.; Schuschnig, M.; Martens, S. Reconstitution defines the roles of p62, NBR1 and TAX1BP1 in ubiquitin condensate formation and autophagy initiation. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Bauer, B.; Qiu, Y.; Schuschnig, M.; Klotz, S.; Anrather, D.; Juretschke, T.; Beli, P.; Gelpi, E.; Martens, S. Tau fibrils evade autophagy by excessive p62 coating and TAX1BP1 exclusion. Sci. Adv. 2024, 10, eadm8449. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, S.; Eftekharzadeh, B.; Tepper, K.; Zoltowska, K.M.; Bennett, R.E.; Dujardin, S.; Laskowski, P.R.; MacKenzie, D.; Kamath, T.; Commins, C.; et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018, 37, e98049. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Shah, H.V.; Kanfer, G.; Pickrell, A.M.; Holtzclaw, L.A.; Ward, M.E.; Youle, R.J. Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. Mol. Cell 2020, 80, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Saha, I.; Yuste-Checa, P.; Padilha, M.D.S.; Guo, Q.; Körner, R.; Holthusen, H.; Trinkaus, V.A.; Dudanova, I.; Fernández-Busnadiego, R.; Baumeister, W.; et al. The AAA+ chaperone VCP disaggregates Tau fibrils and generates aggregate seeds in a cellular system. Nat. Commun. 2023, 14, 1–17. [Google Scholar] [CrossRef]
- Giong, H.-K.; Hyeon, S.J.; Lee, J.-G.; Cho, H.-J.; Park, U.; Stein, T.D.; Lee, J.; Yu, K.; Ryu, H.; Lee, J.-S. Tau accumulation is cleared by the induced expression of VCP via autophagy. Acta Neuropathol. 2024, 148, 1–19. [Google Scholar] [CrossRef]
- Zwang, T.J.; Woost, B.; Bailey, J.; Hoglund, Z.; Richardson, D.S.; E Bennett, R.; Hyman, B.T. Spatial characterization of tangle-bearing neurons and ghost tangles in the human inferior temporal gyrus with three-dimensional imaging. Brain Commun. 2023, 5, fcad130. [Google Scholar] [CrossRef]
- de Calignon, A.; Spires-Jones, T.L.; Pitstick, R.; Carlson, G.A.; Hyman, B.T. Tangle-Bearing Neurons Survive Despite Disruption of Membrane Integrity in a Mouse Model of Tauopathy. J. Neuropathol. Exp. Neurol. 2009, 68, 757–761. [Google Scholar] [CrossRef]
- S. Du et al., “Clearance of protein aggregates during cell division,” , 2024. 20 May. [CrossRef]
- Bhaskar, K.; Maphis, N.; Xu, G.; Varvel, N.H.; Kokiko-Cochran, O.N.; Weick, J.P.; Staugaitis, S.M.; Cardona, A.; Ransohoff, R.M.; Herrup, K.; et al. Microglial derived tumor necrosis factor-α drives Alzheimer's disease-related neuronal cell cycle events. Neurobiol. Dis. 2013, 62, 273–285. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ishikawa, F. Proteostasis failure and cellular senescence in long-term cultured postmitotic rat neurons. Aging Cell 2019, 19, e13071. [Google Scholar] [CrossRef]
- Musi, N.; Valentine, J.M.; Sickora, K.R.; Baeuerle, E.; Thompson, C.S.; Shen, Q.; Orr, M.E. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell 2018, 17, e12840. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 2000, 1, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.-M.; Yen, Y.-H.; Yuan, F.; Zhang, S.-C.; Chong, C.-M. Neuronal Senescence in the Aged Brain. Aging Dis. 2023, 14, 1618–1632. [Google Scholar] [CrossRef] [PubMed]
- Herdy, J.R.; Mertens, J.; Gage, F.H. Neuronal senescence may drive brain aging. Science 2024, 384, 1404–1406. [Google Scholar] [CrossRef]
- Sah, E.; Krishnamurthy, S.; Ahmidouch, M.Y.; Gillispie, G.J.; Milligan, C.; Orr, M.E. The Cellular Senescence Stress Response in Post-Mitotic Brain Cells: Cell Survival at the Expense of Tissue Degeneration. Life 2021, 11, 229. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; di Fagagna, F.D. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2020, 22, 75–95. [Google Scholar] [CrossRef]
