Submitted:
01 October 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials AND Methods
2.1. Study Population and Design
2.2. Biological Samples and DNA Extraction
2.3. Whole-Exome Sequencing (WES)
2.4. Bioinformatics Pipeline to HLA Typing
2.5. Statistical Analysis
2.6. Hardware and Software Environment
2.7. Ethics Statement
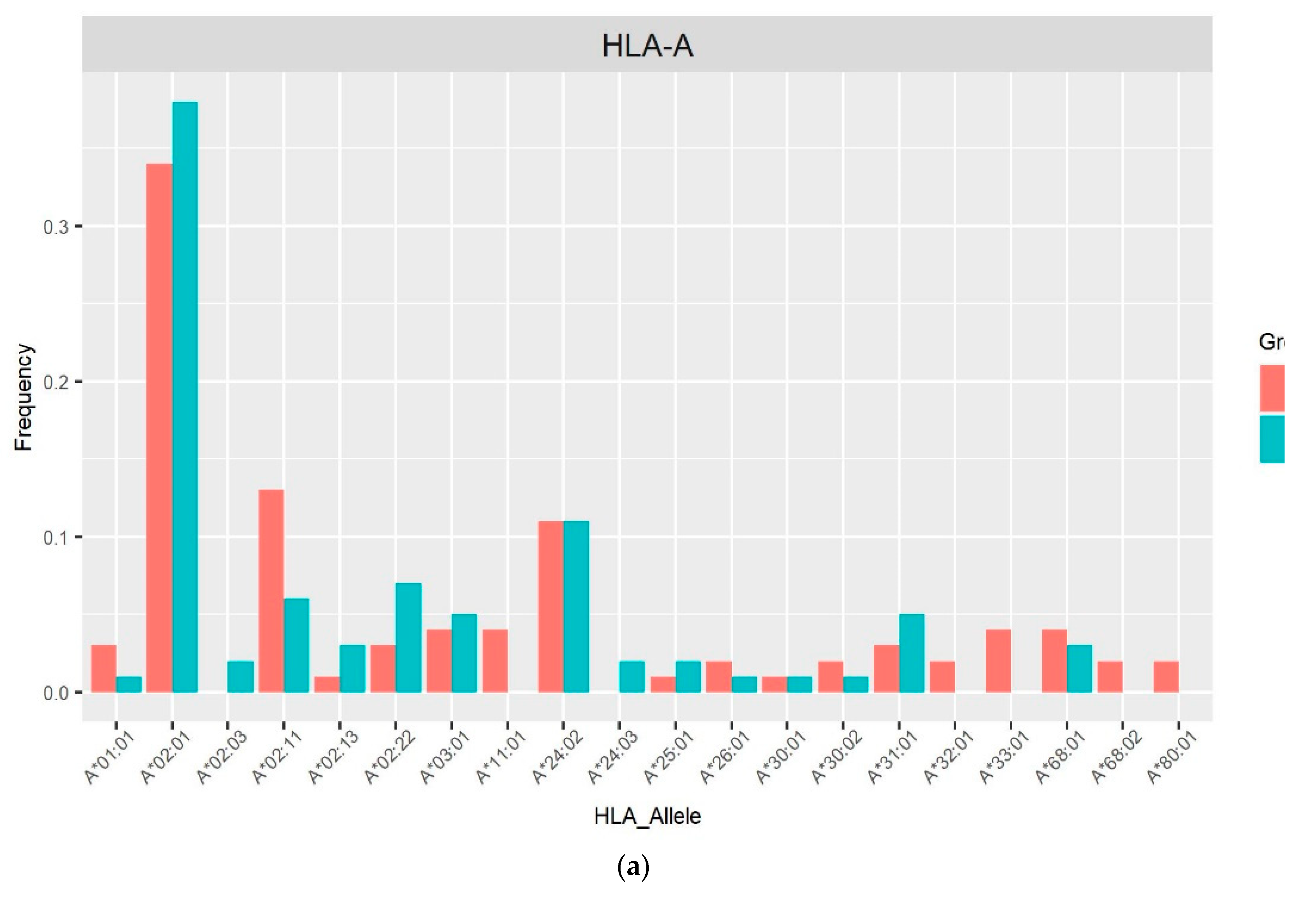
