Submitted:
02 October 2024
Posted:
04 October 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Results
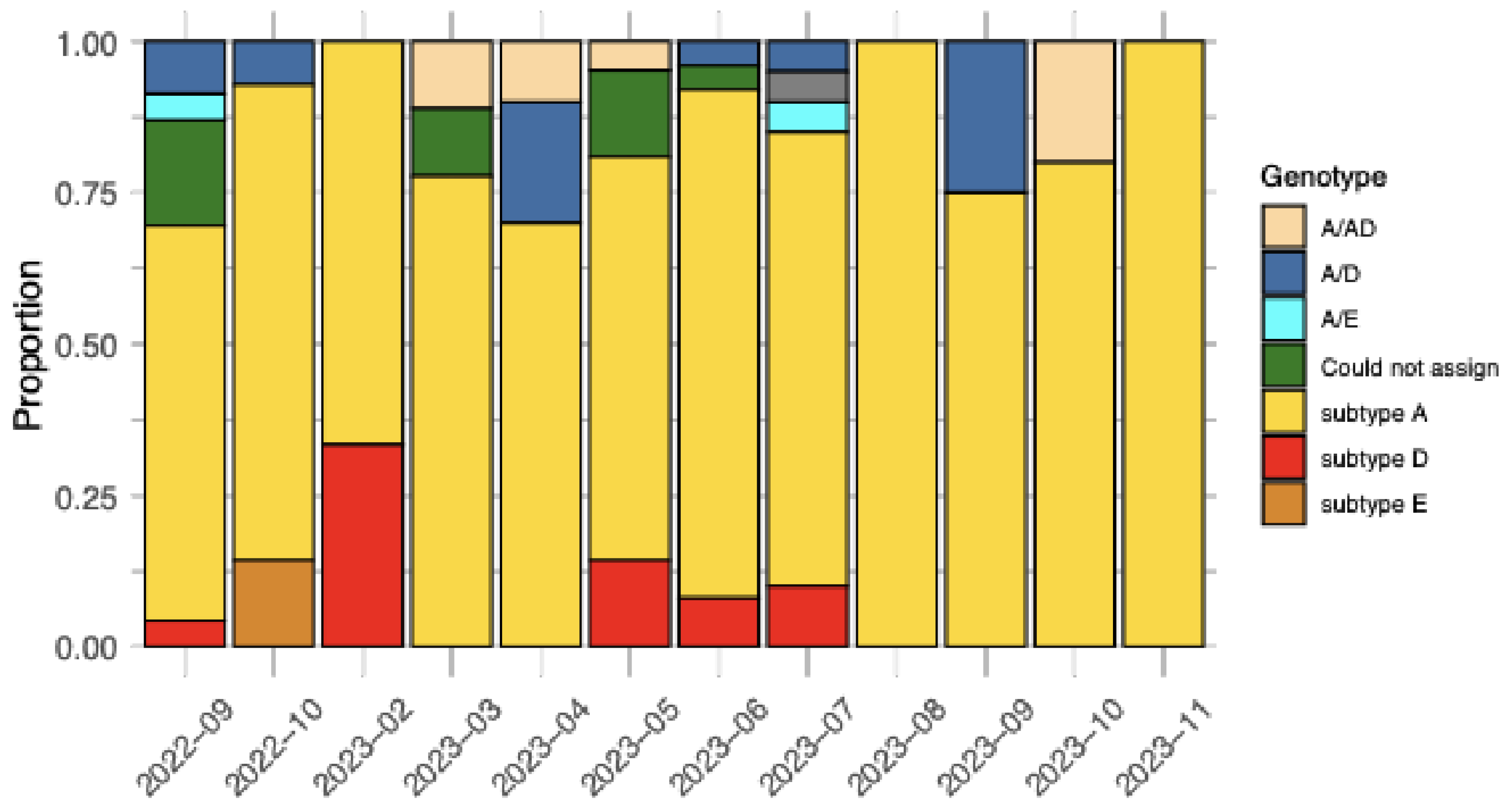
2.1. Sequencing of Samples
2.2. Recombination Detection and Breakpoint Identification
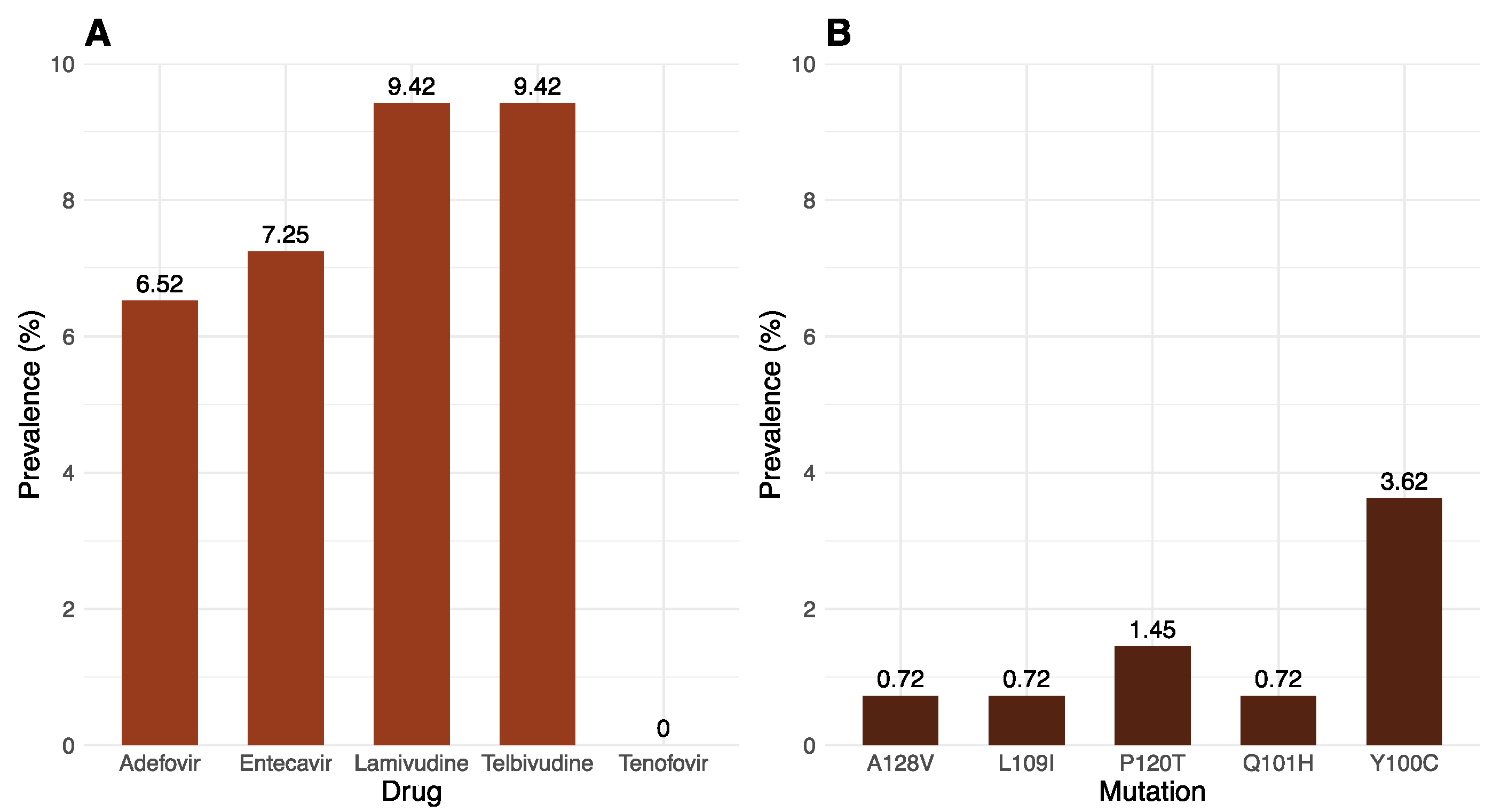
2.3. Drug Resistance and Immune-Evasion Profiling
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. HBV DNA Extraction
4.3. Primer Design
4.4. Tiling-Based Polymerase Chain Reaction
4.5. Raw-Read Assessment and Genotyping
4.6. Recombination Analysis
4.7. Genotyping and the Identification of Drug-Resistance and Immune-Escape Mutations
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sequence | RDP | GeneConv | BootScan | MaxChi | Chimaera | Sister scan | TOPAL |
|---|---|---|---|---|---|---|---|
| K048723* | 1.08x10-15 | 5.51x10-10 | 4.61x10-02 | 7.31x10-09 | 1.15x10-07 | 1.58x10-04 | 6.45x10-13 |
| K048737* | 1.66x10-11 | 3.93x10-11 | NS | 6.41x10-07 | 2.02x10-05 | 1.79x10-08 | 8.29x10-11 |
| K048753* | 3.09x10-10 | 3.43x10-06 | NS | 5.64x10-06 | 4.17x10-05 | 1.26x10-09 | 8.07x10-09 |
| K048756* | 1.59x10-11 | 2.21x10-08 | 1.95x10-02 | 1.00x10-10 | 1.00x10-10 | 1.45x10-13 | 1.50x10-21 |
| K056892 | 2.30x10-07 | 1.76x10-05 | 7.24x10-05 | 2.94x10-04 | 2.01x10-04 | 8.16x10-06 | 9.75x10-07 |
| K056897 | 1.89x10-21 | 1.24x10-19 | 1.67x10-07 | 2.60x10-12 | 6.11x10-13 | 1.21x10-15 | 3.58x10-25 |
| K056900* | 1.47x10-16 | 8.36x10-13 | 2.77x10-13 | 4.40x10-11 | 4.37x10-10 | 1.64x10-13 | 3.32x10-15 |
| K056911 | 2.61x10-10 | 3.51x10-09 | 5.81x10-05 | NS | NS | 3.33x10-02 | 1.75x10-04 |
| K056916* | 2.16x10-10 | 1.92x10-08 | NS | 1.74x10-03 | 1.54x10-03 | 1.14x10-04 | 3.00x10-07 |
| K056934* | 7.56x10-16 | 1.39x10-05 | 1.06x10-04 | 3.93x10-03 | NS | NS | 1.74x10-04 |
| K056939 | 1.86x10-06 | 1.17x10-03 | 7.42x10-05 | 2.05x10-05 | 1.41x10-04 | 3.82x10-08 | 5.02x10-04 |
| K056944* | 4.52x10-18 | 3.36x10-15 | NS | 1.04x10-10 | 5.47x10-11 | 1.38x10-04 | 1.77x10-23 |
| K056945* | 3.42x10-20 | 1.26x10-17 | 1.83x10-02 | 1.88x10-18 | 1.48x10-05 | 2.92x10-26 | 2.53x10-27 |
| K056950* | 5.16x10-08 | 1.87x10-06 | NS | 7.51x10-06 | 2.34x10-06 | 8.20x10-03 | 4.75x10-09 |
| K057256* | 1.47x10-17 | 9.40x10-17 | 4.14x10-04 | 4.17x10-11 | 1.31x10-11 | 4.29x10-12 | 4.52x10-26 |
| K057265 | 7.53x10-11 | 1.72x10-09 | 1.03x10-03 | 4.03x10-08 | 9.05x10-05 | 1.10x10-02 | 1.10x10-10 |
| K057280* | 6.92x10-21 | 2.63x10-22 | 2.12x10-19 | 3.78x10-06 | 3.29x10-06 | 1.28x10-07 | 8.01x10-21 |
