Submitted:
08 October 2024
Posted:
09 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
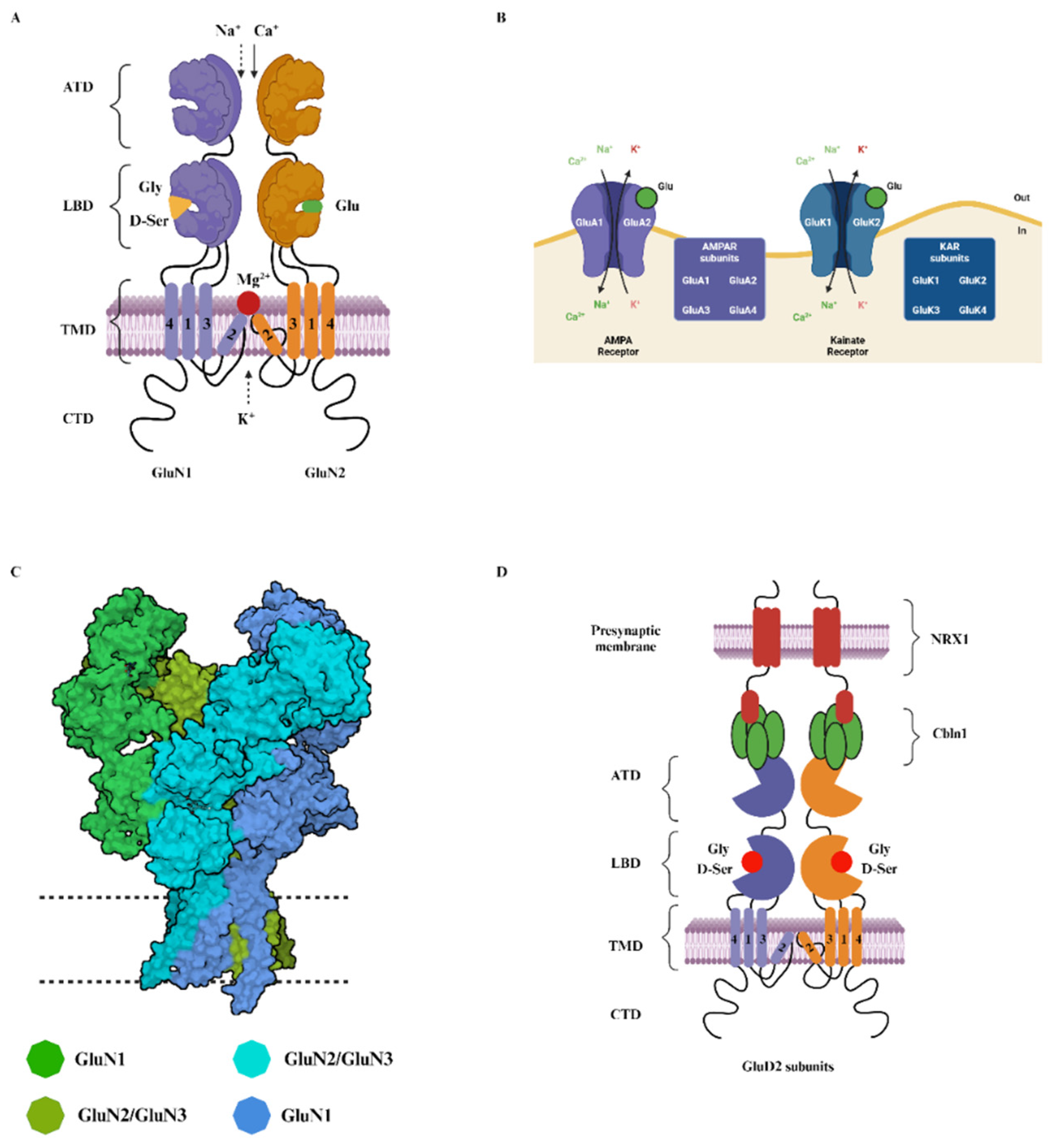
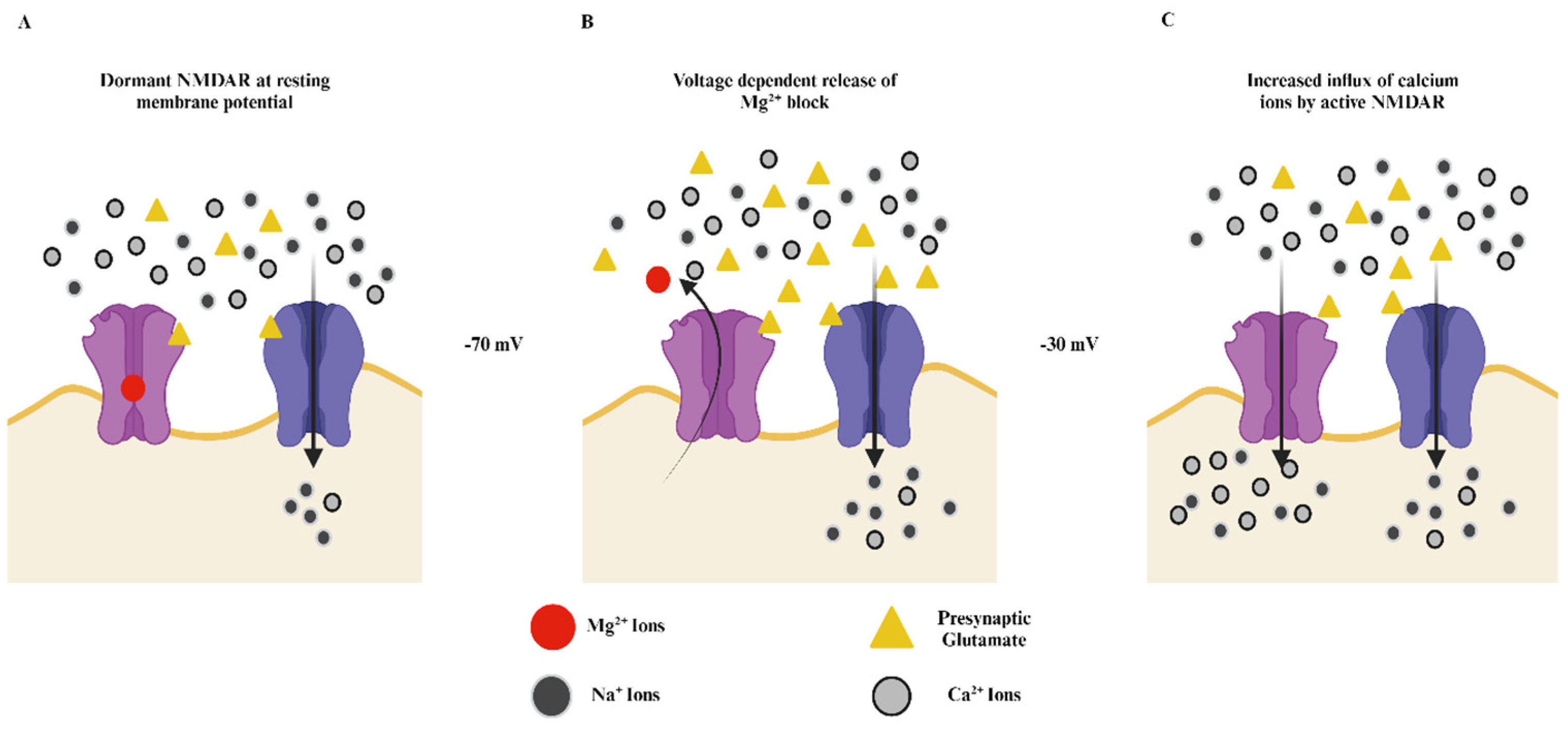
2. Structure and Function of NMDA Receptors
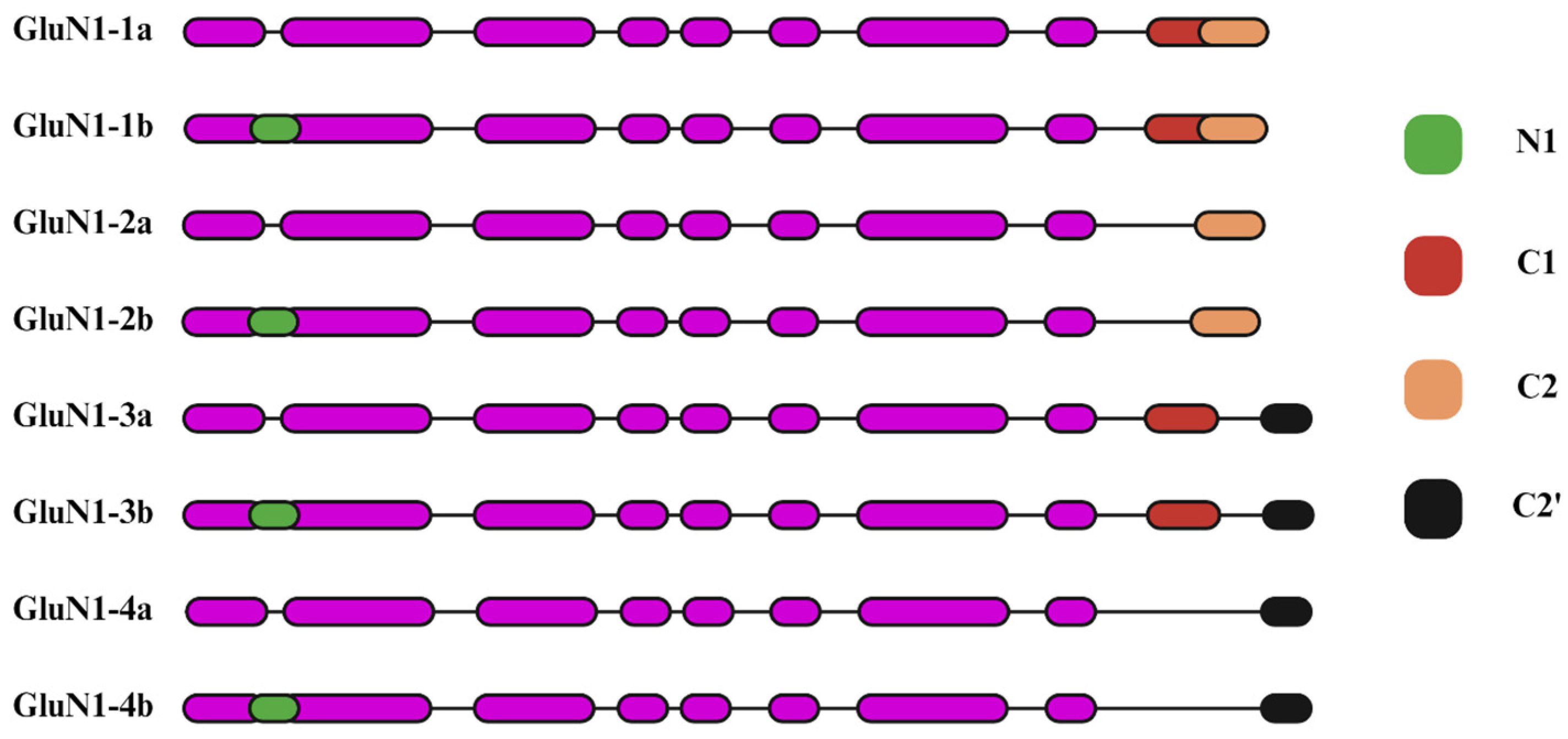
2.1. Glycine-Binding GluN1 Subunit
2.2. The GluN2 Diversity and Its Role in the NMDA Receptors
3. NMDA Receptors in Neurodevelopmental Disorders: Variants of GluN2 Subunits
4. Insights into NMDA Receptor Pathophysiology from Mouse Models of Schizophrenia and ASD
