Submitted:
04 September 2024
Posted:
05 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Specimens, Patients, and Clinical Data
Whole-Exome Sequencing
Read Processing, Quality Control, and Somatic Variant Calling
Somatic Copy-Number Inference
Data Analysis and Visualization
3. Results
3.1. Samples and Clinical Data
3.2. Overview of the Somatic Mutational Signature of IECs
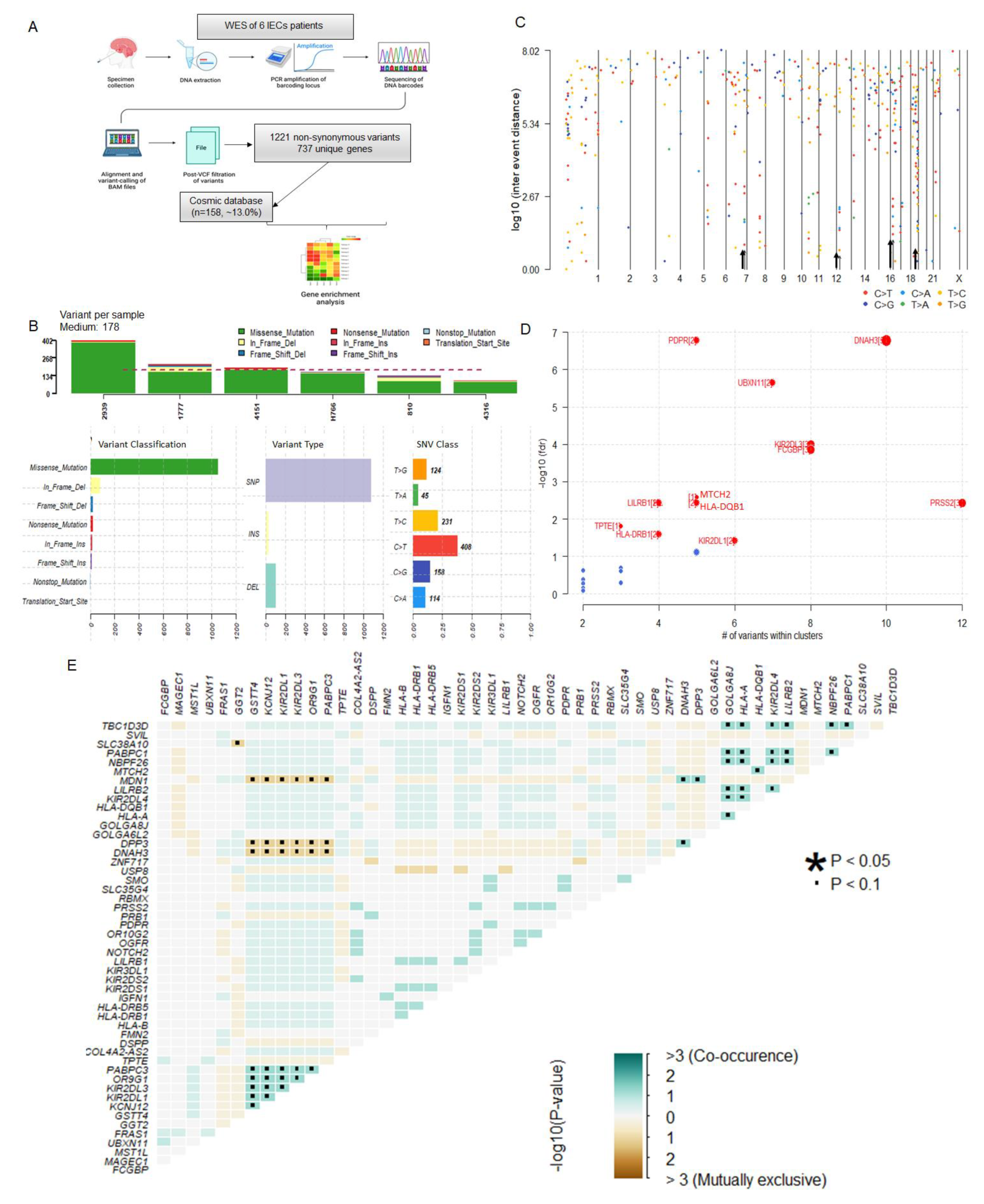
3.3. IECs Are Characterized by an Altered Immune Repertoire
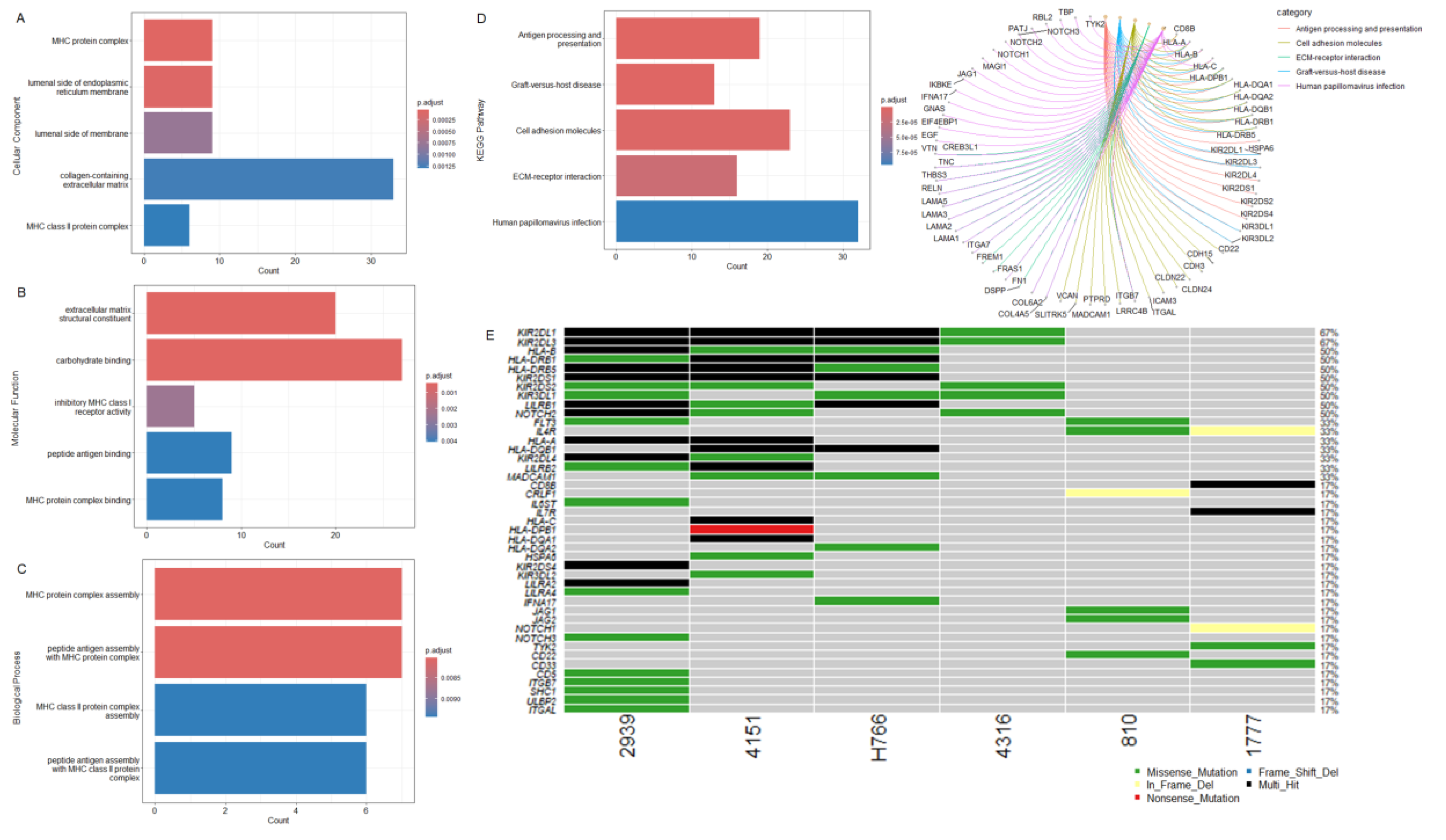
3.4. Oncogenic Driver Candidates of IECs
| Hugo Symbol | Status | Cancer Type | Oncogenic Function | References |
|---|---|---|---|---|
| DSPP | Upregulation |
Head Neck SCC |
ER stress invasion Metastasis Angiogenesis EMT |
[43], [44] |
| FCGBP | Aberrant regulation |
Glioma Colorectal SCC |
Immune infiltration | [45], [46], [47] |