- Ashraf, H.M.; Fernandez, B.; Spencer, S.L. The intensities of canonical senescence biomarkers integrate the duration of cell-cycle withdrawal. Nat. Commun. 2023, 14, 1–13. [Google Scholar] [CrossRef]
- Zhang, L.; Pitcher, L.E.; Yousefzadeh, M.J.; Niedernhofer, L.J.; Robbins, P.D.; Zhu, Y. Cellular senescence: a key therapeutic target in aging and diseases. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef]
- Muthamil, S.; Kim, H.; Jang, H.; Lyu, J.; Shin, U.C.; Go, Y.; Park, S.; Lee, H.G.; Park, J.H. Biomarkers of Cellular Senescence and Aging: Current State-of-the-Art, Challenges and Future Perspectives. Adv. Biol. 2024, 8, e2400079. [Google Scholar] [CrossRef]
- Admasu, T.D.; Rae, M.J.; Stolzing, A. Dissecting primary and secondary senescence to enable new senotherapeutic strategies. Ageing Res. Rev. 2021, 70, 101412. [Google Scholar] [CrossRef]
- Mayford, M.; Siegelbaum, S.A.; Kandel, E.R. Synapses and Memory Storage. Cold Spring Harb. Perspect. Biol. 2012, 4, a005751–a005751. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D. The Enigma of Working Memory: Changing Views. Neurosci. 2022, 28, 420–424. [Google Scholar] [CrossRef]
- Sanhedrai, H.; Havlin, S.; Dvir, H. Mechanistic description of spontaneous loss of memory persistent activity based on neuronal synaptic strength. Heliyon 2023, 10, e23949. [Google Scholar] [CrossRef] [PubMed]
- Häusser, M.; Raman, I.M.; Otis, T.; Smith, S.L.; Nelson, A.; du Lac, S.; Loewenstein, Y.; Mahon, S.; Pennartz, C.; Cohen, I.; et al. The Beat Goes On: Spontaneous Firing in Mammalian Neuronal Microcircuits. J. Neurosci. 2004, 24, 9215–9219. [Google Scholar] [CrossRef] [PubMed]
- Kirova, A.-M.; Bays, R.B.; Lagalwar, S. Working Memory and Executive Function Decline across Normal Aging, Mild Cognitive Impairment, and Alzheimer’s Disease. BioMed Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Stopford, C.L.; Thompson, J.C.; Neary, D.; Richardson, A.M.; Snowden, J.S. Working memory, attention, and executive function in Alzheimer’s disease and frontotemporal dementia. Cortex 2012, 48, 429–446. [Google Scholar] [CrossRef]
- Herdy, J.R.; Traxler, L.; Agarwal, R.K.; Karbacher, L.; Schlachetzki, J.C.; Boehnke, L.; Zangwill, D.; Galasko, D.; Glass, C.K.; Mertens, J.; et al. Increased post-mitotic senescence in aged human neurons is a pathological feature of Alzheimer’s disease. Cell Stem Cell 2022, 29, 1637–1652. [Google Scholar] [CrossRef]
- Dehkordi, S.K.; Walker, J.; Sah, E.; Bennett, E.; Atrian, F.; Frost, B.; Woost, B.; Bennett, R.E.; Orr, T.C.; Zhou, Y.; et al. Profiling senescent cells in human brains reveals neurons with CDKN2D/p19 and tau neuropathology. Nat. Aging 2021, 1, 1107–1116. [Google Scholar] [CrossRef]
- Ota, Y.; Zanetti, A.T.; Hallock, R.M. The Role of Astrocytes in the Regulation of Synaptic Plasticity and Memory Formation. Neural Plast. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Han, X.; Zhang, T.; Liu, H.; Mi, Y.; Gou, X. Astrocyte Senescence and Alzheimer’s Disease: A Review. Front. Aging Neurosci. 2020, 12, 148. [Google Scholar] [CrossRef]