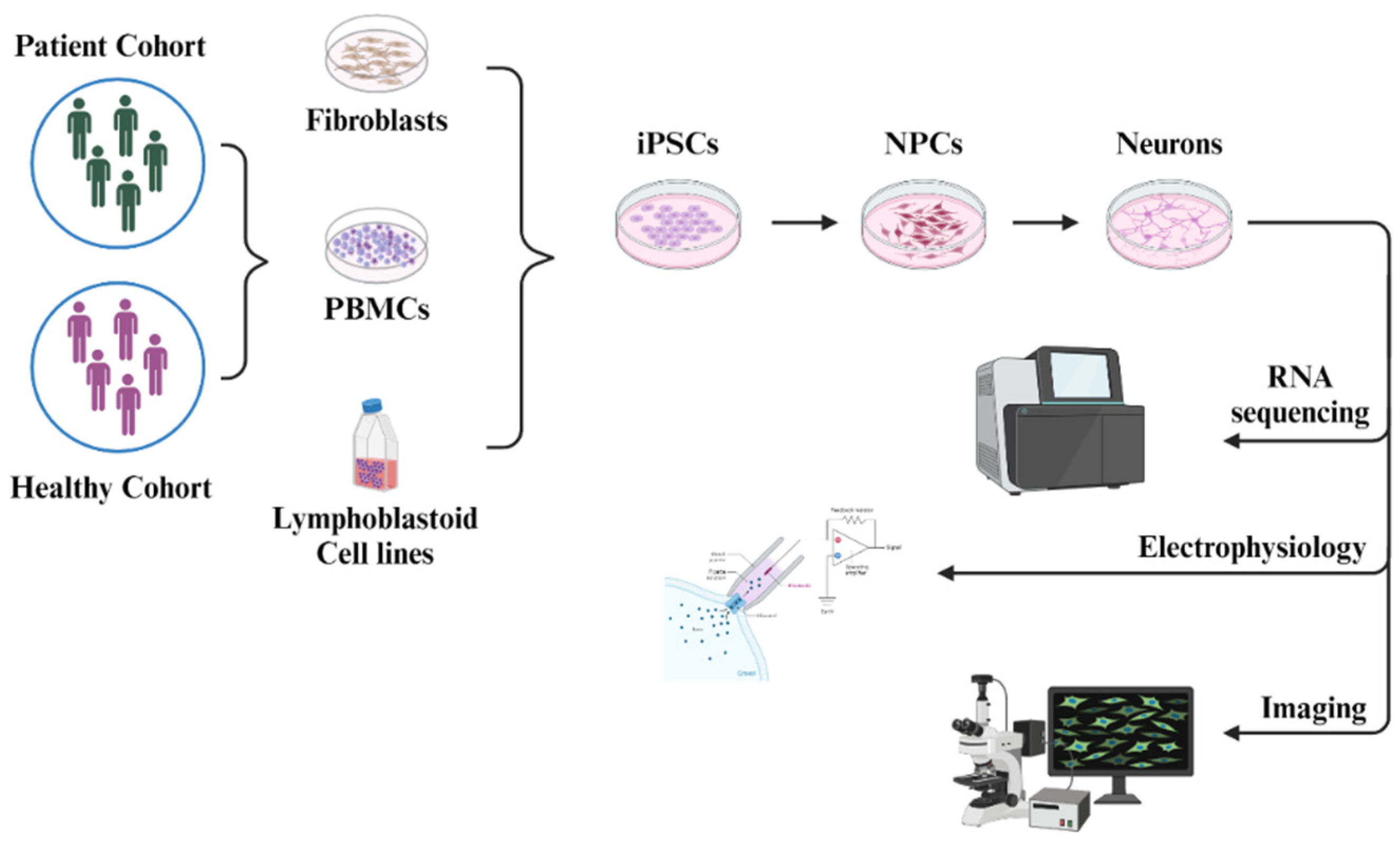
5. Human iPSC-Derived Neurons in the Research of NMDA Receptor Pathophysiology
5.1. iPSC Models for Studying NMDARs in ASD
5.2. iPSC Models for Studying NMDARs in Schizophrenia
5.3. iPSC Models for Studying NMDARs in Epilepsy
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Crupi R, Impellizzeri D, Cuzzocrea S. Role of Metabotropic Glutamate Receptors in Neurological Disorders. Front Mol Neurosci. 2019; 12: 20.