| FMN2 | NR | Osteosarcoma | Regulation of tumor suppressor pathway mediated by p14ARF | [48] |
| FRAS1 | Upregulation | SCC |
Cell growth Metastasis |
[49] |
| GGT2 | Upregulation |
Endometrial Gastrointestinal SCC Melanoma |
Metabolic signaling Cell growth |
[50], [51], [52] |
| GSTT4 | Mutation |
Acute leukemia SCC |
Cellular detoxification | [53], [54], [55], [56] |
| HLA-B |
Mutation Downregulation |
Breast Colon Rectal SCC Melanoma |
Immune infiltration | [57], [58], [59], [60] |
| HLA-DRB1 |
Upregulation Fucosylation |
Melanoma Lung SCC |
Immune infiltration | [61], [62], [63], [64], [65] |
| HLA-DRB5 | Upregulation |
Melanoma Lung SCC |
Immune infiltration | [63], [64], [65] |
| IGFN1 | Aberrant expression Mutation |
NSCL Renal |
Metastasis | [66], [67] |
| KCNJ12 | Mutation |
Glioblastoma Prostate SCC |
Ion channel | [68], [69], [70] |
| KIR2DL1 | Mutation |
Glioblastoma Melanoma |
Immune infiltration | [71], [72] |
| KIR2DL3 | Mutation |
Glioblastoma Melanoma |
Immune infiltration | [71], [72] |
| KIR2DS1 | Mutation |
Glioblastoma Melanoma |
Immune infiltration | [71], [72] |
| KIR2DS2 | Mutation |
Glioblastoma Melanoma |
Immune infiltration | [71], [72] |
| KIR3DL1 | Mutation |
Glioblastoma Melanoma |
Immune infiltration | [71], [72] |
|
LILRB1 |
Upregulation |
Melanoma Multiple cancers |
Immune infiltration | [73], [74], [75] |
| MAGEC1 | Aberrant expression | Cholangiocarcinoma Hepatocellular |
Immune infiltration Apoptosis Cell cycle |
[76], [77] |
| MST1L | Downregulation | Breast | Cell differentiation Adhesion Migration Apoptosis |
[78] |
| NOTCH2 |
Aberrant expression Mutation |
SCC Melanoma Multiple cancers |
Immune infiltration Stem-like proliferation |
[79], [80], [81], [82], [83] |
| OGFR |
Aberrant expression Mutation |