References
- Coste, M., et al., Burden and impacts of chronic hepatitis B infection in rural Senegal: study protocol of a cross-sectional survey in the area of Niakhar (AmBASS ANRS 12356). BMJ Open, 2019. 9(7): p. e030211. [CrossRef]
- Global, regional, and national burden of hepatitis B, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol, 2022. 7(9): p. 796-829. [CrossRef]
- Carman, W.F. , The clinical significance of surface antigen variants of hepatitis B virus. J Viral Hepat, 1997. 4 Suppl 1: p. 11-20. [CrossRef]
- Inoue, J., T. Nakamura, and A. Masamune, Roles of Hepatitis B Virus Mutations in the Viral Reactivation after Immunosuppression Therapies. Viruses, 2019. 11(5). [CrossRef]
- Kurbanov, F., Y. Tanaka, and M. Mizokami, Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res, 2010. 40(1): p. 14-30. [CrossRef]
- Kramvis, A. , Molecular characteristics and clinical relevance of African genotypes and subgenotypes of hepatitis B virus. S Afr Med J, 2018. 108(8b): p. 17-21. [CrossRef]
- Maepa, M.B., et al., Hepatitis B Virus Research in South Africa. Viruses, 2022. 14(9). [CrossRef]
- Abe, H., et al., Genetic Diversity of Hepatitis B and C Viruses Revealed by Continuous Surveillance from 2015 to 2021 in Gabon, Central Africa. Microorganisms, 2023. 11(8). [CrossRef]
- Giandhari, J., et al., Early transmission of SARS-CoV-2 in South Africa: An epidemiological and phylogenetic report. Int J Infect Dis, 2021. 103: p. 234-241. [CrossRef]
- Ingasia, L.A.O., et al., Global and regional dispersal patterns of hepatitis B virus genotype E from and in Africa: A full-genome molecular analysis. PLOS ONE, 2020. 15(10): p. e0240375. [CrossRef]
- Al-Matary, A.M. and F.A.S. Al Gashaa, Comparison of different rapid screening tests and ELISA for HBV, HCV, and HIV among healthy blood donors and recipients at Jibla University Hospital Yemen. J Med Life, 2022. 15(11): p. 1403-1408. [CrossRef]
- Parekh, B.S., et al., Diagnosis of Human Immunodeficiency Virus Infection. Clin Microbiol Rev, 2019. 32(1). [CrossRef]
- Solmone, M., et al., Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J Virol, 2009. 83(4): p. 1718-26. [CrossRef]
- Soleimani, B., et al., Comparison Between Core Set Selection Methods Using Different Illumina Marker Platforms: A Case Study of Assessment of Diversity in Wheat. Frontiers in Plant Science, 2020. 11. [CrossRef]