- Watkins JC, Jane DE. The glutamate story. Br J Pharmacol. 2006; 147: 100-8.
- Michael Hollmann AOS-G, Scott W. Rogers & Stephan Heinemann. Cloning by functional expression of a member of the glutamte receptor family. Nature. 1989; 342: 643-8.
- Seeburg, PH. The TiPS/TINS Lecture: The molecular biology of mammalian glutamate receptors. TiPS Reviews. 1993; 14: 297-303.
- Heinemann MHaS. Cloned Glutamate Receptors. Annu Rev Neurosci. 1994; 17: 31-108.
- SEIJI OZAWA. Glutamate receptors in the mammalian central nervous system. Prog Neurobiol. 1998; 54: 581-618.
- Hiroyuki Sugiyama II, Chikara Hirono. A new type of glutamate receptor linked to inositol phospholipid metabolism. Nature. 1987; 325: 531-3.
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010; 50: 295-322.
- Petronella Kettunen PK, Dietmar Hess, Abdeljabbar El Manira. Signaling Mechanisms of Metabotropic Glutamate Receptor 5 Subtype and Its Endogenous Role in a Locomotor Network. J Neurosci. 2002; 22: 1868–73.
- Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003; 98: 325-54.
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010; 62: 405-96.
- Elisa Carrillo1 CUG, 2, Vladimir Berka1, Vasanthi Jayaraman1,2*. Delta glutamate receptors are functional glycine- and d-serine–gated cation channels in situ. Sci Adv. 2021; 7: 1-9.
- Jane, D. AMPA glutatmate receptors. xPharm: The Comprehensive Pharmacology Reference. 2007:1-19.
- Yang Y, Calakos N. Presynaptic long-term plasticity. Front Synaptic Neurosci. 2013; 5: 8.
- Gan Q, Salussolia CL, Wollmuth LP. Assembly of AMPA receptors: mechanisms and regulation. J Physiol. 2015; 593: 39-48.
- Sabine, M. Schmid MH. To Gate or not to Gate: Are the Delta Subunits in the Glutamate Receptor Family Functional Ion Channels?. Mol Neurobiol. 2008; 37: 126-41.
- Orth A, Tapken D, Hollmann M. The delta subfamily of glutamate receptors: characterization of receptor chimeras and mutants. Eur J Neurosci. 2013; 37: 1620-30.
- Peter Naur* KBH, Anders S. Kristensen‡, Shashank M. Dravid‡, Darryl S. Pickering§, Lars Olsen*, Bente Vestergaard*, Jan Egebjerg†, Michael Gajhede*, Stephen F. Traynelis‡, and Jette S. Kastrup*¶. Ionotropic glutamate-like receptor δ2 binds D-serine and glycine. PNAS. 2007; 104: 14116–21.
- Gao J, Maison SF, Wu X, Hirose K, Jones SM, Bayazitov I, et al. Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol Cell Biol. 2007; 27: 4500-12.
- Yuzaki, M. The delta2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum. 2004; 3: 89-93.
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One. 2012; 7: e32969.
- Bowie, D. Ionotropic Glutamate Receptors & CNS Disorders. CNS Neurol Disord Drug Targets. 2008; 7: 129-143.
- Balazs R, Bridges RJ, Cotman CW, Cotman CA. Glutamate and Glutamate Receptors in Neurological Diseases. Excitatory Amino Acid Transmission in Health and Disease, 2005.
- Ragnarsson L, Dodd PR, Hynd MR. Role of Ionotropic Glutamate Receptors in Neurodegenerative and Other Disorders. In: Kostrzewa RM, editor. Handbook of Neurotoxicity. New York, NY: Springer New York; 2014. p. 1039-70.
- Chen TS, Huang TH, Lai MC, Huang CW. The Role of Glutamate Receptors in Epilepsy. Biomedicines. 2023; 11: 738.
- Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, et al. Structure, Function, and Pharmacology of Glutamate Receptor Ion Channels. Pharmacol Rev. 2021; 73: 298-487.
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005; 15: 603-15.
- Douglas Forrest M, * Holly D. Soares,*, Lily Ng DCL, * Morgan Sheng,+ Colin 1. Stewart,* James I. Morgan,* John A. Connor,* and Tom Curran*. Targeted Disruption of NMDA Receptor 1 Gene Abolishes NMDA Response and Results in Neonatal Death. Neuron. 1994; 13: 325-38.
- Gil-da-Costa R, Stoner GR, Fung R, Albright TD. Nonhuman primate model of schizophrenia using a noninvasive EEG method. PNAS. 2013; 110: 15425-30.
- Philip D Campbell, MG. Zebrafish as a tool to study schizophrenia-associated copy number variants. Disease Models & Mechanisms. 2020; 13: 1-24.
- Lim CS, Kim MJ, Choi JE, Islam MA, Lee YK, Xiong Y, et al. Dysfunction of NMDA receptors in neuronal models of an autism spectrum disorder patient with a DSCAM mutation and in Dscam-knockout mice. Mol Psychiatry. 2021; 26: 7538-7549.
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, et al. Organization of NMDA receptors at extrasynaptic locations. NeuroScience. 2010; 167: 68-87.
- Ralf Mohrmann HHaKG. Developmental regulation of subunit composition of extrasynaptic NMDA receptors in neocortical neurones. Dev Neurosci. 2000; 11: 1203-1208.
- Chang HR, Kuo CC. The activation gate and gating mechanism of the NMDA receptor. J Neurosci. 2008; 28: 1546-1556.
- Jeffrey, A. Dzubay CEJ. Kinetics of NMDA Channel Opening. The Journal of NeuroScience. 1996; 16: 4129–4134.
- Kawamoto EM, Vivar C, Camandola S. Physiology and pathology of calcium signaling in the brain. Front Pharmacol. 2012; 3: 61.
- Mauceri D, Freitag HE, Oliveira AM, Bengtson CP, Bading H. Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron. 2011; 71: 117-30.