- Esposito, Z.; Belli, L.; Toniolo, S.; Sancesario, G.; Bianconi, C.; Martorana, A. Amyloid β, Glutamate, Excitotoxicity in Alzheimer's Disease: Are We on the Right Track? CNS Neurosci. Ther. 2013, 19, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoo, I.D.; Lim, J.; Moon, J.-S. Pathological phenotypes of astrocytes in Alzheimer’s disease. Exp. Mol. Med. 2024, 56, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Reddy, P.H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimer's Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Crowe, E.P.; Bitto, A.; Moh, M.; Katsetos, C.D.; Garcia, F.U.; Johnson, F.B.; Trojanowski, J.Q.; Sell, C.; Torres, C. Astrocyte Senescence as a Component of Alzheimer’s Disease. PLOS ONE 2012, 7, e45069. [Google Scholar] [CrossRef] [PubMed]
- Limbad, C.; Oron, T.R.; Alimirah, F.; Davalos, A.R.; Tracy, T.E.; Gan, L.; Desprez, P.-Y.; Campisi, J. Astrocyte senescence promotes glutamate toxicity in cortical neurons. PLOS ONE 2020, 15, e0227887. [Google Scholar] [CrossRef]
- Bitto, A.; Sell, C.; Crowe, E.; Lorenzini, A.; Malaguti, M.; Hrelia, S.; Torres, C. Stress-induced senescence in human and rodent astrocytes. Exp. Cell Res. 2010, 316, 2961–2968. [Google Scholar] [CrossRef]
- Gaikwad, S.; Puangmalai, N.; Bittar, A.; Montalbano, M.; Garcia, S.; McAllen, S.; Bhatt, N.; Sonawane, M.; Sengupta, U.; Kayed, R. Tau oligomer induced HMGB1 release contributes to cellular senescence and neuropathology linked to Alzheimer’s disease and frontotemporal dementia. Cell Rep. 2021, 36, 109419–109419. [Google Scholar] [CrossRef]
- Simons, M.; Nave, K.-A. Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb. Perspect. Biol. 2015, 8, a020479. [Google Scholar] [CrossRef]
- Kuhn, S.; Gritti, L.; Crooks, D.; Dombrowski, Y. Oligodendrocytes in Development, Myelin Generation and Beyond. Cells 2019, 8, 1424. [Google Scholar] [CrossRef]
- Garton, T.; Gadani, S.P.; Gill, A.J.; Calabresi, P.A. Neurodegeneration and demyelination in multiple sclerosis. Neuron 2024, 112, 3231–3251. [Google Scholar] [CrossRef]
- Costa, V.G.C.; Araújo, S.E.-S.; Alves-Leon, S.V.; Gomes, F.C.A. Central nervous system demyelinating diseases: glial cells at the hub of pathology. Front. Immunol. 2023, 14. [Google Scholar] [CrossRef]
- LaCroix, M.S.; Mirbaha, H.; Shang, P.; Zandee, S.; Foong, C.; Prat, A.; White, C.L.; Stuve, O.; Diamond, M.I. Tau seeding in cases of multiple sclerosis. Acta Neuropathol. Commun. 2022, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ahmad, T.K.; Gozda, K.; Truong, J.; Kong, J.; Namaka, M. Implications of white matter damage in amyotrophic lateral sclerosis. Mol. Med. Rep. 2017, 16, 4379–4392. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Jordan, J.D.; Zhang, Q. Myelin Pathology in Alzheimer's Disease: Potential Therapeutic Opportunities. Aging Dis. 2024, 15, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Torii, T.; Miyamoto, Y.; Nakata, R.; Higashi, Y.; Shinmyo, Y.; Kawasaki, H.; Miyasaka, T.; Misonou, H. Identification of Tau protein as a novel marker for maturation and pathological changes of oligodendrocytes. Glia 2022, 71, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Oguchi, K.; Okabe, S.; Kuno, J.; Terada, S.; Ohshima, T.; Sato-Yoshitake, R.; Takei, Y.; Noda, T.; Hirokawa, N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 1994, 369, 488–491. [Google Scholar] [CrossRef]
- Seiberlich, V.; Bauer, N.G.; Schwarz, L.; Ffrench-Constant, C.; Goldbaum, O.; Richter-Landsberg, C. Downregulation of the microtubule associated protein Tau impairs process outgrowth and myelin basic protein mRNA transport in oligodendrocytes. Glia 2015, 63, 1621–1635. [Google Scholar] [CrossRef]