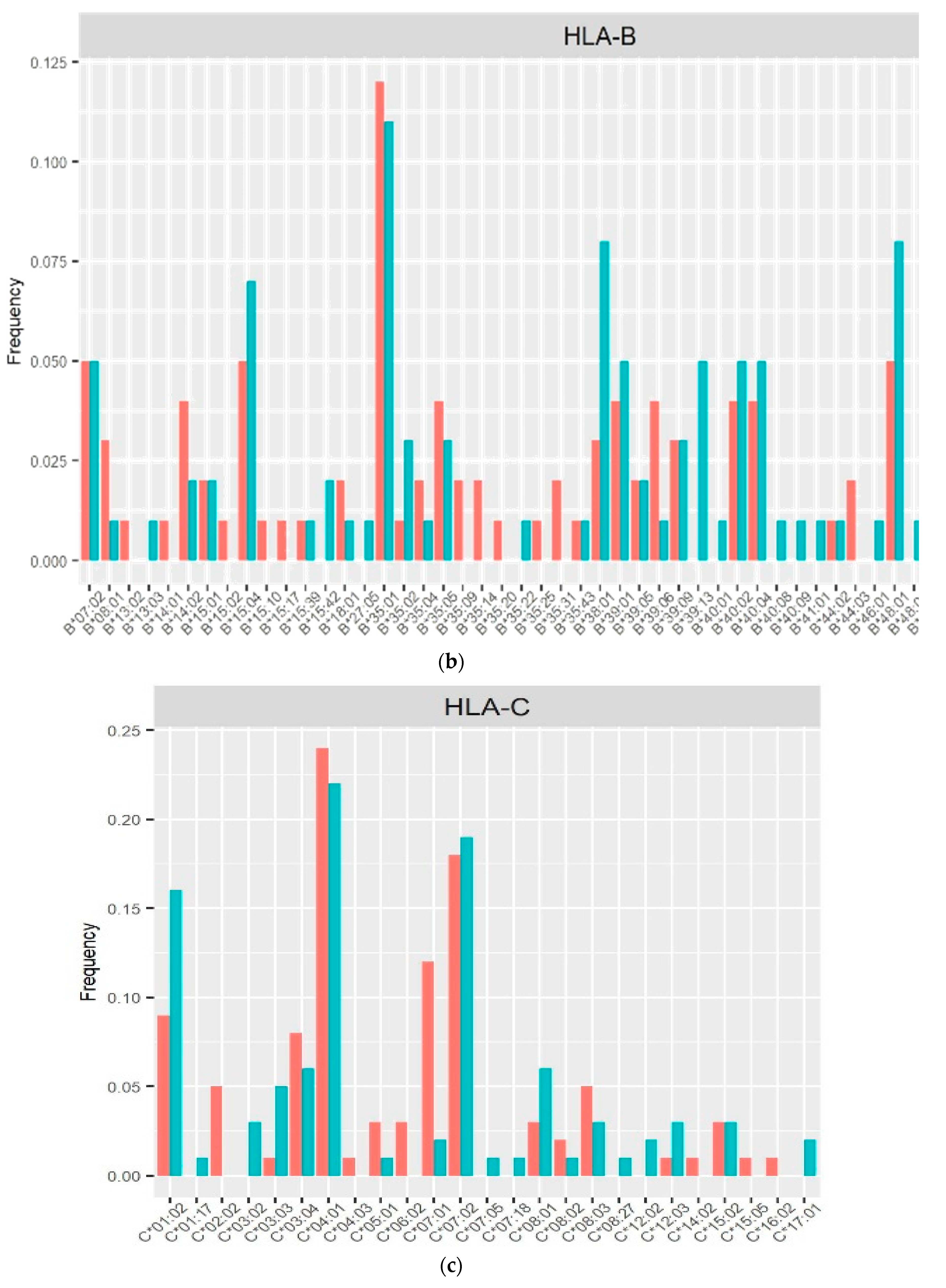
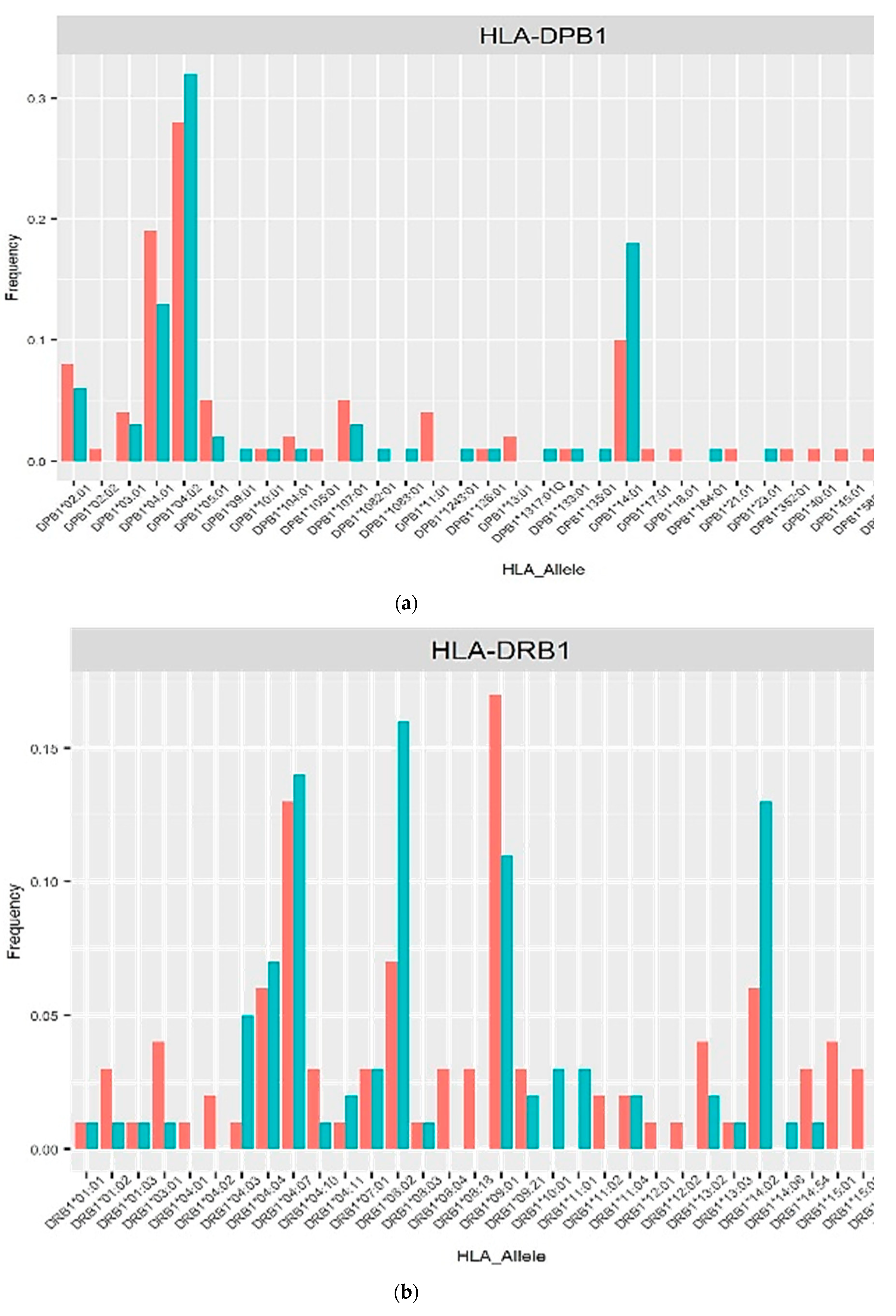
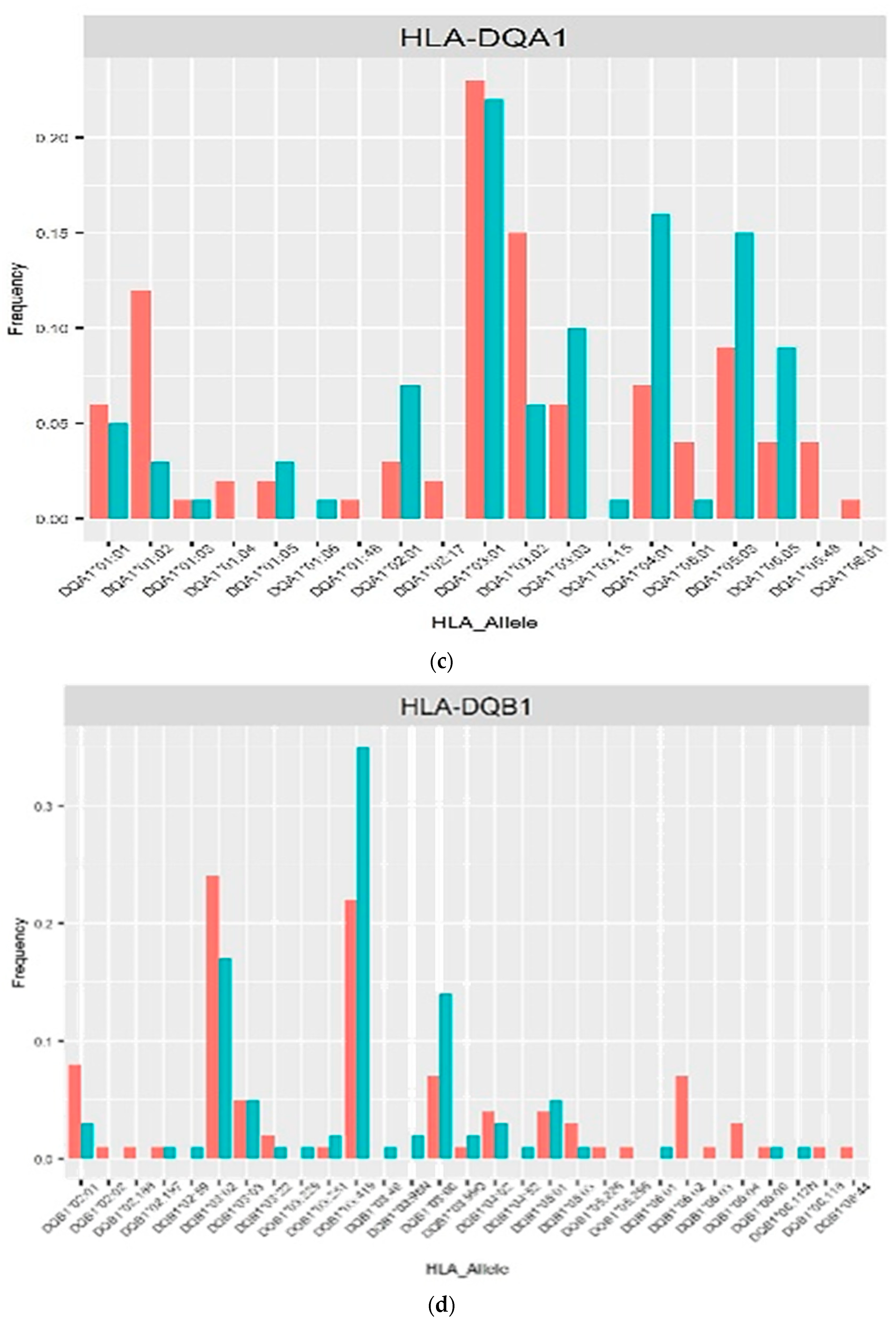
3. Results
4. Discussion
5. Conclusions
Conflict of Interests
Supplementary Materials
Author Contributions
Funding
Acknowledgments
References
- UNAIDS (2023). UNAIDS 2023 Data. Available online at: https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf (accessed October 7, 2023).