- Papadia S, Stevenson P, Hardingham NR, Bading H, Hardingham GE. Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci. 2005; 25: 4279-87.
- Saneyoshi T, Fortin DA, Soderling TR. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr Opin Neurobiol. 2010; 20: 108-15.
- Sprengel R, Eltokhi A. Ionotropic Glutamate Receptors (and Their Role in Health and Disease). NeuroScience in the 21st Century. 2022. p. 57-86.
- Suzuki Y, Nakamoto C, Watanabe-Iida I, Watanabe M, Takeuchi T, Sasaoka T, et al. Quantitative analysis of NMDA receptor subunits proteins in mouse brain. Neurochem Int. 2023; 165: 105517.
- Li H, Rajani V, Han L, Chung D, Cooke JE, Sengar AS, et al. Alternative splicing of GluN1 gates glycine site-dependent nonionotropic signaling by NMDAR receptors. PNAS. 2021; 118: e2026411118.
- R. Suzanne Zukin MVLB. Alternatively spliced isoforms of the NMDAR I receptor subunit. Trends in NeuroSciences. 1995; 18: 306-13.
- Lin Y, Skeberdis VA, Francesconi A, Bennett MV, Zukin RS. Postsynaptic density protein-95 regulates NMDA channel gating and surface expression. J Neurosci. 2004; 24: 10138-10148.
- Incontro S, Diaz-Alonso J, Iafrati J, Vieira M, Asensio CS, Sohal VS, et al. The CaMKII/NMDA receptor complex controls hippocampal synaptic transmission by kinase-dependent and independent mechanisms. Nat Commun. 2018; 9: 2069.
- Michael, D. Ehlers ETF, Richard J. O’Brien, Richard L. Huganir. Splice Variant-Specific Interaction of the NMDA Receptor Subunit NR1 with Neuronal Intermediate Filaments. J Neurosci. 1998; 18: 720-30.
- Guylaine, M. Durand mvlb, R. Suzanne zukin. Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. PNAS. 1993; 90: 6731-5.
- Ling zhang xz, Marie-christine paupard, Alice p. Wang, Linda santchi, Linda k. Friedman, r. Suzanne zukin, Michael v. L. Bennett. Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. PNAS. 1994; 91: 10883-10887.
- Gavin rumbaugh kp, Jian feng wang, Stefano vicini. Exon 5 and Spermine Regulate Deactivation of NMDA Receptor Subtypes. J Neurophysiol. 2000; 83: 1300-1306.
- Vance KM, Hansen KB, Traynelis SF. GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol. 2012; 590: 3857-3875.
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun. 2011; 2: 294.
- Vieira M, Yong XLH, Roche KW, Anggono V. Regulation of NMDA glutamate receptor functions by the GluN2 subunits. J Neurochem. 2020; 154: 121-143.
- Hannah Monyer NB, David J. Laurie, Bert Sakmann, Peter H. Seeburg. Developmental and Regional Expression in the Rat Brain and Functional Properties of Four NMDA Receptors. Neuron. 1994; 12: 529-540.
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994; 347: 150-160.
- Sheng M, Cummings, J., Roldan, L. A., Jan, Y. N., Jan, L. Y. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994; 368: 144–147.
- Wenzel A, Fritschy JM, Mohler H, Benke D. NMDA receptor heterogeneity during postnatal development of the rat brain: differential expression of the NR2A, NR2B, and NR2C subunit proteins. J Neurochem. 1997; 68: 469-478.
- Xi D, Keeler B, Zhang W, Houle JD, Gao WJ. NMDA receptor subunit expression in GABAergic interneurons in the prefrontal cortex: application of laser microdissection technique. J Neurosci Methods. 2009; 176: 172-181.
- Perszyk RE, DiRaddo JO, Strong KL, Low CM, Ogden KK, Khatri A, et al. GluN2D-Containing N-methyl-d-Aspartate Receptors Mediate Synaptic Transmission in Hippocampal Interneurons and Regulate Interneuron Activity. Mol Pharmacol. 2016; 90: 689-702.
- Stephen, G. Brickley CM, M. H. Selina Mok, Masayoshi Mishina, Stuart G. Cull-Candy. NR2B and NR2D Subunits Coassemble in Cerebellar Golgi Cells to Form a Distinct NMDA Receptor Subtype Restricted to Extrasynaptic Sites. J Neurosci. 2003; 23: 4958–4966.
- Charu Misra SGB, Mark Farrant, Stuart G. Cull-Candy. Identification of subunits contributing to synaptic and extrasynaptic NMDA receptors in Golgi cells of the rat cerebellum. J Physiol. 2000; 524: 147-162.
- Andres Barria, RM. Subunit-Specific NMDA Receptor Trafficking to Synapses. Neuron. 2002; 35: 345-353.
- Kenneth, R. Tovar GLW. The Incorporation of NMDA Receptors with a Distinct Subunit Composition at Nascent Hippocampal Synapses In Vitro. J Neurosci. 1999; 19: 4180–4188.
- Herbrechter R, Hube N, Buchholz R, Reiner A. Splicing and editing of ionotropic glutamate receptors: a comprehensive analysis based on human RNA-Seq data. Cell Mol Life Sci. 2021; 78: 5605-5630.
- Warming H, Pegasiou CM, Pitera AP, Kariis H, Houghton SD, Kurbatskaya K, et al. A primate-specific short GluN2A-NMDA receptor isoform is expressed in the human brain. Mol Brain. 2019; 12: 64.
- Hiroyuki Meguro HM, Kazuaki Araki, Etsuko Kushiya, Tatsuya Kutsuwada, Makoto Yamazaki, Toshiro Kumanishi, Masaaki Arakawa, Kenji Sakimura, Masayoshi Mishina Functional Characterization of heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature 1992; 357: 70–74.
- Hannah Monyer RS, Ralf Schoepfer, Anne Herb, Miyoko Higuchi, Hilda Lomeli, Nail Burnashev, Bert Sakmann, Peter H. Seeburg. Heteromeric NMDA Receptors: Molecular and Functional Distinction of Subtypes. Science. 1992; 256: 1217-1221.
- Tabish M, Ticku MK. Alternate splice variants of mouse NR2B gene. Neurochem Int. 2004; 44: 339-343.
- Takahiro Ishii KM, Hidemitsu Sugihara, Kazuhiro Sakurada, Hiroshi Kadotani, Mineto Yokoi, Chihiro Akazawa, Ryuichi Shigemoto, Noboru Mizuno, Masayuki Masu, Shigetada Nakanishi. Molecular Characterization of the Family of the N-Methyl-D-Aspartate Receptor Subunits. JBC. 1993; 268: 2836-2843.
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010; 42: 1021-1026.
- Brian, J. O’Roak LV, Wenqing Fu, Jarrett D. Egertson, Ian B. Stanaway, Ian G. Phelps, Gemma Carvill, Akash Kumar, Choli Lee, Katy Ankenman, Jeff Munson, Joseph B. Hiatt, Emily H. Turner, Roie Levy, Diana R. O’Day, Niklas Krumm, Bradley P. Coe, Beth K. Martin, Elhanan Borenstein, Deborah A. Nickerson, Heather C. Mefford, Dan Doherty, Joshua M. Akey, Raphael Bernier, Evan E. Eichler, Jay Shendure. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science. 2012; 338: 1619-1622.