- Adewale, B.A., Will long-read sequencing technologies replace short-read sequencing technologies in the next 10 years? Afr J Lab Med, 2020. 9(1): p. 1340. [CrossRef]
- Quiñones-Mateu, M.E., et al., Deep sequencing: becoming a critical tool in clinical virology. J Clin Virol, 2014. 61(1): p. 9-19. [CrossRef]
- Liu, W.C., et al., Aligning to the sample-specific reference sequence to optimize the accuracy of next-generation sequencing analysis for hepatitis B virus. Hepatol Int, 2016. 10(1): p. 147-57. [CrossRef]
- Makondo, E., T.G. Bell, and A. Kramvis, Genotyping and molecular characterization of hepatitis B virus from human immunodeficiency virus-infected individuals in southern Africa. PLoS One, 2012. 7(9): p. e46345. [CrossRef]
- Caballero, A., et al., Complex Genotype Mixtures Analyzed by Deep Sequencing in Two Different Regions of Hepatitis B Virus. PLOS ONE, 2016. 10(12): p. e0144816. [CrossRef]
- Han, Y., et al., Analysis of hepatitis B virus genotyping and drug resistance gene mutations based on massively parallel sequencing. J Virol Methods, 2013. 193(2): p. 341-7. [CrossRef]
- Jones, L.R., et al., Hepatitis B virus resistance substitutions: long-term analysis by next-generation sequencing. Arch Virol, 2016. 161(10): p. 2885-91. [CrossRef]
- Lowe, C.F., et al., Implementation of Next-Generation Sequencing for Hepatitis B Virus Resistance Testing and Genotyping in a Clinical Microbiology Laboratory. J Clin Microbiol, 2016. 54(1): p. 127-33. [CrossRef]
- McNaughton, A.L., et al., Illumina and Nanopore methods for whole genome sequencing of hepatitis B virus (HBV). Sci Rep, 2019. 9(1): p. 7081. [CrossRef]
- Radukic, M.T., et al., Nanopore sequencing of native adeno-associated virus (AAV) single-stranded DNA using a transposase-based rapid protocol. NAR Genom Bioinform, 2020. 2(4): p. lqaa074. [CrossRef]
- Ranasinghe, D., et al., Comparison of different sequencing techniques for identification of SARS-CoV-2 variants of concern with multiplex real-time PCR. PLOS ONE, 2022. 17(4): p. e0265220. [CrossRef]
- Loman, N.J. Quick, and J.T. Simpson, A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods, 2015. 12(8): p. 733-735. [CrossRef]
- Krzywinski, M., et al., Circos: an information aesthetic for comparative genomics. Genome Res, 2009. 19(9): p. 1639-45. [CrossRef]
- Hossain, M.G. and K. Ueda, A meta-analysis on genetic variability of RT/HBsAg overlapping region of hepatitis B virus (HBV) isolates of Bangladesh. Infect Agent Cancer, 2019. 14: p. 33. [CrossRef]
