Submitted:
05 September 2024
Posted:
09 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Material
- CD4: Clone 4B12, mouse, ready-to-use (RTU), Leica. Antigen retrieval was achieved using Bond ER2 solution at an alkaline pH for 20 minutes.
- CD8: Clone 4B11, mouse, RTU, Leica. Antigen retrieval was conducted using Bond ER2 solution at an alkaline pH for 30 minutes.
- CD20: Clone L26, mouse, RTU, Leica. Antigen retrieval was performed using Bond ER1 solution at an alkaline pH for 20 minutes.
- CD31: Clone 1A10, mouse, RTU, Leica. Antigen retrieval was accomplished using Bond ER2 solution at an alkaline pH for 10 minutes.
2.2. Image Selection
2.3. Algorithm
2.4. Statistical Assessment
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- BlueBooksOnline Available online:. Available online: https://tumourclassification.iarc.who.int/chaptercontent/64/35 (accessed on 20 May 2024).
- Guo, A.; Liu, X.; Li, H.; Cheng, W.; Song, Y. The Global, Regional, National Burden of Cutaneous Squamous Cell Carcinoma (1990–2019) and Predictions to 2035. European Journal of Cancer Care 2023, 2023, e5484597. [Google Scholar] [CrossRef]
- Caudill, J.; Thomas, J.E.; Burkhart, C.G. The Risk of Metastases from Squamous Cell Carcinoma of the Skin. Int J Dermatol 2023, 62, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Fijałkowska, M.; Koziej, M.; Antoszewski, B. Detailed Head Localization and Incidence of Skin Cancers. Sci Rep 2021, 11, 12391. [Google Scholar] [CrossRef] [PubMed]
- Tokez, S.; Wakkee, M.; Louwman, M.; Noels, E.; Nijsten, T.; Hollestein, L. Assessment of Cutaneous Squamous Cell Carcinoma (cSCC) In Situ Incidence and the Risk of Developing Invasive cSCC in Patients With Prior cSCC In Situ vs the General Population in the Netherlands, 1989-2017. JAMA Dermatology 2020, 156, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Deng, C.-X. Effect of Stromal Cells in Tumor Microenvironment on Metastasis Initiation. Int J Biol Sci 2018, 14, 2083–2093. [Google Scholar] [CrossRef]
- A, S.; V, T.; H, P.; A, M.; S, G.; M, M.; Ep, K. Evaluation of T-Lymphocyte Subpopulations in Actinic Keratosis, in Situ and Invasive Squamous Cell Carcinoma of the Skin. Journal of cutaneous pathology 2018, 45. [Google Scholar] [CrossRef]
- Nishida, H.; Kondo, Y.; Kusaba, T.; Kawamura, K.; Oyama, Y.; Daa, T. CD8/PD-L1 Immunohistochemical Reactivity and Gene Alterations in Cutaneous Squamous Cell Carcinoma. PLoS One 2023, 18, e0281647. [Google Scholar] [CrossRef]
- Schütz, S.; Solé-Boldo, L.; Lucena-Porcel, C.; Hoffmann, J.; Brobeil, A.; Lonsdorf, A.S.; Rodríguez-Paredes, M.; Lyko, F. Functionally Distinct Cancer-Associated Fibroblast Subpopulations Establish a Tumor Promoting Environment in Squamous Cell Carcinoma. Nat Commun 2023, 14, 5413. [Google Scholar] [CrossRef]
- van Kempen, L.C.L.T.; Rijntjes, J.; Claes, A.; Blokx, W.A.M.; Gerritsen, M.-J.P.; Ruiter, D.J.; van Muijen, G.N.P. Type I Collagen Synthesis Parallels the Conversion of Keratinocytic Intraepidermal Neoplasia to Cutaneous Squamous Cell Carcinoma. J Pathol 2004, 204, 333–339. [Google Scholar] [CrossRef]
- Tzoutzos, K.; Batistatou, A.; Kitsos, G.; Liasko, R.; Stefanou, D. Study of Microvascular Density and Expression of Vascular Endothelial Growth Factor and Its Receptors in Cancerous and Precancerous Lesions of the Eyelids. Anticancer Res 2014, 34, 4977–4983. [Google Scholar]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-Associated Stromal Cells as Key Contributors to the Tumor Microenvironment. Breast Cancer Res 2016, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Saeidi, V.; Doudican, N.; Carucci, J.A. Understanding the Squamous Cell Carcinoma Immune Microenvironment. Front Immunol 2023, 14, 1084873. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Ding, J.; Chen, Y. Role of CD8+ T Lymphocyte Cells: Interplay with Stromal Cells in Tumor Microenvironment. Acta Pharm Sin B 2021, 11, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.L.; Reinert, T.; Pauletto, M.M.; Sartori, G.; Graudenz, M.; Barrios, C.H. Implications of Tumor-Infiltrating Lymphocytes in Early-Stage Triple-Negative Breast Cancer: Clinical Oncologist Perspectives. Translational Breast Cancer Research 2024, 5. [Google Scholar] [CrossRef]
- Dwivedi, M.; Tiwari, S.; Kemp, E.H.; Begum, R. Implications of Regulatory T Cells in Anti-Cancer Immunity: From Pathogenesis to Therapeutics. Heliyon 2022, 8, e10450. [Google Scholar] [CrossRef]
- Department of Pathology, Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania; Bungărdean, R. -M.; Stoia, M.-A.; Department of Internal Medicine, Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania; Pop, B.; Department of Pathology, Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania; Department of Pathology, Prof. Dr. Ion Chiricuţă Oncology Institute, Cluj-Napoca, Romania; Crişan, M.; Department of Histology, Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania Morphological Aspects of Basal Cell Carcinoma Vascularization. Rom J Morphol Embryol 2023, 64, 15–23. [Google Scholar] [CrossRef]