- EM Kenny PC, S Furlong, E Heron, G Kenny, C Fahey, E Kelleher, S Ennis, D Tropea, R Anney, AP Corvin, G Donohoe, L Gallagher, M Gill, DW Morris. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry. 2014; 19: 872–879.
- Myers RA, Casals F, Gauthier J, Hamdan FF, Keebler J, Boyko AR, et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 2011; 7(2): e1001318.
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012; 367: 1921-1929.
- Strehlow V, Heyne HO, Vlaskamp DRM, Marwick KFM, Rudolf G, de Bellescize J, et al. GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain. 2019; 142: 80-92.
- Garcia-Recio A, Santos-Gomez A, Soto D, Julia-Palacios N, Garcia-Cazorla A, Altafaj X, et al. GRIN database: A unified and manually curated repertoire of GRIN variants. Hum Mutat. 2021; 42: 8-18.
- Liu XR, Xu XX, Lin SM, Fan CY, Ye TT, Tang B, et al. GRIN2A Variants Associated With Idiopathic Generalized Epilepsies. Front Mol Neurosci. 2021; 14: 720984.
- Harrison PJ, Bannerman DM. GRIN2A (NR2A): a gene contributing to glutamatergic involvement in schizophrenia. Mol Psychiatry. 2023; 28: 3568-3572.
- Shepard N, Baez-Nieto D, Iqbal S, Kurganov E, Budnik N, Campbell AJ, et al. Differential functional consequences of GRIN2A mutations associated with schizophrenia and neurodevelopmental disorders. Sci Rep. 2024; 14: 2798.
- Tarabeux J, Kebir O, Gauthier J, Hamdan FF, Xiong L, Piton A, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry. 2011; 1: e55.
- Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022; 604: 509-16.
- Camp CR, Vlachos A, Klockner C, Krey I, Banke TG, Shariatzadeh N, et al. Loss of Grin2a causes a transient delay in the electrophysiological maturation of hippocampal parvalbumin interneurons. Commun Biol. 2023; 6: 952.
- Kellner S, Berlin S. Rescuing tri-heteromeric NMDA receptor function: the potential of pregnenolone-sulfate in loss-of-function GRIN2B variants. Cell Mol Life Sci. 2024; 81: 235.
- Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci. 2016; 132: 115-121.
- Epi KC, Epilepsy Phenome/Genome P, Allen AS, Berkovic SF, Cossette P, Delanty N, et al. De novo mutations in epileptic encephalopathies. Nature. 2013; 501: 217-221.
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011; 43: 585-589.
- Bahry JA, Fedder-Semmes KN, Sceniak MP, Sabo SL. An Autism-Associated de novo Mutation in GluN2B Destabilizes Growing Dendrites by Promoting Retraction and Pruning. Front Cell Neurosci. 2021; 15: 692232.
- Sceniak MP, Fedder KN, Wang Q, Droubi S, Babcock K, Patwardhan S, et al. An autism-associated mutation in GluN2B prevents NMDA receptor trafficking and interferes with dendrite growth. J Cell Sci. 2019; 132: jcs232892.
- Weickert CS, Fung SJ, Catts VS, Schofield PR, Allen KM, Moore LT, et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry. 2013; 18: 1185-1192.
- Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008; 33(9):2175-86.
- S. Akbarian NJS, D. Bradley,’ A. Tafazzoli, l D. Trinh, l W. P. Hetrick, S. G. Potkin,, C. A. Sandman WEB, Jr., E. G. Jones. Selective Alterations in Gene Expression for NMDA Receptor Subunits in Prefrontal Cortex of Schizophrenics The Journal of NeuroScience. 1996; 16: 19-30.
- Catts VS, Lai YL, Weickert CS, Weickert TW, Catts SV. A quantitative review of the postmortem evidence for decreased cortical N-methyl-D-aspartate receptor expression levels in schizophrenia: How can we link molecular abnormalities to mismatch negativity deficits? Biol Psychol. 2016; 116: 57-67.
- Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, et al. GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet. 2016; 99: 802-816.
- Yu Y, Lin Y, Takasaki Y, Wang C, Kimura H, Xing J, et al. Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl Psychiatry. 2018; 8: 12.
- Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018; 174: 999-1014.
- Matthaei, KI. Genetically manipulated mice: a powerful tool with unsuspected caveats. J Physiol. 2007; 582:481-488.
- Dali Li ZQ, Yanjiao Shao, Yuting Chen, Yuting Guan, Meizhen Liu, Yongmei Li, Na Gao, Liren Wang, Xiaoling Lu, Yongxiang Zhao, Mingyao Liu. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013; 31: 681-683.
- Wohr M, Scattoni ML. Behavioural methods used in rodent models of autism spectrum disorders: current standards and new developments. Behav Brain Res. 2013; 251: 5-17.
- Amy, R. Mohn RRG, Marc G. Caron, Beverly H. Koller. Mice with Reduced NMDA Receptor Expression Display Behaviors Related to Schizophrenia. Cell. 1999; 98: 427-36.
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, et al. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010; 13: 76-83.
- Xu W, Morishita W, Buckmaster PS, Pang ZP, Malenka RC, Sudhof TC. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 2012; 73: 990-1001.
- Yuqing Li RSE, Chong Chen, Sonal Jhaveri, Susumu Tonegawa. Whisker-Related Neuronal Patterns Fail to Develop in the Trigeminal Brainstem Nuclei of NMDARI Knockout Mice. Cell. 1994; 76: 427-37.
- Tatsuya Kutsuwada KS, Toshiya Manabe, Chitoshi Takayama,, Nobuo Katakura EK, Rie Natsume, Masahiko Watanabe, Yoshiro Inoue, Takeshi Yagi, Shinichi Aizawa, Masaaki Arakawa, Tomoyuki Takahashi, Yoshio Nakamura, Hisashi Mori, Masayoshi Mishina. Impairment of Suckling Response, Trigeminal Neuronal Pattern Formation, and Hippocampal LTD in NMDA Receptor E2 Subunit Mutant Mice. Neuron. 1996; 16: 333–344.
- Bialon M, Wasik A. Advantages and Limitations of Animal Schizophrenia Models. Int J Mol Sci. 2022; 23(11).
- Yavas E, Young AMJ. Repeated phencyclidine disrupts nicotinic acetylcholine regulation of dopamine release in nucleus accumbens: Implications for models of schizophrenia. Neurochem Int. 2020; 140: 104836.
- Cao T, Tang M, Jiang P, Zhang B, Wu X, Chen Q, et al. A Potential Mechanism Underlying the Therapeutic Effects of Progesterone and Allopregnanolone on Ketamine-Induced Cognitive Deficits. Front Pharmacol. 2021; 12: 612083.
- Chen G, Lin X, Li G, Jiang D, Lib Z, Jiang R, et al. Risperidone reverses the spatial object recognition impairment and hippocampal BDNF-TrkB signalling system alterations induced by acute MK-801 treatment. Biomed Rep. 2017; 6: 285-290.
- Sobolevsky AI, Yelshansky MV. The trapping block of NMDA receptor channels in acutely isolated rat hippocampal neurones. J Physiol. 2000; 526: 493-506.
- Wallach J, Kang H, Colestock T, Morris H, Bortolotto ZA, Collingridge GL, et al. Pharmacological Investigations of the Dissociative ‘Legal Highs’ Diphenidine, Methoxphenidine and Analogues. PLoS One. 2016; 11: e0157021.