- Astbury, S., et al., Extraction-free direct PCR from dried serum spots permits HBV genotyping and RAS identification by Sanger and minION sequencing. bioRxiv, 2019: p. 552539. [CrossRef]
- Sauvage, V., et al., Early MinION™ nanopore single-molecule sequencing technology enables the characterization of hepatitis B virus genetic complexity in clinical samples. PLOS ONE, 2018. 13(3): p. e0194366. [CrossRef]
- Dopico, E., et al., Genotyping Hepatitis B virus by Next-Generation Sequencing: Detection of Mixed Infections and Analysis of Sequence Conservation. Int J Mol Sci, 2024. 25(10). [CrossRef]
- Gionda, P.O., et al., Analysis of the complete genome of HBV genotypes F and H found in Brazil and Mexico using the next generation sequencing method. Ann Hepatol, 2022. 27 Suppl 1: p. 100569. [CrossRef]
- Chotiyaputta, W. and A.S. Lok, Hepatitis B virus variants. Nat Rev Gastroenterol Hepatol, 2009. 6(8): p. 453-62. [CrossRef]
- Kimbi, G.C., A. Kramvis, and M.C. Kew, Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J Gen Virol, 2004. 85(Pt 5): p. 1211-1220. [CrossRef]
- Jose-Abrego, A., et al., Hepatitis B Virus (HBV) Genotype Mixtures, Viral Load, and Liver Damage in HBV Patients Co-infected With Human Immunodeficiency Virus. Front Microbiol, 2021. 12: p. 640889. [CrossRef]
- Kafeero, H.M., et al., Mapping hepatitis B virus genotypes on the African continent from 1997 to 2021: a systematic review with meta-analysis. Sci Rep, 2023. 13(1): p. 5723. [CrossRef]
- Tyler, B., et al., Hepatitis B Virus Genotype E/A Recombinants from Blood Donors in Beira, Mozambique. Microbiol Resour Announc, 2023. 12(6): p. e0018223. [CrossRef]
- Chen, B.F., et al., High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology, 2006. 130(4): p. 1153-68. [CrossRef]
- Beggel, B., et al., Genotyping hepatitis B virus dual infections using population-based sequence data. J Gen Virol, 2012. 93(Pt 9): p. 1899-1907. [CrossRef]
- Neumann-Fraune, M., et al., Hepatitis B virus drug resistance tools: one sequence, two predictions. Intervirology, 2014. 57(3-4): p. 232-6. [CrossRef]
- Mokaya, J., et al., A systematic review of hepatitis B virus (HBV) drug and vaccine escape mutations in Africa: A call for urgent action. PLOS Neglected Tropical Diseases, 2018. 12(8): p. e0006629. [CrossRef]
- Spearman, C.W., et al., South African guideline for the management of chronic hepatitis B: 2013. S Afr Med J, 2013. 103(5 Pt 2): p. 337-49.