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat Rev Genet 2004; 5(12):889-99. [CrossRef]
- Kim, J. Y., Lee, S. Y., Kim, G. G., Song, H. I., Jang, M. M., Lee, C. S., Hong, J. Y., Shin, M. G., & Choi, H. J. Validation and application of new NGS-based HLA genotyping to clinical diagnostic practice. HLA 2023, 101(5), 496–506. [CrossRef]
- Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P,Marsh SGE. IPD-IMGT/HLA Database. Nucleic Acids Res 2020; 48(D1):D948-D955. [CrossRef]
- Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010.Tissue Antigens. 2010; 75(4):291-455.
- Luo, M. Natural Immunity against HIV-1: Progression of Understanding after Association Studies. Viruses. 2022 Jun 8;14(6):1243. Erratum in: Viruses. 2023 ;15(6). [CrossRef]
- Dashti M, Malik MZ, Nizam R, Jacob S, Al-Mulla F and Thanaraj TA (2024), Evaluation of HLA typing content of next-generation sequencing datasets from family trios and individuals of arab ethnicity. Front. Genet. 15:1407285. [CrossRef]
- Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, Trachtenberg EA, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens 2009; 74:393-403.
- Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens 2012; 80:1-11.
- Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nuc Acids Res 1988; 16:1215.
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:884–90.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [CrossRef]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [CrossRef]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: Precision HLA typing from next-generation sequencing data. Bioinformatics. 2014; 30(23):3310–6.
- Orenbuch R, Filip I, Comito D, Shaman J, Pe’er I, Rabadan R. arcasHLA: high-resolution HLA typing from RNAseq. Bioinformatics. 2020; 36(1):33-40. [CrossRef]
- Fan Y, Song YQ. PyHLA: tests for the association between HLA alleles and diseases. BMC Bioinformatics. 2017; 18:90. [CrossRef]
- Liu, P., Yao, M., Gong, Y., Song, Y., Chen, Y., Ye, Y., Liu, X., Li, F., Dong, H., Meng, R., Chen, H., & Zheng, A. (2021). Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms. Frontiers in immunology, 12, 652258. [CrossRef]
- Sanchez-Mazas, A. A review of HLA allele and SNP associations with highly prevalent infectious diseases in human populations. Swiss Med Wkly 2020; 150:w20214. [CrossRef]
- Amanzo-Vargas MP, Arellano-Veintemilla T, González-Lagos E, Echevarría J, Mejía F, Graña A, Gotuzzo E. Socio-Demographic, Clinical, and Mortality Differences between HIV-Infected and HIV/HTLV-1 Co-Infected Patients in Peru. Pathogens 2023; 12(7):869. [CrossRef]
- Gonzalez-Galarza FF, McCabe A, Santos EJ, Jones J, Takeshita LY, Ortega-Rivera ND, Del Cid-Pavon GM, Ramsbottom K, Ghattaoraya GS, Alfirevic A, Middleton D and Jones AR. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acid Res 2020, 48:D783-8.
- Arnaiz-Villena A, Gonzalez-Alcos V, Serrano-Vela JI, Reguera R, Barbolla L, Parga-Lozano C, Gómez-Prieto P, Abd-El-Fatah-Khalil S, Moscoso J. HLA genes in Uros from Titikaka Lake, Peru: origin and relationship with other Amerindians and worldwide populations. Int J Immunogenet. 2009; 36(3):159-67. [CrossRef]
- Olvera A, Pérez-Álvarez S, Ibarrondo J, Ganoza C, Lama JR, Lucchetti A, Cate S, Hildebrand W, Bernard N, Gomez L, et al. The HLA-C*04: 01/KIR2DS4 gene combination and human leukocyte antigen alleles with high population frequency drive rate of HIV disease progression. AIDS 2015, 29:507–517. [CrossRef]
- Blais, M. E., Dong, T., & Rowland-Jones, S. HLA-C as a mediator of natural killer and T-cell activation: spectator or key player?. Immunology 2011, 133(1), 1–7. https://doi.org/10.1111/j.1365-2567.2011.03422.x. [CrossRef]
- International HIV Controllers Study, Pereyra, F., Jia, X., McLaren, P. J., Telenti, A., de Bakker, P. I., Walker, B. D., Ripke, S., Brumme, C. J., Pulit, S. L., Carrington, M., Kadie, C. M., Carlson, J. M., Heckerman, D., Graham, R. R., Plenge, R. M., Deeks, S. G., Gianniny, L., Crawford, G., Sullivan, J., … Zhao, M. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010, 330(6010): 1551–1557. [CrossRef]
- M.D. Strettell, L.J. Thomson, P.T. Donaldson, M. Bunce, C.M. O’Neill, R. Williams. HLA-C genes and susceptibility to type 1 autoimmune hepatitis. Hepatology 1997, 26: 1023–1026.