- Vales K, Holubova K. Minireview: Animal model of schizophrenia from the perspective of behavioral pharmacology: Effect of treatment on cognitive functions. Neurosci Lett. 2021; 761: 136098.
- Salmi M, Bolbos R, Bauer S, Minlebaev M, Burnashev N, Szepetowski P. Transient microstructural brain anomalies and epileptiform discharges in mice defective for epilepsy and language-related NMDA receptor subunit gene Grin2a. Epilepsia. 2018; 59: 1919-1930.
- Kazutaka Ikeda KA, Chitoshi Takayama, Yoshiro Inoue, Takeshi Yagi, Shinichi Aizawa, Masayoshi Mishina. Reduced spontaneous activity of mice defective in the e4 subunit of the NMDA receptor channel. Brain Res Mol Brain Res. 1995; 33: 61-71.
- Alexander, K. Ebralidze DJR, 2 Susumu Tonegawa,1 and N. Traverse Slater2. Modification of NMDA Receptor Channels and Synaptic Transmission by Targeted Disruption of the NR2C Gene. J Neurosci. 1996; 16: 5014-5025.
- Saumya Das* YFS, Thomas Rothe†‡, Louis S. Premkumar§, Mari Takasu*, James E. Crandallk¶, Pieter Dikkes#, David A. Conner$, Posina V. Rayudu‡, Wing Cheung‡, H.-S. Vincent Chen‡, Stuart A. Lipton‡¶ & Nobuki Nakanishi*¶. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 1998; 393: 377-381.
- Takuji Iwasato RSE, † Patricio T. Huerta,* Dong Feng Chen,* Toshikuni Sasaoka,* Emel Ulupinar,† and Susumu Tonegawa*. NMDA Receptor-Dependent Refinement of Somatotopic Maps. Neuron. 1997; 19: 1201–1210.
- Lee LJ, Lo FS, Erzurumlu RS. NMDA receptor-dependent regulation of axonal and dendritic branching. J Neurosci. 2005; 25: 2304-2311.
- Duncan GE, Moy SS, Perez A, Eddy DM, Zinzow WM, Lieberman JA, et al. Deficits in sensorimotor gating and tests of social behavior in a genetic model of reduced NMDA receptor function. Behav Brain Res. 2004; 153: 507-519.
- Moy SS, Perez A, Koller BH, Duncan GE. Amphetamine-induced disruption of prepulse inhibition in mice with reduced NMDA receptor function. Brain Res. 2006; 1089: 186-194.
- Bickel S, Lipp HP, Umbricht D. Impaired attentional modulation of auditory evoked potentials in N-methyl-D-aspartate NR1 hypomorphic mice. Genes Brain Behav. 2007; 6: 558-568.
- Dzirasa K, Ramsey AJ, Takahashi DY, Stapleton J, Potes JM, Williams JK, et al. Hyperdopaminergia and NMDA receptor hypofunction disrupt neural phase signaling. J Neurosci. 2009; 29: 8215-8224.
- Halene TB, Ehrlichman RS, Liang Y, Christian EP, Jonak GJ, Gur TL, et al. Assessment of NMDA receptor NR1 subunit hypofunction in mice as a model for schizophrenia. Genes Brain Behav. 2009; 8: 661-675.
- Wang W, Rein B, Zhang F, Tan T, Zhong P, Qin L, et al. Chemogenetic Activation of Prefrontal Cortex Rescues Synaptic and Behavioral Deficits in a Mouse Model of 16p11.2 Deletion Syndrome. J Neurosci. 2018; 38: 5939-5948.
- do Nascimento P, Oliveira Silva DF, de Morais T, de Rezende AA. Zinc Status and Autism Spectrum Disorder in Children and Adolescents: A Systematic Review. Nutrients. 2023; 15: 3663.
- Sauer AK, Hagmeyer S, Grabrucker AM. Prenatal Zinc Deficient Mice as a Model for Autism Spectrum Disorders. Int J Mol Sci. 2022; 23: 6082.
- Faber S, Zinn GM, Kern JC, 2nd, Kingston HM. The plasma zinc/serum copper ratio as a biomarker in children with autism spectrum disorders. Biomarkers. 2009; 14: 171-180.
- Lee K, Jung Y, Vyas Y, Skelton I, Abraham WC, Hsueh YP, et al. Dietary zinc supplementation rescues fear-based learning and synaptic function in the Tbr1(+/-) mouse model of autism spectrum disorders. Mol Autism. 2022; 13:13.
- Shin W, Kim K, Serraz B, Cho YS, Kim D, Kang M, et al. Early correction of synaptic long-term depression improves abnormal anxiety-like behavior in adult GluN2B-C456Y-mutant mice. PLoS Biol. 2020; 18: e3000717.
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010; 42: 489-91.
- Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012; 8: e1002521.
- Delling JP, Boeckers TM. Comparison of SHANK3 deficiency in animal models: phenotypes, treatment strategies, and translational implications. J Neurodev Disord. 2021; 13: 55.
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011; 472: 437-442.
- Fourie C, Vyas Y, Lee K, Jung Y, Garner CC, Montgomery JM. Dietary Zinc Supplementation Prevents Autism Related Behaviors and Striatal Synaptic Dysfunction in Shank3 Exon 13-16 Mutant Mice. Front Cell Neurosci. 2018; 12: 374.
- Ey E, Leblond CS, Bourgeron T. Behavioral profiles of mouse models for autism spectrum disorders. Autism Res. 2011; 4: 5-16.
- Tordjman S, Drapier D, Bonnot O, Graignic R, Fortes S, Cohen D, et al. Animal models relevant to schizophrenia and autism: validity and limitations. Behav Genet. 2007; 37: 61-78.
- Tania M, Khan MA, Xia K. Recent advances in animal model experimentation in autism research. Acta Neuropsychiatr. 2014; 26: 264-271.
- Feng G, Jensen FE, Greely HT, Okano H, Treue S, Roberts AC, et al. Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. PNAS. 2020; 117: 24022-24031.
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126: 663-676.
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131: 861-872.
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010; 143: 527-539.
- Vahsen BF, Gray E, Candalija A, Cramb KML, Scaber J, Dafinca R, et al. Human iPSC co-culture model to investigate the interaction between microglia and motor neurons. Sci Rep. 2022; 12: 12606.
- Nascimento JM, Saia-Cereda VM, Zuccoli GS, Reis-de-Oliveira G, Carregari VC, Smith BJ, et al. Proteomic signatures of schizophrenia-sourced iPSC-derived neural cells and brain organoids are similar to patients’ postmortem brains. Cell Biosci. 2022; 12: 189.
- Fan Y, Li Y, Yang X, Zhang H, Wang B, Guan J, et al. Generation and characterization of PBMCs-derived human induced pluripotent stem cell (iPSC) line SDQLCHi051-A from an autism spectrum disorder patient with compound CHD8 gene mutations. Stem Cell Res. 2023; 69: 103114.
- Longobardi E, Miceli F, Secondo A, Cicatiello R, Izzo A, Tinto N, et al. Generation of an iPSC line (UNINAi001-A) from a girl with neonatal-onset epilepsy and non-syndromic intellectual disability carrying the homozygous KCNQ3 p.PHE534ILEfs*15 variant and of an iPSC line (UNINAi002-A) from a non-carrier, unaffected brother. Stem Cell Res. 2021; 53: 102311.