- Torii, T. Abnormal expression of Tau in damaged oligodendrocytes of HLD1 mice. Neural Regen. Res. 2023, 19, 1405–1406. [Google Scholar] [CrossRef]
- De Rossi, P.; Buggia-Prévot, V.; Clayton, B.L.L.; Vasquez, J.B.; van Sanford, C.; Andrew, R.J.; Lesnick, R.; Botté, A.; Deyts, C.; Salem, S.; et al. Predominant expression of Alzheimer’s disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol. Neurodegener. 2016, 11, 1–21. [Google Scholar] [CrossRef]
- Sottejeau, Y.; Bretteville, A.; Cantrelle, F.-X.; Malmanche, N.; Demiaute, F.; Mendes, T.; Delay, C.; Dos Alves, H.A.; Flaig, A.; Davies, P.; et al. Tau phosphorylation regulates the interaction between BIN1’s SH3 domain and Tau’s proline-rich domain. Acta Neuropathol. Commun. 2015, 3, 1–12. [Google Scholar] [CrossRef]
- Gautier, H.O.B.; Evans, K.A.; Volbracht, K.; James, R.; Sitnikov, S.; Lundgaard, I.; James, F.; Lao-Peregrin, C.; Reynolds, R.; Franklin, R.J.M.; et al. Neuronal activity regulates remyelination via glutamate signalling to oligodendrocyte progenitors. Nat. Commun. 2015, 6, 8518. [Google Scholar] [CrossRef] [PubMed]
- Viney, T.J.; Sarkany, B.; Ozdemir, A.T.; Hartwich, K.; Schweimer, J.; Bannerman, D.; Somogyi, P. Spread of pathological human Tau from neurons to oligodendrocytes and loss of high-firing pyramidal neurons in aging mice. Cell Rep. 2022, 41, 111646. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Changolkar, L.; Riddle, D.M.; Kats, A.; Stieber, A.; Weitzman, S.A.; Zhang, B.; Li, Z.; Roberson, E.D.; Trojanowski, J.Q.; et al. Human tau pathology transmits glial tau aggregates in the absence of neuronal tau. J. Exp. Med. 2019, 217. [Google Scholar] [CrossRef] [PubMed]
- Sasmita, A.O.; Depp, C.; Nazarenko, T.; Sun, T.; Siems, S.B.; Ong, E.C.; Nkeh, Y.B.; Böhler, C.; Yu, X.; Bues, B.; et al. Oligodendrocytes produce amyloid-β and contribute to plaque formation alongside neurons in Alzheimer’s disease model mice. Nat. Neurosci. 2024, 1–7. [Google Scholar] [CrossRef]
- Zhang, P.; Kishimoto, Y.; Grammatikakis, I.; Gottimukkala, K.; Cutler, R.G.; Zhang, S.; Abdelmohsen, K.; Bohr, V.A.; Sen, J.M.; Gorospe, M.; et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 2019, 22, 719–728. [Google Scholar] [CrossRef]
- Rouillard, M.E.; Hu, J.; Sutter, P.A.; Kim, H.W.; Huang, J.K.; Crocker, S.J. The Cellular Senescence Factor Extracellular HMGB1 Directly Inhibits Oligodendrocyte Progenitor Cell Differentiation and Impairs CNS Remyelination. Front. Cell. Neurosci. 2022, 16, 833186. [Google Scholar] [CrossRef]
- Roy, E.R.; Chiu, G.; Li, S.; Propson, N.E.; Kanchi, R.; Wang, B.; Coarfa, C.; Zheng, H.; Cao, W. Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques. Immunity 2022, 55, 879–894. [Google Scholar] [CrossRef]
- Jin, M.; Xu, R.; Wang, L.; Alam, M.M.; Ma, Z.; Zhu, S.; Martini, A.C.; Jadali, A.; Bernabucci, M.; Xie, P.; et al. Type-I-interferon signaling drives microglial dysfunction and senescence in human iPSC models of Down syndrome and Alzheimer’s disease. Cell Stem Cell 2022, 29, 1135–1153. [Google Scholar] [CrossRef]
- Lana-Elola, E.; Watson-Scales, S.D.; Fisher, E.M.C.; Tybulewicz, V.L.J. Down syndrome: searching for the genetic culprits. Dis. Model. Mech. 2011, 4, 586–595. [Google Scholar] [CrossRef]
- Hong, S.; Beja-Glasser, V.F.; Nfonoyim, B.M.; Frouin, A.; Li, S.; Ramakrishnan, S.; Merry, K.M.; Shi, Q.; Rosenthal, A.; Barres, B.A.; et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 2016, 352, 712–716. [Google Scholar] [CrossRef]