- Cross, S.S. Fractals in Pathology. The Journal of Pathology 1997, 182, 1–8. [Google Scholar] [CrossRef]
- da Silva, L.G.; da Silva Monteiro, W.R.S.; de Aguiar Moreira, T.M.; Rabelo, M.A.E.; de Assis, E.A.C.P.; de Souza, G.T. Fractal Dimension Analysis as an Easy Computational Approach to Improve Breast Cancer Histopathological Diagnosis. Appl Microsc 2021, 51, 6. [Google Scholar] [CrossRef]
- Di Ieva, A. Fractal Analysis in Clinical Neurosciences: An Overview. Adv Neurobiol 2024, 36, 261–271. [Google Scholar] [CrossRef]
- Di Ieva, A.; Al-Kadi, O.S. Computational Fractal-Based Analysis of Brain Tumor Microvascular Networks. Adv Neurobiol 2024, 36, 525–544. [Google Scholar] [CrossRef]
- Miola, A.C.; Castilho, M.A.; Schmitt, J.V.; Marques, M.E.A.; Miot, H.A. Contribution to Characterization of Skin Field Cancerization Activity: Morphometric, Chromatin Texture, Proliferation, and Apoptosis Aspects. An Bras Dermatol 2019, 94, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Bedin, V.; Adam, R.L.; de Sá, B.C.; Landman, G.; Metze, K. Fractal Dimension of Chromatin Is an Independent Prognostic Factor for Survival in Melanoma. BMC Cancer 2010, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Piantanelli, A.; Maponi, P.; Scalise, L.; Serresi, S.; Cialabrini, A.; Basso, A. Fractal Characterisation of Boundary Irregularity in Skin Pigmented Lesions. Med Biol Eng Comput 2005, 43, 436–442. [Google Scholar] [CrossRef]
- Popecki, P.; Kozakiewicz, M.; Ziętek, M.; Jurczyszyn, K. Fractal Dimension Analysis of Melanocytic Nevi and Melanomas in Normal and Polarized Light-A Preliminary Report. Life (Basel) 2022, 12, 1008. [Google Scholar] [CrossRef]
- Quatresooz, P.; Pierard-Franchimont, C.; Paquet, P.; Pierard, G.E. Angiogenic Fast-Growing Melanomas and Their Micrometastases. Eur J Dermatol 2010, 20, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color Transfer between Images. IEEE Computer Graphics and Applications 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Şerbănescu, M.S.; Pleşea, I.E. A Hardware Approach for Histological and Histopathological Digital Image Stain Normalization. Rom J Morphol Embryol 2015, 56, 735–741. [Google Scholar]
- Reyes-Aldasoro, C. c.; Williams, L. j.; Akerman, S.; Kanthou, C.; Tozer, G. m. An Automatic Algorithm for the Segmentation and Morphological Analysis of Microvessels in Immunostained Histological Tumour Sections. Journal of Microscopy 2011, 242, 262–278. [Google Scholar] [CrossRef]
- Reyes-Aldasoro, C.C.; Griffiths, M.K.; Savas, D.; Tozer, G.M. CAIMAN: An Online Algorithm Repository for Cancer Image Analysis. Computer Methods and Programs in Biomedicine 2011, 103, 97–103. [Google Scholar] [CrossRef]
- Pleşea, R.M.; Şerbănescu, M.S.; Ciovică, D.V.; Roşu, G.C.; Moldovan, V.T.; Bungărdean, R.M.; Popescu, N.A.; Pleşea, I.E. The Study of Tumor Architecture Components in Prostate Adenocarcinoma Using Fractal Dimension Analysis. Rom J Morphol Embryol 2019, 60, 501–519. [Google Scholar]
- Șerbănescu, M.-S. Fractal Dimension Box-Counting Algorithm Optimization Through Integral Images. In Proceedings of the 7th International Conference on Advancements of Medicine and Health Care through Technology; Vlad, S., Roman, N.M., Eds.; Springer International Publishing: Cham, 2022; pp. 95–101. [Google Scholar]
- Stoiculescu, A.; Pleşea, I.E.; Pop, O.T.; Alexandru, D.O.; Man, M.; Serbănescu, M.; Pleşea, R.M. Correlations between Intratumoral Interstitial Fibrillary Network and Tumoral Architecture in Prostatic Adenocarcinoma. Rom J Morphol Embryol 2012, 53, 941–950. [Google Scholar] [PubMed]
- Pleşea, I.E.; Stoiculescu, A.; Serbănescu, M.; Alexandru, D.O.; Man, M.; Pop, O.T.; Pleşea, R.M. Correlations between Intratumoral Vascular Network and Tumoral Architecture in Prostatic Adenocarcinoma. Rom J Morphol Embryol 2013, 54, 299–308. [Google Scholar] [PubMed]
- Mitroi, G.; Pleşea, R.M.; Pop, O.T.; Ciovică, D.V.; Şerbănescu, M.S.; Alexandru, D.O.; Stoiculescu, A.; Pleşea, I.E. Correlations between Intratumoral Interstitial Fibrillary Network and Vascular Network in Srigley Patterns of Prostate Adenocarcinoma. Rom J Morphol Embryol 2015, 56, 1319–1328. [Google Scholar] [PubMed]
- Bărbălan, A.; Nicolaescu, A.C.; Măgăran, A.V.; Mercuţ, R.; Bălăşoiu, M.; Băncescu, G.; Şerbănescu, M.S.; Lazăr, O.F.; Săftoiu, A. Immunohistochemistry Predictive Markers for Primary Colorectal Cancer Tumors: Where Are We and Where Are We Going? Rom J Morphol Embryol 2018, 59, 29–42. [Google Scholar]