- Stern S, Sarkar A, Stern T, Mei A, Mendes APD, Stern Y, et al. Mechanisms Underlying the Hyperexcitability of CA3 and Dentate Gyrus Hippocampal Neurons Derived from Patients with Bipolar Disorder. Biol Psychiatry. 2020; 88: 139-149.
- Zhou YY, Zeng F. Integration-free methods for generating induced pluripotent stem cells. Genomics Proteomics Bioinformatics. 2013; 11: 284-287.
- Nayak R, Rosh I, Rabinski T, Falik D, Mendel Percia M, Stern S. Generation and characterization of iPSC lines (UOHi003-A, UOHi002-A) from a patient with SHANK3 mutation and her healthy mother. Stem Cell Res. 2022; 64: 102899.
- Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development. 2018; 145: dev156166.
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013; 14: 383-400.
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994; 12: 529-540.
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013; 78: 785-798.
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014; 83: 69-86.
- Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep. 2018; 23: 2509-2523.
- Zhang WB, Ross PJ, Tu Y, Wang Y, Beggs S, Sengar AS, et al. Fyn Kinase regulates GluN2B subunit-dominant NMDA receptors in human induced pluripotent stem cell-derived neurons. Sci Rep. 2016; 6: 23837.
- Frega M, Linda K, Keller JM, Gumus-Akay G, Mossink B, van Rhijn JR, et al. Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nat Commun. 2019; 10: 4928.
- Neagoe I, Liu C, Stumpf A, Lu Y, He D, Francis R, et al. The GluN2B subunit represents a major functional determinant of NMDA receptors in human induced pluripotent stem cell-derived cortical neurons. Stem Cell Res. 2018; 28: 105-114.
- Ruden JB, Dixit M, Zepeda JC, Grueter BA, Dugan LL. Robust Expression of Functional NMDA Receptors in Human Induced Pluripotent Stem Cell-Derived Neuronal Cultures Using an Accelerated Protocol. Front Mol Neurosci. 2021; 14: 777049.
- Bell S, Maussion G, Jefri M, Peng H, Theroux JF, Silveira H, et al. Disruption of GRIN2B Impairs Differentiation in Human Neurons. Stem Cell Reports. 2018; 11: 183-196.
- Ben-Reuven L, Reiner O. Modeling the autistic cell: iPSCs recapitulate developmental principles of syndromic and nonsyndromic ASD. Dev Growth Differ. 2016; 58: 481-491.
- Lim CS, Yang JE, Lee YK, Lee K, Lee JA, Kaang BK. Understanding the molecular basis of autism in a dish using hiPSCs-derived neurons from ASD patients. Mol Brain. 2015; 8: 57.
- Tripathi MK, Ojha SK, Kartawy M, Hamoudi W, Choudhary A, Stern S, et al. The NO Answer for Autism Spectrum Disorder. Adv Sci. 2023; 10: e2205783.
- Brant B, Stern T, Shekhidem HA, Mizrahi L, Rosh I, Stern Y, et al. IQSEC2 mutation associated with epilepsy, intellectual disability, and autism results in hyperexcitability of patient-derived neurons and deficient synaptic transmission. Mol Psychiatry. 2021; 26: 7498-7508.
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002; 81: 903-910.
- Kreienkamp, HJ. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb Exp Pharmacol. 2008; 186: 365-380.
- Arons MH, Thynne CJ, Grabrucker AM, Li D, Schoen M, Cheyne JE, et al. Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. J Neurosci. 2012; 32: 14966-14978.
- Amal H, Barak B, Bhat V, Gong G, Joughin BA, Wang X, et al. Shank3 mutation in a mouse model of autism leads to changes in the S-nitroso-proteome and affects key proteins involved in vesicle release and synaptic function. Mol Psychiatry. 2020; 25: 1835-1848.
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012; 74: 285-299.
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012; 485: 237-241.
- Hussein Y, Tripathi U, Choudhary A, Nayak R, Peles D, Rosh I, et al. Early maturation and hyperexcitability is a shared phenotype of cortical neurons derived from different ASD-associated mutations. Transl Psychiatry. 2023; 13: 246.
- Stern S, Zhang L, Wang M, Wright R, Rosh I, Hussein Y, et al. Monozygotic twins discordant for schizophrenia differ in maturation and synaptic transmission. Mol Psychiatry. 2024; 29: 3208-3222.
- Vilain J, Galliot AM, Durand-Roger J, Leboyer M, Llorca PM, Schurhoff F, et al. Environmental risk factors for schizophrenia: a review. Encephale. 2013; 39: 19-28.
- Romanovsky E, Choudhary A, Peles D, Abu-Akel A, Stern S. Uncovering convergence and divergence between autism and schizophrenia using genomic tools and patients’ neurons. Mol Psychiatry 2024. (ahead of print). [CrossRef]
- Dracheva S, Marras SA, Elhakem SL, Kramer FR, Davis KL, Haroutunian V. N-methyl-D-aspartic acid receptor expression in the dorsolateral prefrontal cortex of elderly patients with schizophrenia. Am J Psychiatry. 2001; 158: 1400-1410.
- Le Corre S, Harper CG, Lopez P, Ward P, Catts S. Increased levels of expression of an NMDARI splice variant in the superior temporal gyrus in schizophrenia. Neuroreport. 2000; 11: 983-986.
- Sokolov, BP. Expression of NMDAR1, GluR1, GluR7, and KA1 glutamate receptor mRNAs is decreased in frontal cortex of “neuroleptic-free” schizophrenics: evidence on reversible up-regulation by typical neuroleptics. J Neurochem. 1998; 71: 2454-2464.
- Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, et al. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996; 16: 19-30.
- Rasanen N, Tiihonen J, Koskuvi M, Lehtonen S, Koistinaho J. The iPSC perspective on schizophrenia. Trends Neurosci. 2022; 45: 8-26.
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011; 16: 358-360.
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011; 473: 221-225.
- Balan S, Toyoshima M, Yoshikawa T. Contribution of induced pluripotent stem cell technologies to the understanding of cellular phenotypes in schizophrenia. Neurobiol Dis. 2019; 131: 104162.
- Nakazawa T, Hashimoto R, Takuma K, Hashimoto H. Modeling of psychiatric disorders using induced pluripotent stem cell-related technologies. J Pharmacol Sci. 2019; 140: 321-324.
- Powell SK, O’Shea CP, Shannon SR, Akbarian S, Brennand KJ. Investigation of Schizophrenia with Human Induced Pluripotent Stem Cells. Adv Neurobiol. 2020; 25: 155-206.
- Wen Z, Christian KM, Song H, Ming GL. Modeling psychiatric disorders with patient-derived iPSCs. Curr Opin Neurobiol. 2016; 36: 118-127.
- Choudhary A, Peles D, Nayak R, Mizrahi L, Stern S. Current progress in understanding schizophrenia using genomics and pluripotent stem cells: A meta-analytical overview. Schizophr Res. 2022.
- Fischer, M. Psychoses in the offspring of schizophrenic monozygotic twins and their normal co-twins. Br J Psychiatry. 1971; 118: 43-52.
- Narayan CL, Shikha D, Shekhar S. Schizophrenia in identical twins. Indian J Psychiatry. 2015; 57: 323-324.
- Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, et al. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol Psychiatry. 2018; 830: 492-498.