- Peng, Q.; Malhotra, S.; Torchia, J.A.; Kerr, W.G.; Coggeshall, K.M.; Humphrey, M.B. TREM2- and DAP12-Dependent Activation of PI3K Requires DAP10 and Is Inhibited by SHIP1. Sci. Signal. 2010, 3, ra38–ra38. [Google Scholar] [CrossRef] [PubMed]
- Wißfeld, J.; Mathews, M.; Mossad, O.; Picardi, P.; Cinti, A.; Redaelli, L.; Pradier, L.; Brüstle, O.; Neumann, H. Reporter cell assay for human CD33 validated by specific antibodies and human iPSC-derived microglia. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mócsai, A.; Abram, C.L.; Jakus, Z.; Hu, Y.; Lanier, L.L.; A Lowell, C. Integrin signaling in neutrophils and macrophages uses adaptors containing immunoreceptor tyrosine-based activation motifs. Nat. Immunol. 2006, 7, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, S.; Larionov, S.; Wang, Y.; Dannenberg, H.; Matozaki, T.; Monsonego, A.; Thal, D.R.; Neumann, H. Signal Regulatory Protein-β1. Am. J. Pathol. 2009, 175, 2528–2539. [Google Scholar] [CrossRef] [PubMed]
- Haure-Mirande, J.-V.; Audrain, M.; Fanutza, T.; Kim, S.H.; Klein, W.L.; Glabe, C.; Readhead, B.; Dudley, J.T.; Blitzer, R.D.; Wang, M.; et al. Deficiency of TYROBP, an adapter protein for TREM2 and CR3 receptors, is neuroprotective in a mouse model of early Alzheimer’s pathology. Acta Neuropathol. 2017, 134, 769–788. [Google Scholar] [CrossRef]
- Audrain, M.; Haure-Mirande, J.-V.; Wang, M.; Kim, S.H.; Fanutza, T.; Chakrabarty, P.; Fraser, P.; George-Hyslop, P.H.S.; Golde, T.E.; Blitzer, R.D.; et al. Integrative approach to sporadic Alzheimer’s disease: deficiency of TYROBP in a tauopathy mouse model reduces C1q and normalizes clinical phenotype while increasing spread and state of phosphorylation of tau. Mol. Psychiatry 2018, 24, 1383–1397. [Google Scholar] [CrossRef]
- Haure-Mirande, J.-V.; Wang, M.; Audrain, M.; Fanutza, T.; Kim, S.H.; Heja, S.; Readhead, B.; Dudley, J.T.; Blitzer, R.D.; Schadt, E.E.; et al. Integrative approach to sporadic Alzheimer’s disease: deficiency of TYROBP in cerebral Aβ amyloidosis mouse normalizes clinical phenotype and complement subnetwork molecular pathology without reducing Aβ burden. Mol. Psychiatry 2018, 24, 431–446. [Google Scholar] [CrossRef]
- Yuan, P.; Condello, C.; Keene, C.D.; Wang, Y.; Bird, T.D.; Paul, S.M.; Luo, W.; Colonna, M.; Baddeley, D.; Grutzendler, J. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron 2016, 90, 724–739. [Google Scholar] [CrossRef]
- Rachmian, N.; Medina, S.; Cherqui, U.; Akiva, H.; Deitch, D.; Edilbi, D.; Croese, T.; Salame, T.M.; Ramos, J.M.P.; Cahalon, L.; et al. Identification of senescent, TREM2-expressing microglia in aging and Alzheimer’s disease model mouse brain. Nat. Neurosci. 2024, 27, 1–9. [Google Scholar] [CrossRef]
- Koenigsknecht, J.; Landreth, G. Microglial Phagocytosis of Fibrillar β-Amyloid through a β1Integrin-Dependent Mechanism. J. Neurosci. 2004, 24, 9838–9846. [Google Scholar] [CrossRef]
- Mandrekar, S.; Jiang, Q.; Lee, C.Y.D.; Koenigsknecht-Talboo, J.; Holtzman, D.M.; Landreth, G.E. Microglia Mediate the Clearance of Soluble Aβ through Fluid Phase Macropinocytosis. J. Neurosci. 2009, 29, 4252–4262. [Google Scholar] [CrossRef] [PubMed]
- Condello, C.; Yuan, P.; Schain, A.; Grutzendler, J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-N.; Shiao, Y.-J.; Shie, F.-S.; Guo, B.-S.; Chen, P.-H.; Cho, C.-Y.; Chen, Y.-J.; Huang, F.-L.; Tsay, H.-J. Mechanism mediating oligomeric Aβ clearance by naïve primary microglia. Neurobiol. Dis. 2011, 42, 221–230. [Google Scholar] [CrossRef]