- Qian, J., Chen, Y., Yang, X., Wang, Q., Zhao, J., Deng, X., Ding, Y., Li, S., Liu, Y., Tian, Z., Shen, J., Liao, Q., Wang, Y., Zuo, X., Zhang, X., Li, M., Cui, Y., Yu, X., & Zeng, X. Association Study Identified HLA-DQA1 as a Novel Genetic Risk of Systemic Lupus Erythematosus-Associated Pulmonary Arterial Hypertension. Arthritis Rheumatol 2023, 75(12), 2207–2215. [CrossRef]
- Anzurez, A., Naka, I., Miki, S., Nakayama-Hosoya, K., Isshiki, M., Watanabe, Y., Nakamura-Hoshi, M., Seki, S., Matsumura, T., Takano, T., Onodera, T., Adachi, Y., Moriyama, S., Terahara, K., Tachikawa, N., Yoshimura, Y., Sasaki, H., Horiuchi, H., Miyata, N., Miyazaki, K., … Kawana-Tachikawa, A. Association of HLA-DRB1*09:01 with severe COVID-19. HLA 2021, 98(1): 37–42. [CrossRef]
- Choo S., Y. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med J 2007, 48(1), 11–23. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T. P., Kikuchi, M., Vu, T. Q., Do, Q. H., Tran, T. T., Vo, D. T., Ha, M. T., Vo, V. T., Cao, T. P., Tran, V. D., Oyama, T., Morita, K., Yasunami, M., & Hirayama, K. Protective and enhancing HLA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl Trop Dis 2008, 2(10): e304. [CrossRef]
- Sarri CA, Giannoulis T, Moutou KA, Mamuris Z. HLA class II peptide-binding-region analysis reveals funneling of polymorphism in action. Immunol Lett. 2021; 238: 75–95.
- Roe, D. L., Lewis, R. E., & Cruse, J. M. Association of HLA-DQ and -DR alleles with protection from or infection with HIV-1. Exp Mol Pathol 2000, 68(1): 21–28. [CrossRef]
- Rallón N, Restrepo C, Vicario J, Romero J, Rodríguez C, García-Samaniego J, et al. Human leucocyte antigen (HLA)-DQB1*03:02 and HLA-A*02:01 have opposite patterns in their effects on susceptibility to HIV infection. HIV Med. 2017; 18(8):587-594. [CrossRef]
- Hardie R, Luo M, Bruneau B, Knight E, Nagelkerke N, Kimani J, Wachihi C, Ngugi E, Plummer F. Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS 2008, 22: 807–816. [CrossRef]
- Sandoval J, Salazar-Granara A, Acosta O, Castillo-Herrera W, Fujita R, Pena SDJ, et al. Tracing the genomic ancestry of Peruvians reveals a major legacy of pre-Columbian ancestors. J Hum Genet 2013; 58(9):627-34. [CrossRef]
| Characteristics | PLHIV (n=59) |
HIV-uninfected (n=46) |
p |
|---|---|---|---|
| Sex, n (%) | |||
| Female | 36 (61.02) | 1 (2.17) | 0.0000 *a |
| Male | 23 (38.98) | 45 (97.83) | |
| Age at entry (Mean ± SD) | 41.05 ± 11.08 | 35.78 ± 10.23 | 0.0132 *a |