- Harbiyeli, I.F.C.; Burtea, D.E.; Ivan, E.T.; Streață, I.; Nicoli, E.R.; Uscatu, D.; Șerbănescu, M.-S.; Ioana, M.; Vilmann, P.; Săftoiu, A. Assessing Putative Markers of Colorectal Cancer Stem Cells: From Colonoscopy to Gene Expression Profiling. Diagnostics (Basel) 2022, 12, 2280. [Google Scholar] [CrossRef]
- Șerbănescu, M.-S.; Bungărdean, R.M.; Georgiu, C.; Crișan, M. Nodular and Micronodular Basal Cell Carcinoma Subtypes Are Different Tumors Based on Their Morphological Architecture and Their Interaction with the Surrounding Stroma. Diagnostics (Basel) 2022, 12, 1636. [Google Scholar] [CrossRef]
- Pătru, A.; Şurlin, V.; Mărgăritescu, C.; Ciucă, E.M.; Matei, M.; Şerbănescu, M.S.; Camen, A. Analysis of the Distribution and Expression of Some Tumor Invasiveness Markers in Palate Squamous Cell Carcinomas. Rom J Morphol Embryol 2020, 61, 1259–1278. [Google Scholar] [CrossRef]
- Lertkiatmongkol, P.; Liao, D.; Mei, H.; Hu, Y.; Newman, P.J. Endothelial Functions of Platelet/Endothelial Cell Adhesion Molecule-1 (CD31). Curr Opin Hematol 2016, 23, 253–259. [Google Scholar] [CrossRef]
- Newman, P.J. The Biology of PECAM-1. J Clin Invest 1997, 99, 3–8. [Google Scholar] [CrossRef]
- Mei, H.; Campbell, J.M.; Paddock, C.M.; Lertkiatmongkol, P.; Mosesson, M.W.; Albrecht, R.; Newman, P.J. Regulation of Endothelial Cell Barrier Function by Antibody-Driven Affinity Modulation of Platelet Endothelial Cell Adhesion Molecule-1 (PECAM-1) *. Journal of Biological Chemistry 2014, 289, 20836–20844. [Google Scholar] [CrossRef]
- Cao, G.; O’Brien, C.D.; Zhou, Z.; Sanders, S.M.; Greenbaum, J.N.; Makrigiannakis, A.; DeLisser, H.M. Involvement of Human PECAM-1 in Angiogenesis and in Vitro Endothelial Cell Migration. Am J Physiol Cell Physiol 2002, 282, C1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Andreata, F.; Clément, M.; Benson, R.A.; Hadchouel, J.; Procopio, E.; Even, G.; Vorbe, J.; Benadda, S.; Ollivier, V.; Ho-Tin-Noe, B.; et al. CD31 Signaling Promotes the Detachment at the Uropod of Extravasating Neutrophils Allowing Their Migration to Sites of Inflammation. Elife 2023, 12, e84752. [Google Scholar] [CrossRef] [PubMed]
- Kuriri, F.A.; O’Malley, C.J.; Jackson, D.E. Molecular Mechanisms of Immunoreceptors in Platelets. Thromb Res 2019, 176, 108–114. [Google Scholar] [CrossRef]
- Pavlasova, G.; Mraz, M. The Regulation and Function of CD20: An “Enigma” of B-Cell Biology and Targeted Therapy. Haematologica 2020, 105, 1494–1506. [Google Scholar] [CrossRef] [PubMed]
- Casan, J.M.L.; Wong, J.; Northcott, M.J.; Opat, S. Anti-CD20 Monoclonal Antibodies: Reviewing a Revolution. Hum Vaccin Immunother 2018, 14, 2820–2841. [Google Scholar] [CrossRef]
- Carlson, A.K.; Amin, M.; Cohen, J.A. Drugs Targeting CD20 in Multiple Sclerosis: Pharmacology, Efficacy, Safety, and Tolerability. Drugs 2024, 84, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Su, Y.; Jiao, A.; Wang, X.; Zhang, B. T Cells in Health and Disease. Signal Transduct Target Ther 2023, 8, 235. [Google Scholar] [CrossRef]
- Zhen, A.; Krutzik, S.R.; Levin, B.R.; Kasparian, S.; Zack, J.A.; Kitchen, S.G. CD4 Ligation on Human Blood Monocytes Triggers Macrophage Differentiation and Enhances HIV Infection. J Virol 2014, 88, 9934–9946. [Google Scholar] [CrossRef]
- Jardine, L.; Barge, D.; Ames-Draycott, A.; Pagan, S.; Cookson, S.; Spickett, G.; Haniffa, M.; Collin, M.; Bigley, V. Rapid Detection of Dendritic Cell and Monocyte Disorders Using CD4 as a Lineage Marker of the Human Peripheral Blood Antigen-Presenting Cell Compartment. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Chen, H.; Sameshima, J.; Yokomizo, S.; Sueyoshi, T.; Nagano, H.; Miyahara, Y.; Sakamoto, T.; Fujii, S.; Kiyoshima, T.; Guy, T.; et al. Expansion of CD4+ Cytotoxic T Lymphocytes with Specific Gene Expression Patterns May Contribute to Suppression of Tumor Immunity in Oral Squamous Cell Carcinoma: Single-Cell Analysis and in Vitro Experiments. Front Immunol 2023, 14, 1305783. [Google Scholar] [CrossRef]
- Eizenberg-Magar, I.; Rimer, J.; Zaretsky, I.; Lara-Astiaso, D.; Reich-Zeliger, S.; Friedman, N. Diverse Continuum of CD4+ T-Cell States Is Determined by Hierarchical Additive Integration of Cytokine Signals. Proceedings of the National Academy of Sciences 2017, 114, E6447–E6456. [Google Scholar] [CrossRef]
- Kruse, B.; Buzzai, A.C.; Shridhar, N.; Braun, A.D.; Gellert, S.; Knauth, K.; Pozniak, J.; Peters, J.; Dittmann, P.; Mengoni, M.; et al. CD4+ T Cell-Induced Inflammatory Cell Death Controls Immune-Evasive Tumours. Nature 2023, 618, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Charles A Janeway, J.; Travers, P.; Walport, M.; Shlomchik, M.J. T Cell-Mediated Cytotoxicity. In Immunobiology: The Immune System in Health and Disease. 5th edition; Garland Science, 2001.