- Banati, R.B.; Rothe, G.; Valet, G.; Kreutzberg, G.W. Detection of lysosomal cysteine proteinases in microglia: Flow cytometric measurement and histochemical localization of cathepsin B and L. Glia 1993, 7, 183–191. [Google Scholar] [CrossRef]
- Hu, Y.; Fryatt, G.L.; Ghorbani, M.; Obst, J.; Menassa, D.A.; Martin-Estebane, M.; Muntslag, T.A.; Olmos-Alonso, A.; Guerrero-Carrasco, M.; Thomas, D.; et al. Replicative senescence dictates the emergence of disease-associated microglia and contributes to Aβ pathology. Cell Rep. 2021, 35, 109228. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.-E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Olmos-Alonso, A.; Schetters, S.T.T.; Sri, S.; Askew, K.; Mancuso, R.; Vargas-Caballero, M.; Holscher, C.; Perry, V.H.; Gomez-Nicola, D. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain 2016, 139, 891–907. [Google Scholar] [CrossRef]
- Easley-Neal, C.; Foreman, O.; Sharma, N.; Zarrin, A.A.; Weimer, R.M. CSF1R Ligands IL-34 and CSF1 Are Differentially Required for Microglia Development and Maintenance in White and Gray Matter Brain Regions. Front. Immunol. 2019, 10, 2199. [Google Scholar] [CrossRef]
- Füger, P.; Hefendehl, J.K.; Veeraraghavalu, K.; Wendeln, A.-C.; Schlosser, C.; Obermüller, U.; Wegenast-Braun, B.M.; Neher, J.J.; Martus, P.; Kohsaka, S.; et al. Microglia turnover with aging and in an Alzheimer's model via long-term in vivo single-cell imaging. Nat. Neurosci. 2017, 20, 1371–1376. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef]
- Choi, I.; Wang, M.; Yoo, S.; Xu, P.; Seegobin, S.P.; Li, X.; Han, X.; Wang, Q.; Peng, J.; Zhang, B.; et al. Autophagy enables microglia to engage amyloid plaques and prevents microglial senescence. Nat. Cell Biol. 2023, 25, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Liu, B.; Gan, L. Autophagy prevents microglial senescence. Nat. Cell Biol. 2023, 25, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Karabag, D.; Scheiblich, H.; Griep, A.; Santarelli, F.; Schwartz, S.; Heneka, M.T.; Ising, C. Characterizing microglial senescence: Tau as a key player. J. Neurochem. 2023, 166, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Matsudaira, T.; Nakano, S.; Konishi, Y.; Kawamoto, S.; Uemura, K.; Kondo, T.; Sakurai, K.; Ozawa, T.; Hikida, T.; Komine, O.; et al. Cellular senescence in white matter microglia is induced during ageing in mice and exacerbates the neuroinflammatory phenotype. Commun. Biol. 2023, 6, 1–13. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; Lebrasseur, N.K.; Childs, B.G.; Van De Sluis, B.; Kirkland, J.L.; Van Deursen, J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Bussian, T.J.; Aziz, A.; Meyer, C.F.; Swenson, B.L.; Van Deursen, J.M.; Baker, D.J. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 2018, 562, 578–582. [Google Scholar] [CrossRef]
- Ogrodnik, M.; Evans, S.A.; Fielder, E.; Victorelli, S.; Kruger, P.; Salmonowicz, H.; Weigand, B.M.; Patel, A.D.; Pirtskhalava, T.; Inman, C.L.; et al. Whole-body senescent cell clearance alleviates age-related brain inflammation and cognitive impairment in mice. Aging Cell 2021, 20, e13296. [Google Scholar] [CrossRef]
- Wu, D.; Sun, J.K.-L.; Chow, K.H.-M. Neuronal cell cycle reentry events in the aging brain are more prevalent in neurodegeneration and lead to cellular senescence. PLOS Biol. 2024, 22, e3002559. [Google Scholar] [CrossRef]