- Lee, H.W., et al., Viral evolutionary changes during tenofovir treatment in a chronic hepatitis B patient with sequential nucleos(t)ide therapy. J Clin Virol, 2014. 60(3): p. 313-6. [CrossRef]
- Tegally, H., et al., Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 2021. 592(7854): p. 438-443. [CrossRef]
- Tyson, J.R., et al., Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv, 2020. [CrossRef]
- Katoh, K. and D.M. Standley, MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution, 2013. 30(4): p. 772-780. [CrossRef]
- Martin, D.P., et al., RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol, 2021. 7(1): p. veaa087. [CrossRef]
- Martin, D. and E. Rybicki, RDP: detection of recombination amongst aligned sequences. Bioinformatics, 2000. 16(6): p. 562-3. [CrossRef]
- Padidam, M., S. Sawyer, and C.M. Fauquet, Possible emergence of new geminiviruses by frequent recombination. Virology, 1999. 265(2): p. 218-25. [CrossRef]
- Smith, J.M. , Analyzing the mosaic structure of genes. J Mol Evol, 1992. 34(2): p. 126-9. [CrossRef]
- Martin, D.P., et al., A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses, 2005. 21(1): p. 98-102. [CrossRef]
- Posada, D. and K.A. Crandall, Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A, 2001. 98(24): p. 13757-62. [CrossRef]
- Gibbs, M.J., J.S. Armstrong, and A.J. Gibbs, Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics, 2000. 16(7): p. 573-582. [CrossRef]
- Lam, H.M., O. Ratmann, and M.F. Boni, Improved Algorithmic Complexity for the 3SEQ Recombination Detection Algorithm. Mol Biol Evol, 2018. 35(1): p. 247-251. [CrossRef]
- Kiwelu, I.E., et al., Frequent intra-subtype recombination among HIV-1 circulating in Tanzania. PLoS One, 2013. 8(8): p. e71131. [CrossRef]
- Martin, D.P., et al., RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics, 2010. 26(19): p. 2462-3. [CrossRef]
- Sentandreu, V., et al., Evidence of Recombination in Intrapatient Populations of Hepatitis C Virus. PLOS ONE, 2008. 3(9): p. e3239. [CrossRef]