| Initial CD4 cell count (cells/mm3), n (%) | 634.00 ± 291.50 | 952.67 ± 316.36 | 0.0000 *b |
| < 200 | 4 (6.78) | 0 | |
| 200–499 | 13 (22.03) | 2 (4.35) | |
| ≥ 500 | 42 (71.19) | 44 (95.65) | |
| Birthplace, n (%) | |||
| Central Coast region: Lima | 37 (62.71) | 38 (82.61) | |
| North Coast region | 15 (25.42) | 5 (10.87) | |
| Andean region | 7 (11.86) | 1 (2.17) | |
| Amazon region | 0 | 2 (4.35) | |
| Use of ART, n (%) | |||
| Yes | 59 (100.00) ** | - | |
| No | - | - | |
| ART regimen, n (%) | |||
| 2 PIs + 1 IIs | 1 (1.69) | - | |
| 2 PIs + 2 NRTIs | 5 (8.47) | - | |
| 2 NRTIs + 1 IIs | 24 (40.68) | - | |
| 2 NRTIs + 1 NNRTIs | 26 (44.07) | - | |
| Drugs, n (%) | |||
| NRTIs | |||
| Lamivudine (3TC) | 50 (84.75) | - | |
| Tenofovir (TDF) | 51 (86.44) | - | |
| Emtricitabine (FTC) | 5 (8.47) | - | |
| Abacavir (ABC) | 3 (5.08) | - | |
| Zidovudine (AZT) | 1 (1.69) | - | |
| NNRTIs | |||
| Efavirenz (EFV) | 26 (44.07) | - | |
| PIs | |||
| Ritonavir (RTV) | 6 (10.17) | - | |
| Lopinavir (LPV) | 4 (6.78) | - | |
| Atazanavir (ATV) | 1 (1.69) | - | |
| Darunavir (DRV) | 1 (1.69) | - | |
| IIs | |||
| Dolutegravir (DTG) | 21 (35.59) | - | |
| Raltegravir (RAL) | 4 (6.78) | - | |
| Sexual Orientation, n (%) | |||
| Homosexual | - | 14 (30.43) | |
| Heterosexual | - | 26 (56.52) | |
| Bisexual | - | 6 (13.04) | |
| Sex encounters with sex workers, n (%) | |||
| Yes | - | 46 (100.00) | |
| No | - | - | |
| No. of sex partners in last 12 month | |||
| Less than 5 sexual partners | - | 3 (6.52) | |
| 5 – 15 sexual partners | - | 35 (76.09) | |
| More than 15 sexual partners | - | 8 (17.39) |
| HLA locus | Number of alleles | Alleles by locus, total | |
|---|---|---|---|
| PLHIV (n=56) |
HIV-uninfected (n=44) |
||
| HLA-A | 112 | 88 | 200 |
| HLA-B | 112 | 88 | 200 |
| HLA-C | 110 | 88 | 198 |
| HLA-DPB1 | 112 | 88 | 200 |
| HLA-DQA1 | 108 | 88 | 196 |
| HLA-DQB1 | 112 | 88 | 200 |
| HLA-DRB1 | 112 | 88 | 200 |
| HLA alleles | Allele frequency | Allele effect | |||
|---|---|---|---|---|---|
| PLHIV | HIV-uninfected | OR (95% CI) | p | p adj | |
| Susceptible | |||||
| C*07:01 | 0.1182 | 0.0227 | 10.222 (1.40-74.55) | 0.0219 | 0.0101 |
| DQA1*03:02 | 0.1481 | 0.0568 | 5.2972 (1.48-19.02) | 0.0106 | 0.0051 |
| DRB1*09:01 | 0.1696 | 0.1136 | 4.7880 (1.39-16.44) | 0.0129 | 0.0119 |
| Protective | |||||
| DQB1*03:419 | 0.2232 | 0.3523 | 0.3273 (0.11-0.96) | 0.0412 | 0.0478 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