- Castellani CA, Laufer BI, Melka MG, Diehl EJ, O’Reilly RL, Singh SM. DNA methylation differences in monozygotic twin pairs discordant for schizophrenia identifies psychosis related genes and networks. BMC Med Genomics. 2015; 8: 17.
- Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012; 37: 4-15.
- Rosburg T, Kreitschmann-Andermahr I. The effects of ketamine on the mismatch negativity (MMN) in humans - A meta-analysis. Clin Neurophysiol. 2016; 127: 1387-1394.
- Laurens KR, Murphy J, Dickson H, Roberts RE, Gutteridge TP. Trajectories of Mismatch Negativity and P3a Amplitude Development from Ages 9 to 16 Years in Children with Risk Factors for Schizophrenia. Biol Psychiatry Cogn Neurosci Neuroimaging. 2020; 5: 1085-1094.
- Kim NS, Wen Z, Liu J, Zhou Y, Guo Z, Xu C, et al. Pharmacological rescue in patient iPSC and mouse models with a rare DISC1 mutation. Nat Commun. 2021; 12:1398.
- Burrack N, Yitzhaky A, Mizrahi L, Wang M, Stern S, Hertzberg L. Altered Expression of PDE4 Genes in Schizophrenia: Insights from a Brain and Blood Sample Meta-Analysis and iPSC-Derived Neurons. Genes. 2024; 15: 609.
- Gilleen J, Nottage J, Yakub F, Kerins S, Valdearenas L, Uz T, et al. The effects of roflumilast, a phosphodiesterase type-4 inhibitor, on EEG biomarkers in schizophrenia: A randomised controlled trial. J Psychopharmacol. 2021; 35: 15-22.
- Tiihonen J, Koskuvi M, Lahteenvuo M, Trontti K, Ojansuu I, Vaurio O, et al. Molecular signaling pathways underlying schizophrenia. Schizophr Res. 2021; 232: 33-41.
- Yaka R, He DY, Phamluong K, Ron D. Pituitary adenylate cyclase-activating polypeptide (PACAP(1-38)) enhances N-methyl-D-aspartate receptor function and brain-derived neurotrophic factor expression via RACK1. J Biol Chem. 2003; 278: 9630-8.
- Rosenbrock H, Desch M, Wunderlich G. Development of the novel GlyT1 inhibitor, iclepertin (BI 425809), for the treatment of cognitive impairment associated with schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2023; 273: 1557-1566.
- Panizzutti R, Fisher M, Garrett C, Man WH, Sena W, Madeira C, et al. Association between increased serum d-serine and cognitive gains induced by intensive cognitive training in schizophrenia. Schizophr Res. 2019; 207: 63-69.
- O’Donnell P, Dong C, Murthy V, Asgharnejad M, Du X, Summerfelt A, et al. The D-amino acid oxidase inhibitor luvadaxistat improves mismatch negativity in patients with schizophrenia in a randomized trial. Neuropsychopharmacology. 2023; 48: 1052-1059.
- Goff, D. The Therapeutic Role of d-Cycloserine in Schizophrenia. Adv Pharmacol. 2016; 76: 39-66.
- Wu Q, Huang J, Wu R. Drugs Based on NMDAR Hypofunction Hypothesis in Schizophrenia. Front Neurosci. 2021; 15: 641047.
- Chen W, Liu J, Zhang L, Xu H, Guo X, Deng S, et al. Generation of the SCN1A epilepsy mutation in hiPS cells using the TALEN technique. Sci Rep. 2014; 4: 5404.
- Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. PNAS. 2010; 107: 17668-17673.
- Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D, et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol. 2012; 14: 911-923.
- Yamashita S, Chiyonobu T, Yoshida M, Maeda H, Zuiki M, Kidowaki S, et al. Mislocalization of syntaxin-1 and impaired neurite growth observed in a human iPSC model for STXBP1-related epileptic encephalopathy. Epilepsia. 2016; 57: e81-86.
- Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011; 17: 1657-1662.
- Dolce A, Ben-Zeev B, Naidu S, Kossoff EH. Rett syndrome and epilepsy: an update for child neurologists. Pediatr Neurol. 2013; 48: 337-345.
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011; 20: 2103-2105.
- Maezawa I, Swanberg S, Harvey D, LaSalle JM, Jin LW. Rett syndrome astrocytes are abnormal and spread MeCP2 deficiency through gap junctions. J Neurosci. 2009; 29: 5051-5061.
- Chen S, Xu D, Fan L, Fang Z, Wang X, Li M. Roles of N-Methyl-D-Aspartate Receptors (NMDARs) in Epilepsy. Front Mol Neurosci. 2021; 14: 797253.
- Zehavi Y, Mandel H, Zehavi A, Rashid MA, Straussberg R, Jabur B, et al. De novo GRIN1 mutations: An emerging cause of severe early infantile encephalopathy. Eur J Med Genet. 2017; 60: 317-320.
- Hanada, T. Ionotropic Glutamate Receptors in Epilepsy: A Review Focusing on AMPA and NMDA Receptors. Biomolecules. 2020; 10: 464.
- Sharawat IK, Yadav J, Saini L. Novel GRIN2B mutation: A rare cause of severe epileptic encephalopathy. Neurol India. 2019; 67: 562-563.
- Sun C, Yang M, Qin F, Guo R, Liang S, Hu H. Generation of an induced pluripotent stem cell line SYSUi-003-A from a child with epilepsy carrying GRIN2A mutation. Stem Cell Res. 2020; 43: 101706.
- Rabinski T, Sagiv ST, Hausman-Kedem M, Fattal-Valevski A, Rubinstein M, Avraham KB, et al. Reprogramming of two induced pluripotent stem cell lines from a heterozygous GRIN2D developmental and epileptic encephalopathy (DEE) patient (BGUi011-A) and from a healthy family relative (BGUi012-A). Stem Cell Res. 2021; 51: 102178.
- Shi Z, Liu H, Feng F, Huang Z, Chen WX. Generation of an induced pluripotent stem cell line GWCMCi006-A from a patient with autosomal dominant neurodevelopmental disorder with or without hyperkinetic movements and seizures harboring GRIN1 c.389A > G mutation. Stem Cell Res. 2024; 76: 103371.
- Fedele L, Newcombe J, Topf M, Gibb A, Harvey RJ, Smart TG. Disease-associated missense mutations in GluN2B subunit alter NMDA receptor ligand binding and ion channel properties. Nat Commun. 2018; 9: 957.
- Addis L, Virdee JK, Vidler LR, Collier DA, Pal DK, Ursu D. Epilepsy-associated GRIN2A mutations reduce NMDA receptor trafficking and agonist potency - molecular profiling and functional rescue. Sci Rep. 2017; 7: 66.
- Liu S, Zhou L, Yuan H, Vieira M, Sanz-Clemente A, Badger JD, 2nd, et al. A Rare Variant Identified Within the GluN2B C-Terminus in a Patient with Autism Affects NMDA Receptor Surface Expression and Spine Density. J Neurosci. 2017; 37: 4093-4102.
- Platzer K, Yuan H, Schutz H, Winschel A, Chen W, Hu C, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet. 2017; 54: 460-470.
- Myers SJ, Yuan H, Perszyk RE, Zhang J, Kim S, Nocilla KA, et al. Classification of missense variants in the N-methyl-d-aspartate receptor GRIN gene family as gain- or loss-of-function. Hum Mol Genet. 2023; 32: 2857-2871.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