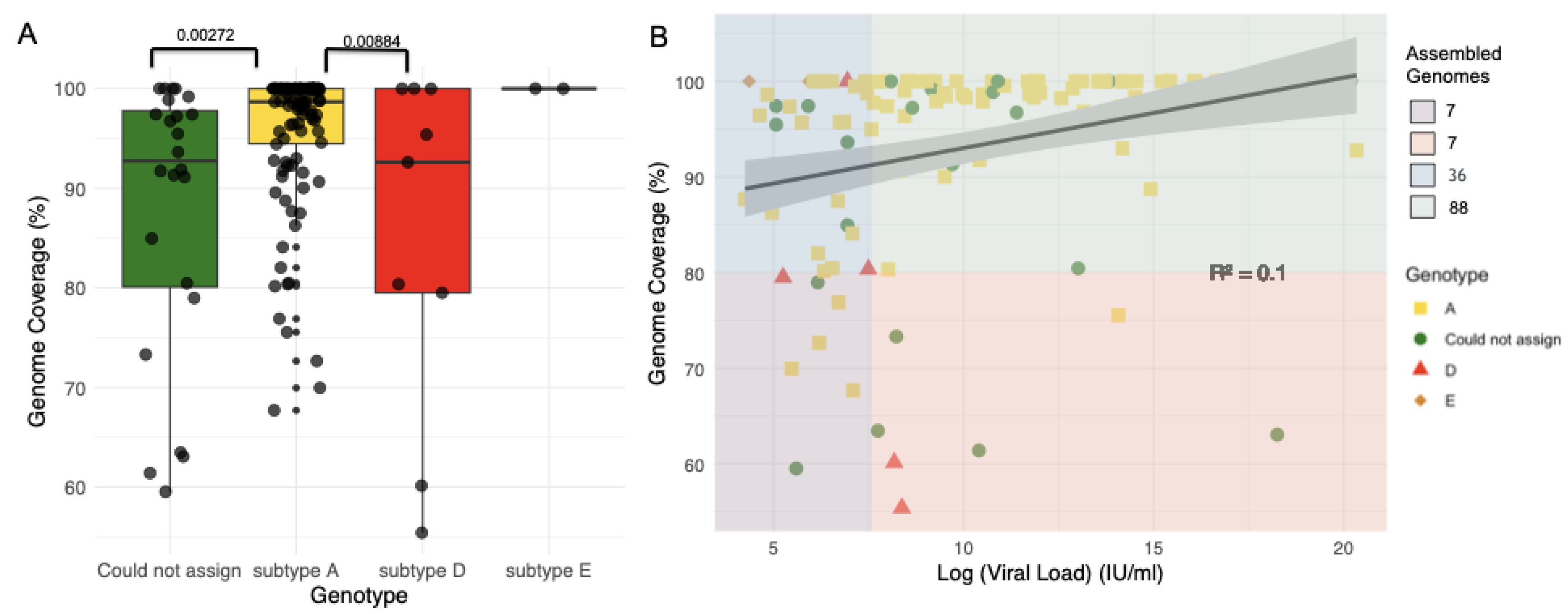
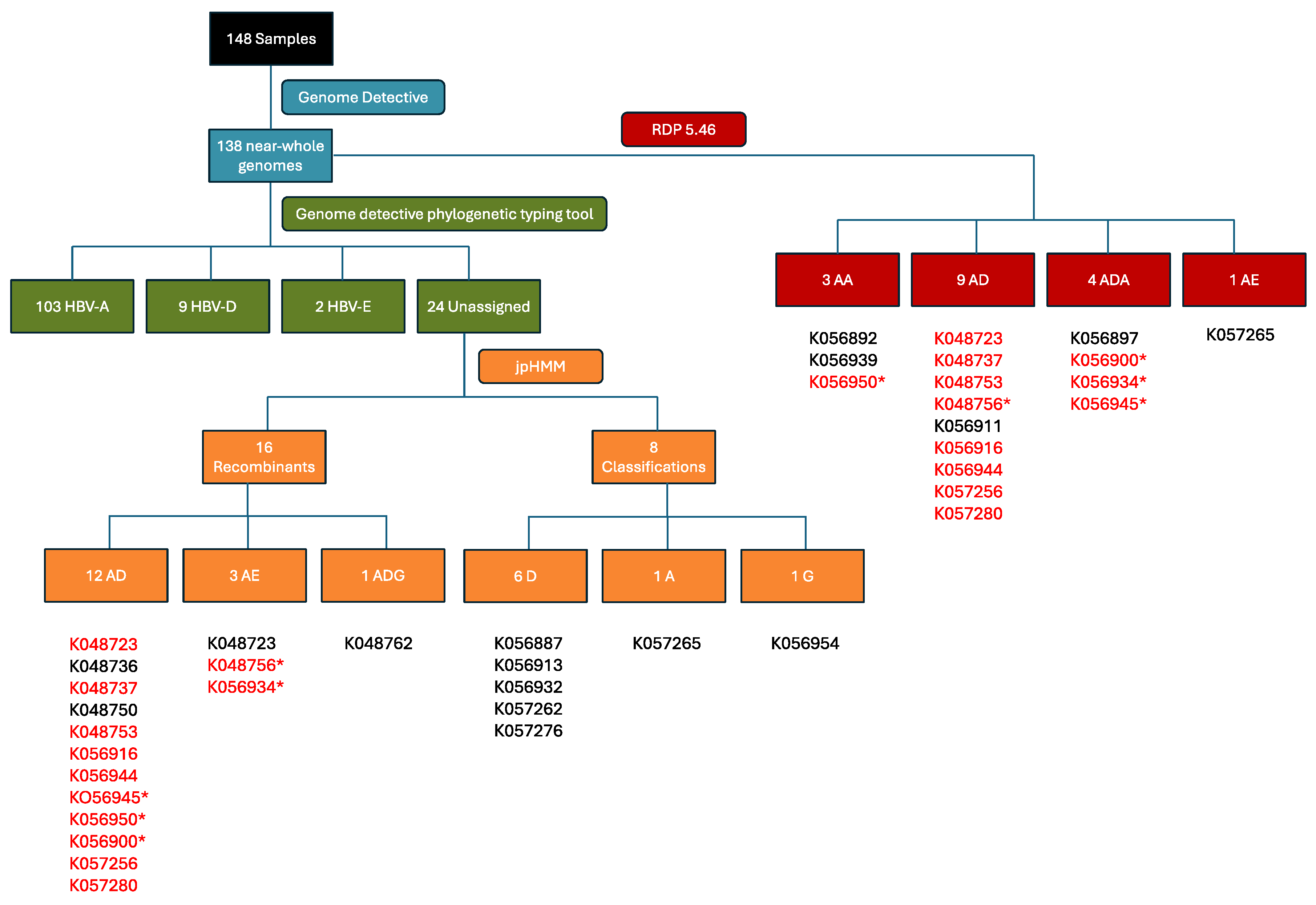
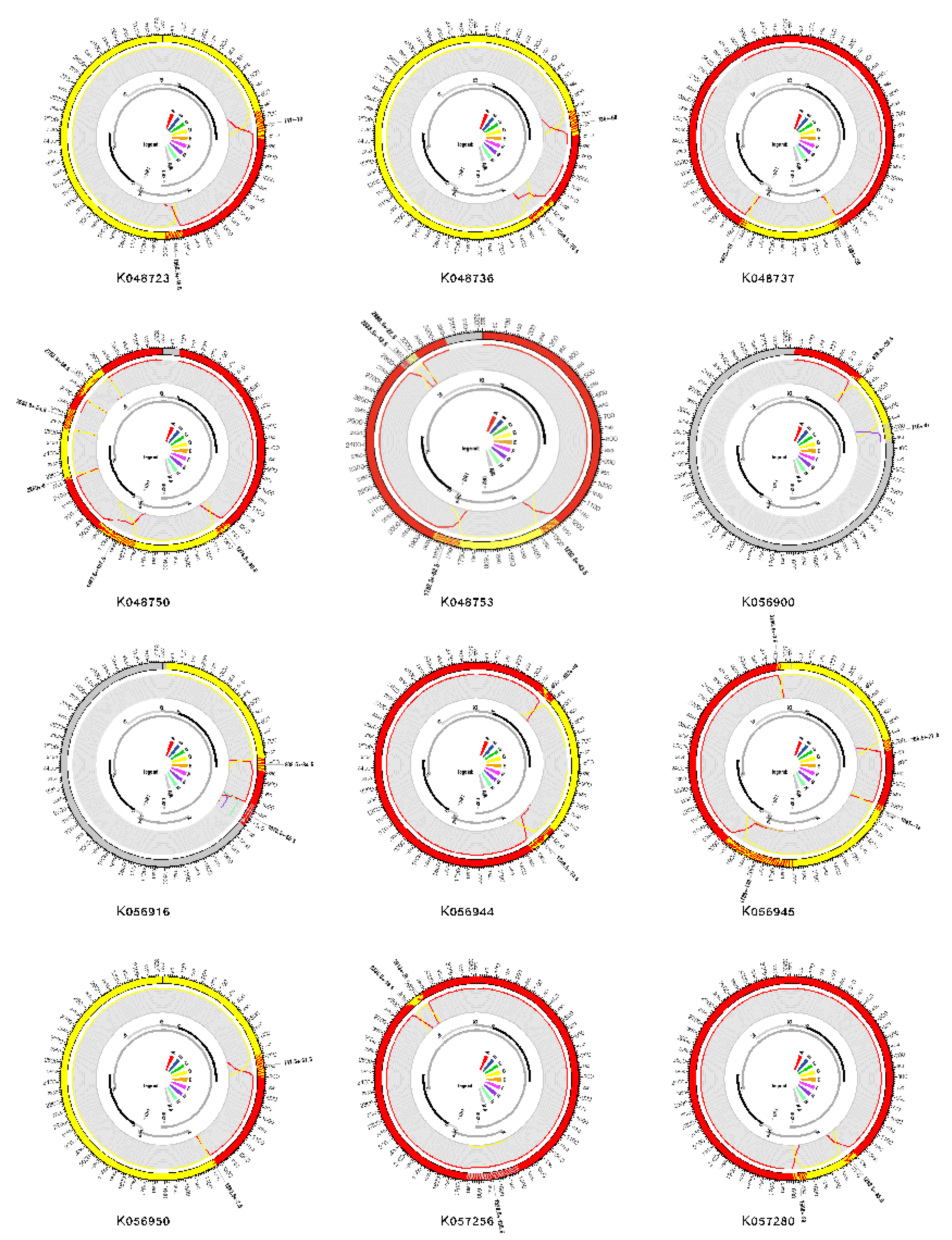
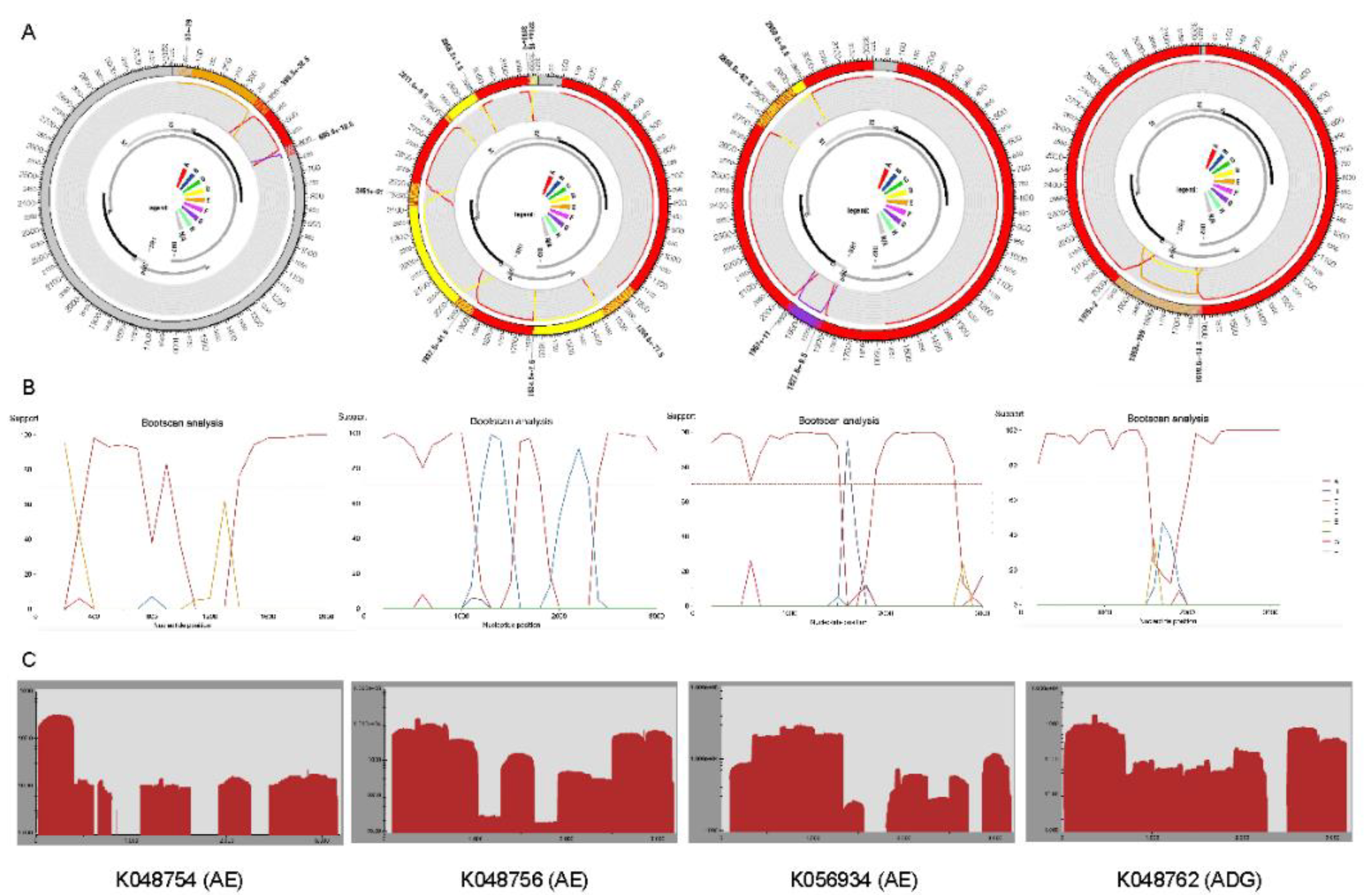

| Characteristic | Total N=138 (%) |
|---|---|
| Gender | |
| Male | 86 (62.32) |
| Female | 52 (37.68) |
| Age | |
| <18 | 2 (1.45) |
| 18 - 24 | 6 (4.35) |
| 25 - 34 | 30 (21.74) |
| 35 - 44 | 44 (31.88) |
| ≥45 | 56 (40.58) |
| HBV viral load (IU/ml) | |
| <2000 | 39 (28.26) |
| 2000 - 20 000 | 31 (22,46) |
| >20 000 | 68 (49.28) |
| Mutation | Adefovir | Entecavir | Lamuvidine | Telbivudine |
|---|---|---|---|---|
| 180M | 0 | 4 | 4 | 0 |
| 204V | 0 | 5 | 5 | 5 |
| 181T | 1 | 0 | 0 | 0 |
| 202K | 0 | 2 | 0 | 0 |
| 250S | 0 | 1 | 0 | 0 |
| 250N | 0 | 1 | 0 | 0 |
| Primer Name | Sequence | Genomic positions (bp) |
|---|---|---|
| SC_1_LEFT | TTC CAC CAA GCT CTG CAA GATC | 11 - 32 |
| SC_1_RIGHT | AGAGGAATATGATAAAACGCCGCA | 384-407 |
| SC_2_LEFT | CATCATCATCAT CACCA CCTCC | 325-346 |