- Sabath, N.; Levy-Adam, F.; Younis, A.; Rozales, K.; Meller, A.; Hadar, S.; Soueid-Baumgarten, S.; Shalgi, R. Cellular proteostasis decline in human senescence. Proc. Natl. Acad. Sci. 2020, 117, 31902–31913. [Google Scholar] [CrossRef]
- Husom, A.D.; A Peters, E.; A Kolling, E.; A Fugere, N.; Thompson, L.V.; A Ferrington, D. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004, 421, 67–76. [Google Scholar] [CrossRef]
- Strucksberg, K.-H.; Tangavelou, K.; Schröder, R.; Clemen, C.S. Proteasomal activity in skeletal muscle: A matter of assay design, muscle type, and age. Anal. Biochem. 2009, 399, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Nago, N.; Murata, S.; Tanaka, K.; Tanahashi, N. Changes in brain proteasome dynamics associated with aging. Genes Cells 2024, 29, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Puangmalai, N.; Sengupta, U.; Bhatt, N.; Gaikwad, S.; Montalbano, M.; Bhuyan, A.; Garcia, S.; McAllen, S.; Sonawane, M.; Jerez, C.; et al. Lysine 63-linked ubiquitination of tau oligomers contributes to the pathogenesis of Alzheimer’s disease. J. Biol. Chem. 2022, 298, 101766. [Google Scholar] [CrossRef] [PubMed]
- Keck, S.; Nitsch, R.; Grune, T.; Ullrich, O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J. Neurochem. 2003, 85, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Stratford, F.L.L.; Trougakos, I.P.; Friguet, B.; Rivett, A.J.; Gonos, E.S. Central Role of the Proteasome in Senescence and Survival of Human Fibroblasts: Induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003, 278, 28026–28037. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Gonos, E.S. Proteasome inhibition induces a senescence-like phenotype in primary human fibroblasts cultures. Biogerontology 2004, 5, 55–61. [Google Scholar] [CrossRef]
- Torres, C.; Lewis, L.; Cristofalo, V.J. Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J. Cell. Physiol. 2006, 207, 845–853. [Google Scholar] [CrossRef]
- Takenaka, Y.; Inoue, I.; Nakano, T.; Ikeda, M.; Kakinuma, Y. Prolonged disturbance of proteostasis induces cellular senescence via temporal mitochondrial dysfunction and subsequent mitochondrial accumulation in human fibroblasts. FEBS J. 2021, 289, 1650–1667. [Google Scholar] [CrossRef]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef]
- Iriki, T.; Iio, H.; Yasuda, S.; Masuta, S.; Kato, M.; Kosako, H.; Hirayama, S.; Endo, A.; Ohtake, F.; Kamiya, M.; et al. Senescent cells form nuclear foci that contain the 26S proteasome. Cell Rep. 2023, 42, 112880. [Google Scholar] [CrossRef]
- Heckenbach, I.; Mkrtchyan, G.V.; Ben Ezra, M.; Bakula, D.; Madsen, J.S.; Nielsen, M.H.; Oró, D.; Osborne, B.; Covarrubias, A.J.; Idda, M.L.; et al. Nuclear morphology is a deep learning biomarker of cellular senescence. Nat. Aging 2022, 2, 742–755. [Google Scholar] [CrossRef]
- Freund, A.; Laberge, R.-M.; Demaria, M.; Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 2012, 23, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Slobodnyuk, K.; Radic, N.; Ivanova, S.; Llado, A.; Trempolec, N.; Zorzano, A.; Nebreda, A.R. Autophagy-induced senescence is regulated by p38α signaling. Cell Death Dis. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.; Xu, C.; Donahue, G.; Shimi, T.; Pan, J.-A.; Zhu, J.; Ivanov, A.; Capell, B.C.; Drake, A.M.; Shah, P.P.; et al. Autophagy mediates degradation of nuclear lamina. Nature 2015, 527, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.T.; Lee, K.B.; Kim, S.Y.; Choi, H.R.; Park, S.C. Autophagy Impairment Induces Premature Senescence in Primary Human Fibroblasts. PLOS ONE 2011, 6, e23367. [Google Scholar] [CrossRef]