- Schmidt, M.E.; Varga, S.M. The CD8 T Cell Response to Respiratory Virus Infections. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.-H.; Lee, S.; Kwak, M.; Kim, B.-S.; Chung, Y. CD8 T-Cell Subsets: Heterogeneity, Functions, and Therapeutic Potential. Exp Mol Med 2023, 55, 2287–2299. [Google Scholar] [CrossRef] [PubMed]
- Brummel, K.; Eerkens, A.L.; de Bruyn, M.; Nijman, H.W. Tumour-Infiltrating Lymphocytes: From Prognosis to Treatment Selection. Br J Cancer 2023, 128, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Buruiană, A.; Gheban, B.-A.; Gheban-Roșca, I.-A.; Georgiu, C.; Crișan, D.; Crișan, M. The Tumor Stroma of Squamous Cell Carcinoma: A Complex Environment That Fuels Cancer Progression. Cancers 2024, 16, 1727. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Brockton, N.T.; Guggisberg, K.; Nakoneshny, S.C.; Kornaga, E.; Klimowicz, A.C.; Tambasco, M.; Dort, J.C. Fractal Analysis of Nuclear Histology Integrates Tumor and Stromal Features into a Single Prognostic Factor of the Oral Cancer Microenvironment. BMC Cancer 2015, 15, 409. [Google Scholar] [CrossRef]
- Goutzanis, L.P.; Papadogeorgakis, N.; Pavlopoulos, P.M.; Petsinis, V.; Plochoras, I.; Eleftheriadis, E.; Pantelidaki, A.; Patsouris, E.; Alexandridis, C. Vascular Fractal Dimension and Total Vascular Area in the Study of Oral Cancer. Head Neck 2009, 31, 298–307. [Google Scholar] [CrossRef]