| SC_2_RIGHT | AAAGCCCTACGAACCACTGAAC | 692-713 |
| SC_3_LEFT | AAATACCTATGGGAGTGGGCCT | 632-653 |
| SC_3_RIGHT | TTGTGTAAATGGAGCGGCAAAG | 1 655-1 676 |
| SC_4_LEFT | AGAAAACTTCCTGTTAACAGACCTATTG | 949-976 |
| SC_4_RIGHT | GGACGACAGAATTATCAGTCCCG | 1 326-1 348 |
| SC_5_LEFT | TCCATACTGCGGAACTCCTAGC | 1 265-1 286 |
| SC_5_RIGHT | TGTAAGACCTTGGGCAGGATTTG | 1 632-1 654 |
| SC_6_LEFT | CTTCTCATCTGCCGGTCCGTGT | 1 559-1580 |
| SC_6_RIGHT | AGAAGTCAGAAGGCAAACGAGA | 1 947-1 970 |
| SC_7_LEFT | GGCTTTGGGGCATGGACATT | 1 890-1 909 |
| SC_7_RIGHT | ATCCACACTCCGAAAGAGACCA | 2 256-2 277 |
| SC_8_LEFT | GACAACTATTGTGGTTTCATATTTCT | 2 193-2 218 |
| SC_8_RIGHT | TTGTTGACACCTATTAATAATGTCCTCA | 2 576-2 594 |
| SC_9_LEFT | TGGGCTTTATTCCTCTACTGTCCC | 2 492-2 515 |
| SC_9_RIGHT | GGGAACAGAAAGATTCGTCCCC | 2 889-2 910 |
| SC_10_LEFT | TTGCGGGTCACCATATTCTTGG | 2 816-2 837 |
| SC_10_RIGHT | GGCCTGAGGATGACTGTCTCTT | 3 189-3 210 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