- Frost, B.; Bardai, F.H.; Feany, M.B. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr. Biol. 2015, 26, 129–136. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, X.; Zhang, Y.; Gao, Z.; Liu, Y.; Hu, J.; Hu, X.; Li, L.; Shi, J.; Gao, N. Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J. Exp. Clin. Cancer Res. 2019, 38, 1–14. [Google Scholar] [CrossRef]
- En, A.; Takauji, Y.; Miki, K.; Ayusawa, D.; Fujii, M. Lamin B receptor plays a key role in cellular senescence induced by inhibition of the proteasome. FEBS Open Bio 2019, 10, 237–250. [Google Scholar] [CrossRef]
- Lukášová, E.; Kovarˇík, A.; Bacˇíková, A.; Falk, M.; Kozubek, S. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem. J. 2017, 474, 281–300. [Google Scholar] [CrossRef]
- Arai, R.; En, A.; Takauji, Y.; Maki, K.; Miki, K.; Fujii, M.; Ayusawa, D. Lamin B receptor (LBR) is involved in the induction of cellular senescence in human cells. Mech. Ageing Dev. 2019, 178, 25–32. [Google Scholar] [CrossRef]
- Arai, R.; En, A.; Ukekawa, R.; Miki, K.; Fujii, M.; Ayusawa, D. Aberrant localization of lamin B receptor (LBR) in cellular senescence in human cells. Biochem. Biophys. Res. Commun. 2016, 473, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- De Sandre-Giovannoli, A.; Bernard, R.; Cau, P.; Navarro, C.; Amiel, J.; Boccaccio, I.; Lyonnet, S.; Stewart, C.L.; Munnich, A.; Le Merrer, M.; et al. Lamin A Truncation in Hutchinson-Gilford Progeria. Science 2003, 300, 2055–2055. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.A.; Madsen, A.S.; Olsen, C.A.; Hirschey, M.D. Metabolic control by sirtuins and other enzymes that sense NAD+, NADH, or their ratio. Biochim. et Biophys. Acta (BBA) - Bioenerg. 2017, 1858, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Lin, H. Sirtuins in Epigenetic Regulation. Chem. Rev. 2015, 115, 2350–2375. [Google Scholar] [CrossRef]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Xu, C.; Wang, L.; Fozouni, P.; Evjen, G.; Chandra, V.; Jiang, J.; Lu, C.; Nicastri, M.; Bretz, C.; Winkler, J.D.; et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 2020, 22, 1170–1179. [Google Scholar] [CrossRef]
- Young, A.R.J.; Arakawa, S.; Samarajiwa, S.A.; Nakashima, T.; Yoshida, S.; Hong, S.; Berry, L.S.; Reichelt, S.; Ferreira, M.; Tavaré, S.; et al. Spatial Coupling of mTOR and Autophagy Augments Secretory Phenotypes. Science 2011, 332, 966–970. [Google Scholar] [CrossRef]
- Tan, J.X.; Finkel, T. Lysosomes in senescence and aging. Embo Rep. 2023, 24, e57265. [Google Scholar] [CrossRef]
- Curnock, R.; Yalci, K.; Palmfeldt, J.; Jäättelä, M.; Liu, B.; Carroll, B. TFEB-dependent lysosome biogenesis is required for senescence. EMBO J. 2023, 42, e111241. [Google Scholar] [CrossRef]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Arakawa, S.; Kanaseki, T.; Miyatsuka, T.; Fujitani, Y.; Watada, H.; Tsujimoto, Y.; Shimizu, S. Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J. 2016, 35, 1991–2007. [Google Scholar] [CrossRef] [PubMed]
- Baron, O.; Boudi, A.; Dias, C.; Schilling, M.; Nölle, A.; Vizcay-Barrena, G.; Rattray, I.; Jungbluth, H.; Scheper, W.; Fleck, R.A.; et al. Stall in Canonical Autophagy-Lysosome Pathways Prompts Nucleophagy-Based Nuclear Breakdown in Neurodegeneration. Curr. Biol. 2017, 27, 3626–3642. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