- Margaritescu, C.; Raica, M.; Pirici, D.; Simionescu, C.; Mogoanta, L.; Stinga, A.C.; Stinga, A.S.; Ribatti, D. Podoplanin Expression in Tumor-Free Resection Margins of Oral Squamous Cell Carcinomas: An Immunohistochemical and Fractal Analysis Study. Histol Histopathol 2010, 25, 701–711. [Google Scholar] [CrossRef]
- Capasso, A.; Viggiano, D.; Lee, M.W.; Palladino, G.; Bilancio, G.; Simeoni, M.; Capolongo, G.; Secondulfo, C.; Ronchi, A.; Caputo, A.; et al. Kidney Transplant Modifies the Architecture and Microenvironment of Basal Cell Carcinomas. Kidney and Blood Pressure Research 2020, 45, 368–377. [Google Scholar] [CrossRef]
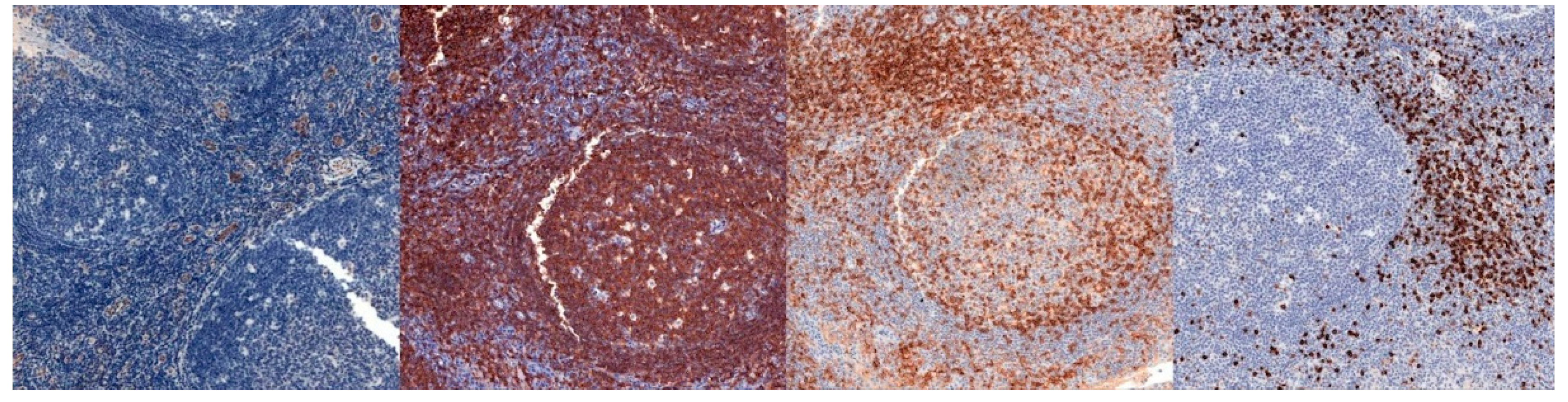
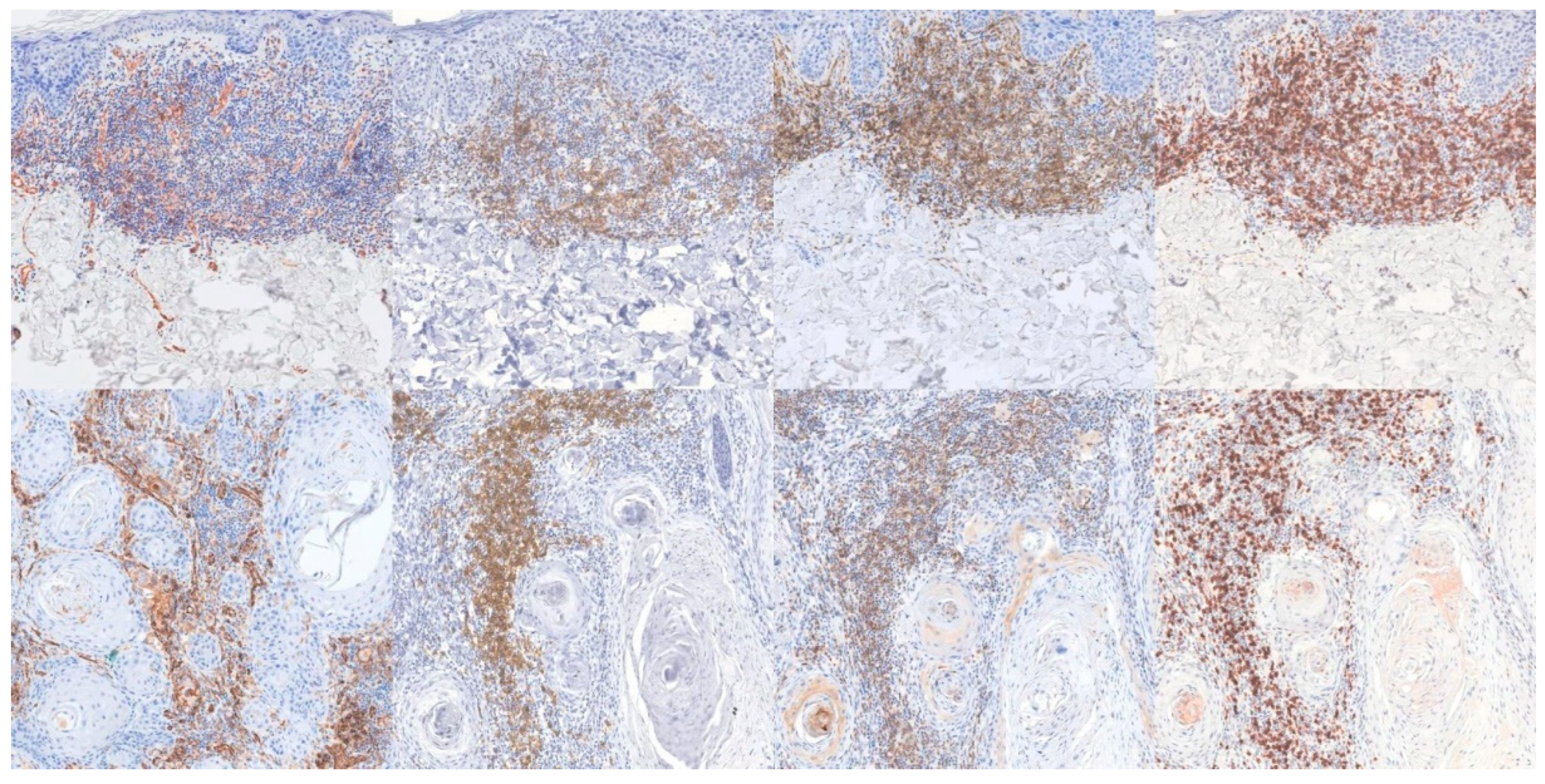
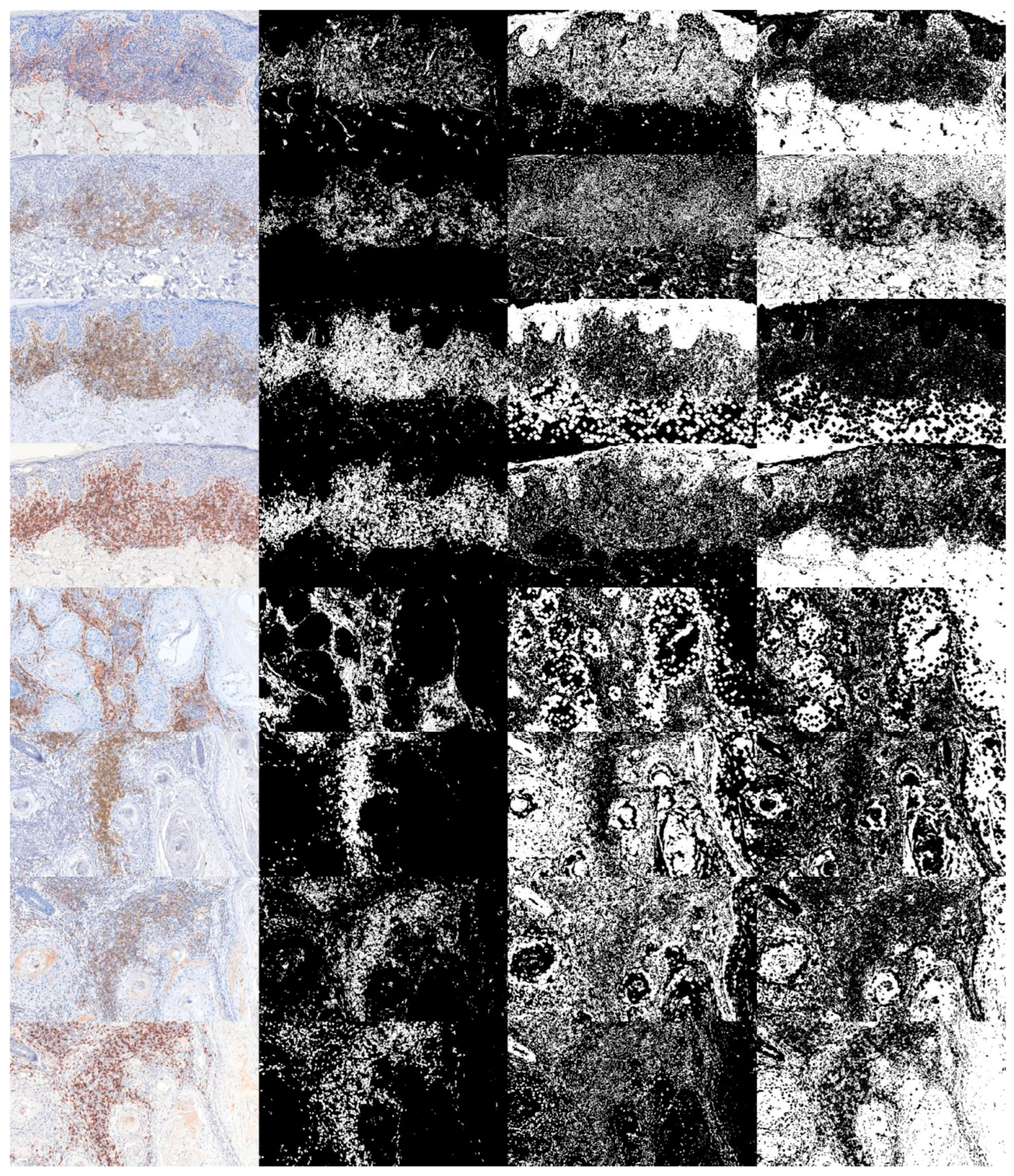
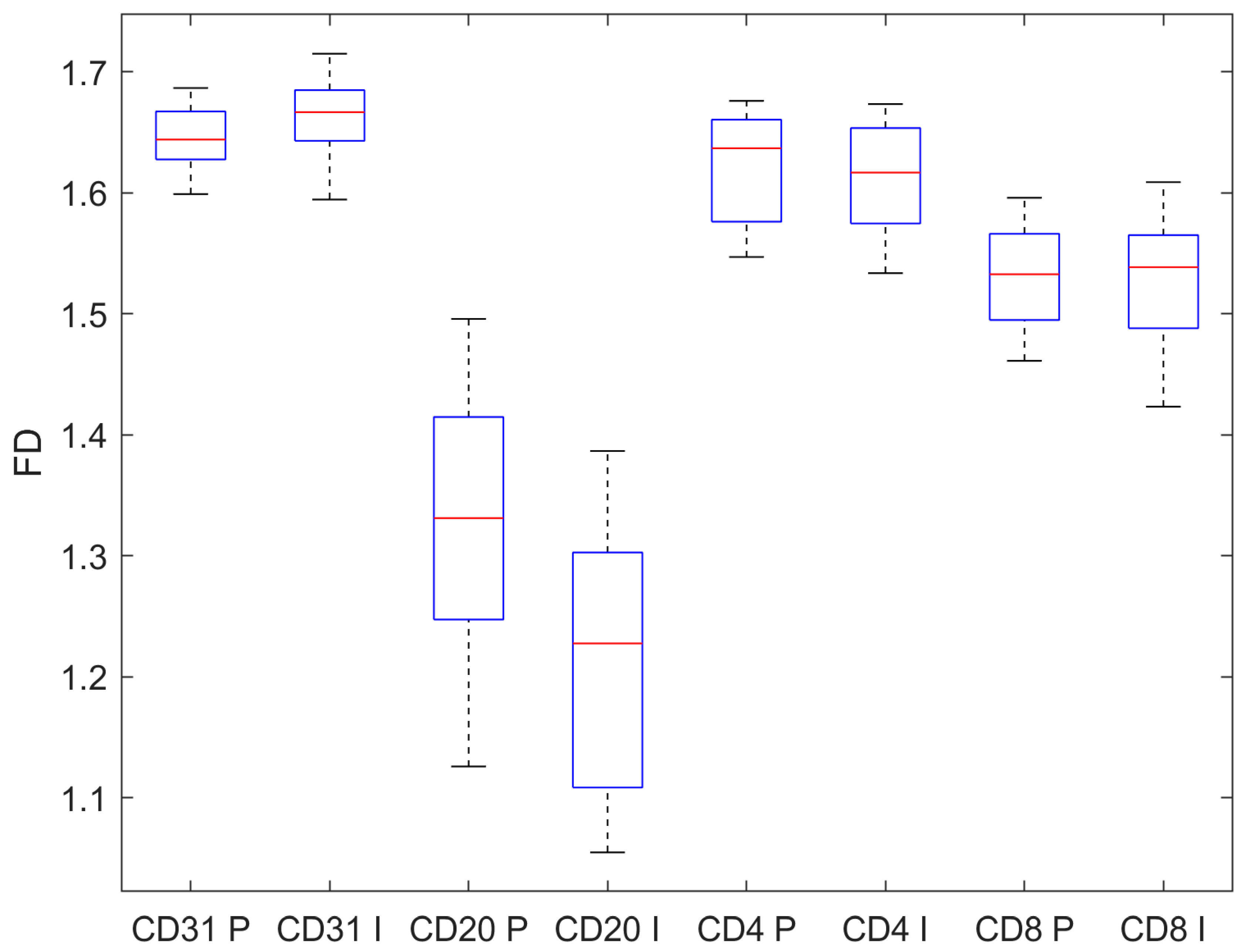
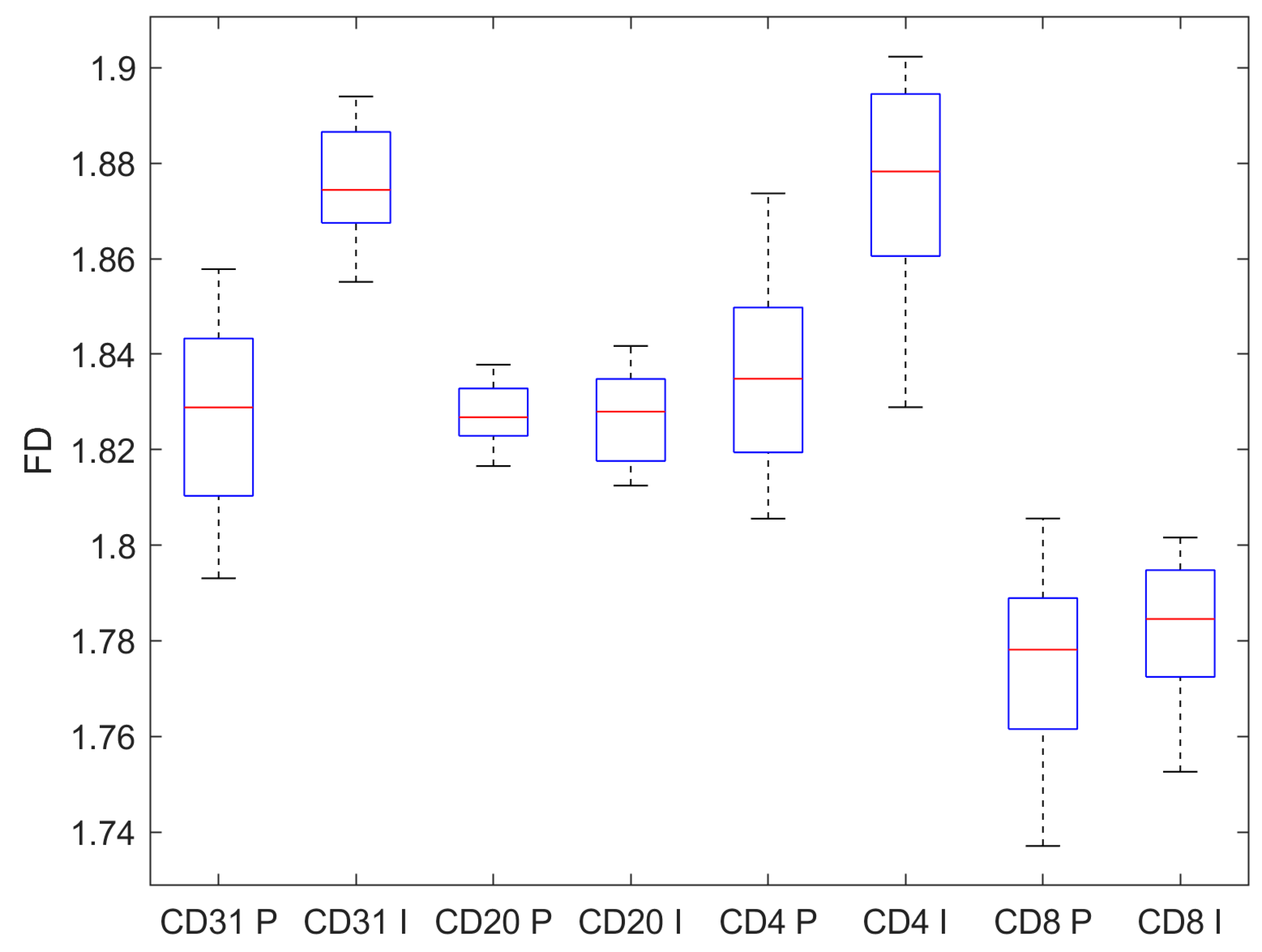
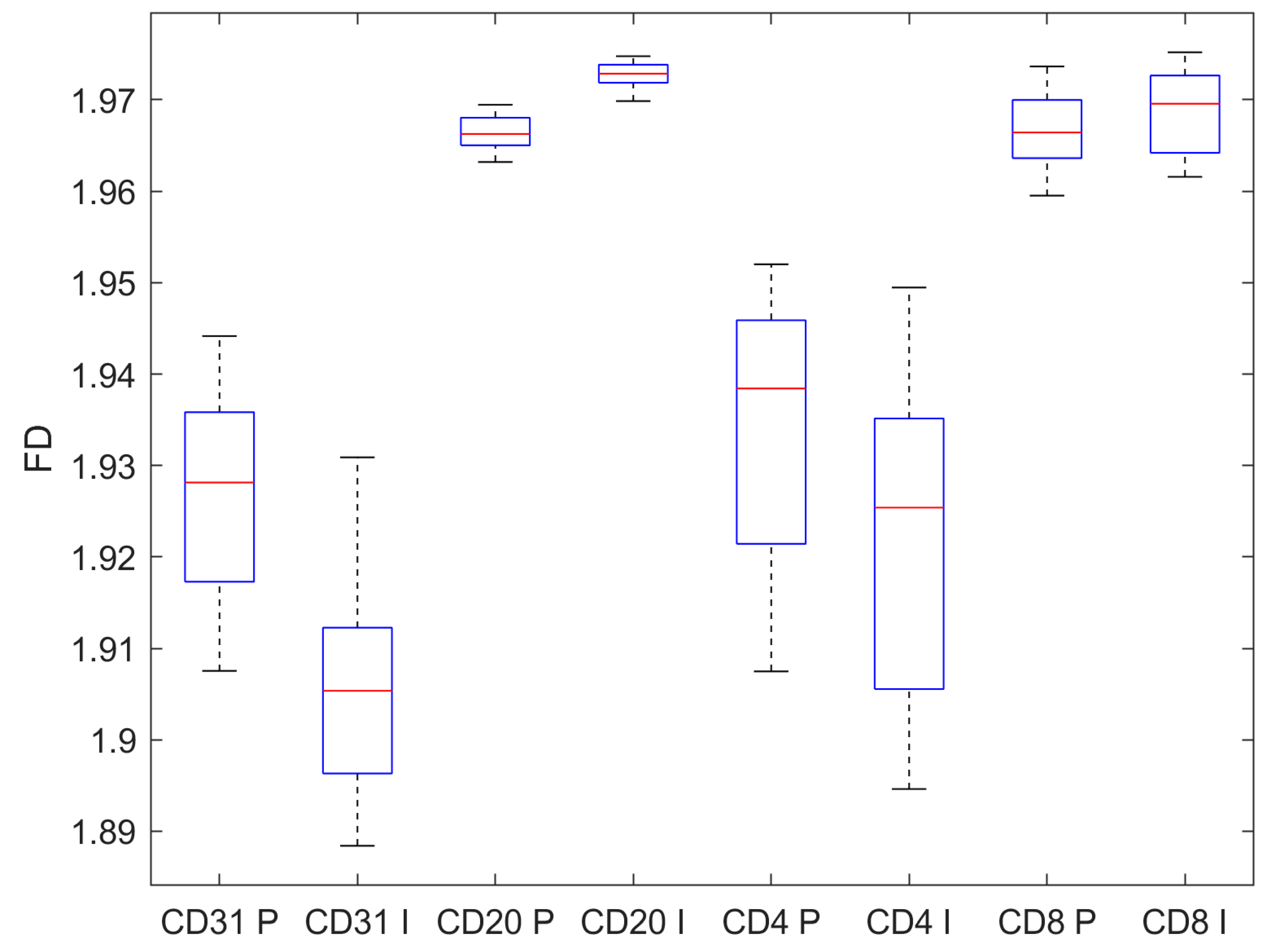
| CD31 | CD20 | CD4 | CD8 | |
|---|---|---|---|---|
| Pre-invasive | 1.645 ± 0.024 | 1.321 ± 0.104 | 1.623 ± 0.041 | 1.530 ± 0.041 |
| Invasive | 1.661 ± 0.035 | 1.220 ± 0.102 | 1.610 ± 0.046 | 1.527 ± 0.053 |
| p, t-test | 0.010 | < 0.001 | 0.133 | 0.738 |
| CD31 | CD20 | CD4 | CD8 | |
|---|---|---|---|---|
| Pre-invasive | 1.827 ± 0.019 | 1.827 ± 0.006 | 1.835 ± 0.020 | 1.774 ± 0.019 |
| Invasive | 1.876 ± 0.012 | 1.827 ± 0.009 | 1.875 ± 0.021 | 1.783 ± 0.014 |
| p, t-test | <0.001 | 0.619 | <0.001 | 0.007 |
| CD31 | CD20 | CD4 | CD8 | |
|---|---|---|---|---|
| Pre-invasive | 1.926 ± 0.011 | 1.966 ± 0.002 | 1.934 ± 0.015 | 1.967 ± 0.004 |
| Invasive | 1.906 ± 0.011 | 1.973 ± 0.001 | 1.922 ± 0.017 | 1.969 ± 0.004 |
| p, t-test | <0.001 | <0.001 | <0.001 | 0.019 |
| CD4/CD8 | CD4+CD8 | (CD4+CD8)/CD20 | |
|---|---|---|---|
| Pre-invasive | 1.061 ± 0.006 | 3.153 ± 0.082 | 2.397 ± 0.132 |
| Invasive | 1.055 ± 0.009 | 3.136 ± 0.099 | 2.581 ± 0.136 |
| p, t-test | 0.010 | 0.368 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