Gynecological Ovarian SCC |
Cell proliferation Cell cycle |
[84], [85], [86] |
| OR10G2 | Aberrant expression | Multiple cancers | Cell differentiation Invasion Metastasis |
[87], [88] |
| OR9G1 | Aberrant expression | Multiple cancers | Cell differentiation Invasion Metastasis |
[87], [88] |
| PABPC3 | Aberrant expression | Pancreas Osteosarcoma |
Immune infiltration Cell proliferation Metastasis |
[89], [90] |
| PDPR | Downregulation | Thyroid | NR | [91] |
| PRB1 | NR | NR | NR | NR |
| PRSS2 | Upregulation | Gastric | EMT Metastasis Microenvironment |
[92], [93] |
| RBMX | Downregulation | Multiple cancers | Tumor development | [94], [95] |
| SLC35G4 | Upregulation | Glioma Breast |
Immune infiltration | [96], [97] |
| TPTE | Upregulation | Lung Prostate |
Immune infiltration | [98], [99] |
| UBXN11 | Upregulation Mutation |
Multiple cancers | Tumor progression | [100] |
| USP8 |
Upregulation Mutation |
SCC Melanoma Multiple cancers |
Immune infiltration Tumor progression Drug resistance |
[101], [102], [103] |
| ZNF717 | Mutation | Colorectal Hepatocellular |
NR | [104], [105] |
3.5. Recurrent Variants in NOTCH2 and USP8
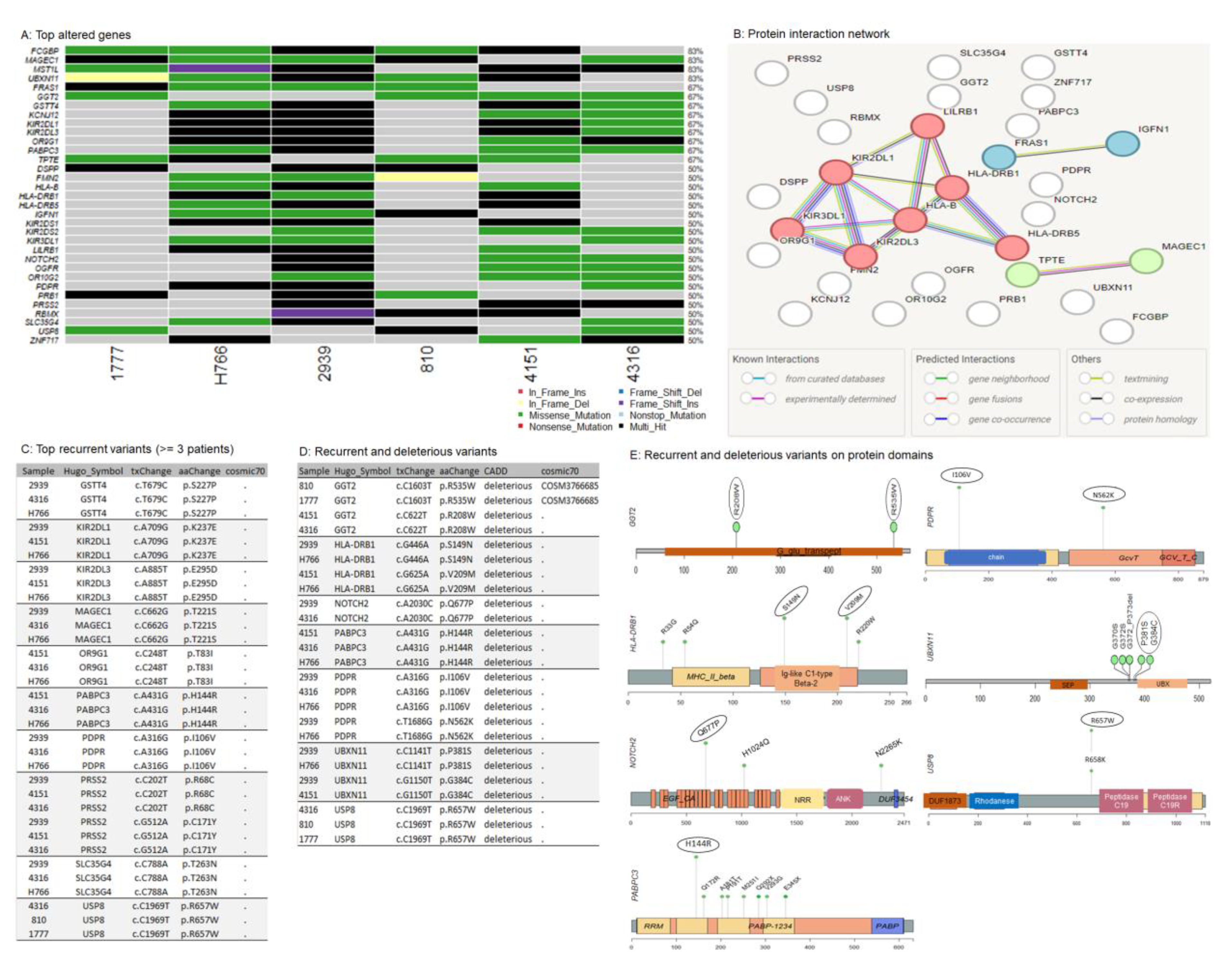
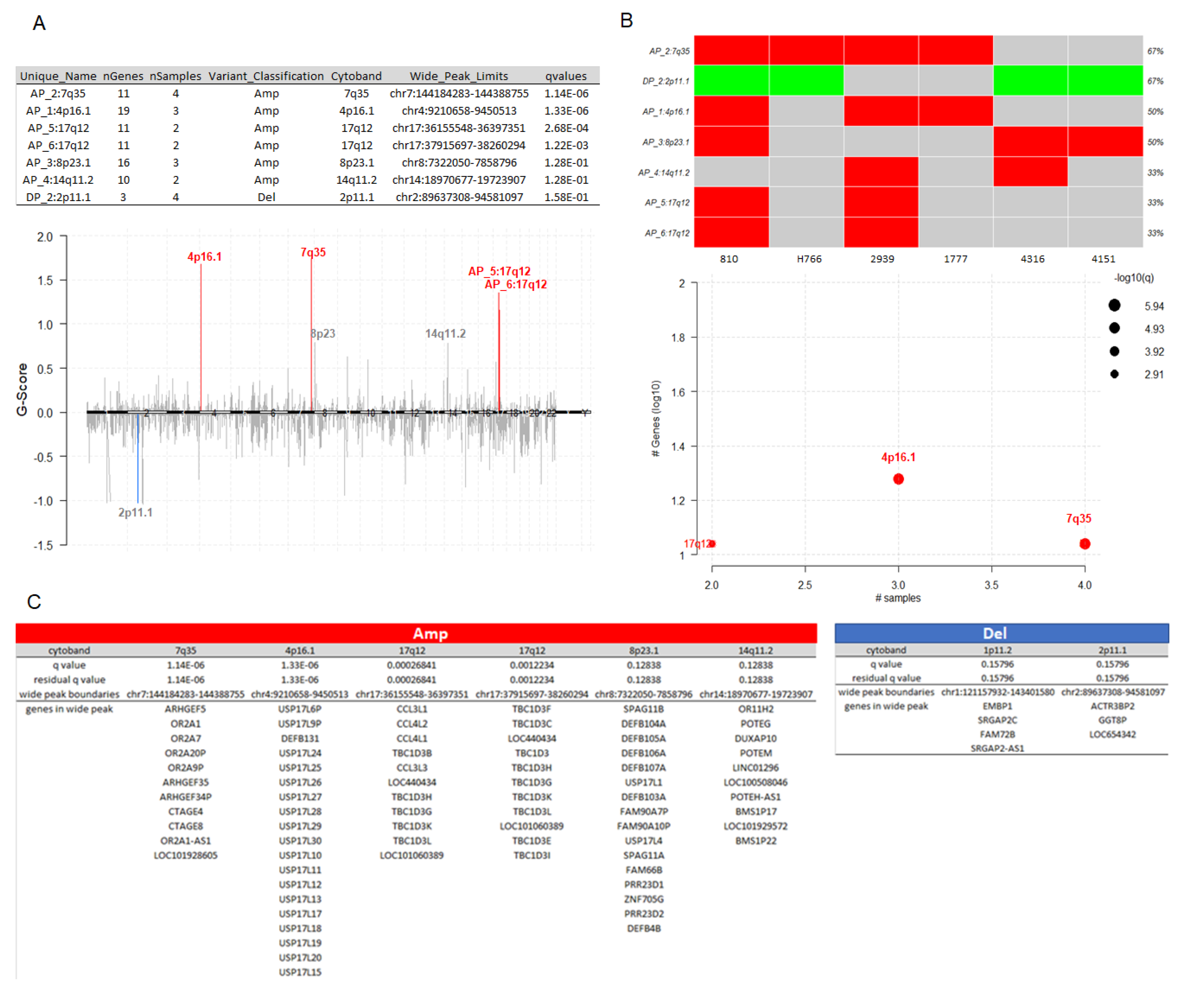
3.6. Somatic Copy-Number Inference Strengthens the Involvement of Immune-Associated Genes in the Pathogenesis of IECs
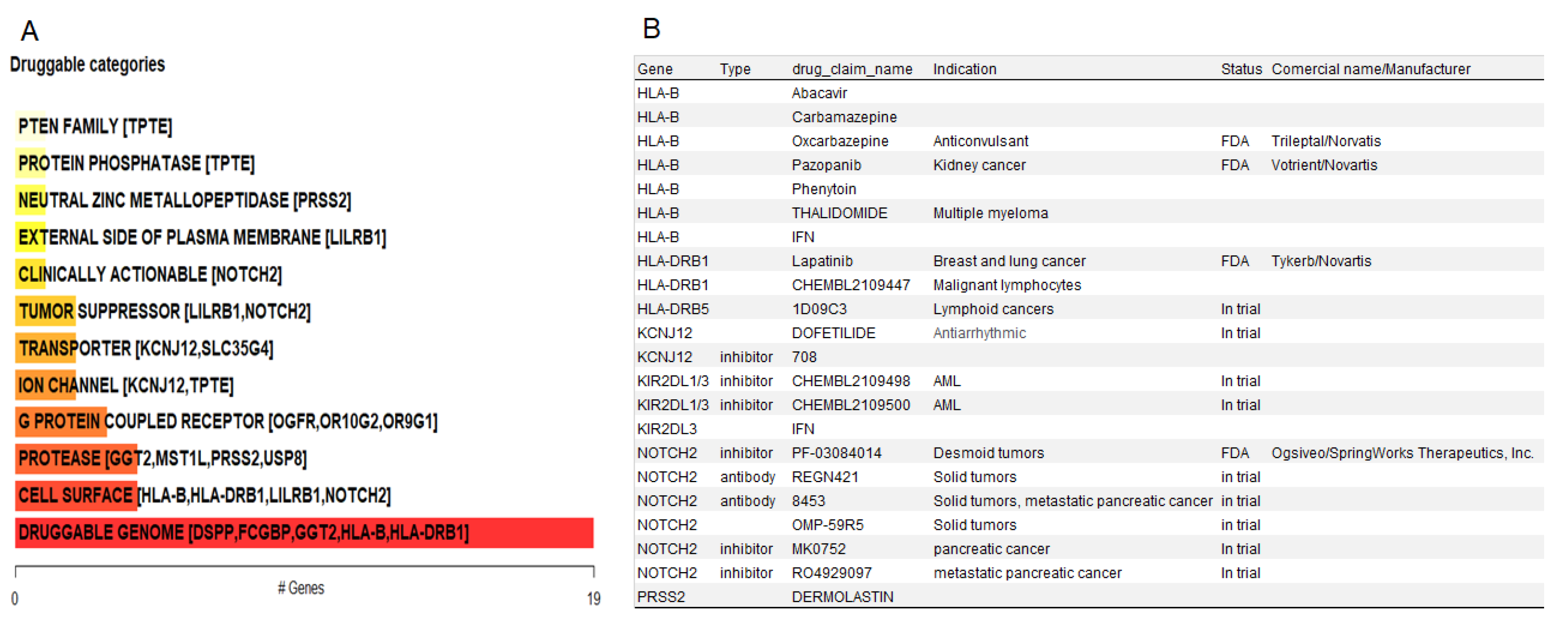
3.7. Potential Actionable Targets in IECs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- J. S. Tytus and J. Pennybacker, ‘PEARLY TUMOURS IN RELATION TO THE CENTRAL NERVOUS SYSTEM’, 1956.
- K. Yamakawa, N. Shitara, S. Genka, S. Manaka, and K. Takakura, ‘Clinical Course and Surgical Prognosis of 33 Cases of Intracranial Epidermoid Tumors’, Neurosurgery, vol. 24, no. 4, pp. 568–573, Apr. 1989. [CrossRef]
- G. Iaconetta, G. A. Carvalho, P. Vorkapic, and M. Samii, ‘Intracerebral Epidermoid Tumor: A Case Report and Review of the Literature’, 2001.
- I. F. Braun, T. P. Naidich, N. E. Leeds, M. Koslow, H. M. Zimmerman, and N. E. Chase, ‘Dense Intracranial Epidermoid Tumors’, Radiology, vol. 122, no. 3, pp. 717–719, Mar. 1977. [CrossRef]
- M. Vinchon, J. P. Lejeune, I. Krivosic, R. Assaker, J. P. Pruvo, and J. L. Christiaens, ‘[Cranio-cerebral dermoid and epidermoid cysts. Classification and pathogenesis].’, Neurochirurgie, vol. 41, no. 1, pp. 29–37, 1995.
- S. Mohanty, R. N. Bhattacharya, S. C. Tandon, and P. K. Shukla, ‘Intracerebral cystic epidermoid’, Acta Neurochir (Wien), vol. 57, no. 1–2, pp. 107–113, Mar. 1981. [CrossRef]
- W. G. Obana and C. B. Wilson, ‘Epidermoid cysts of the brain stem’, J Neurosurg, vol. 74, no. 1, pp. 123–128, Jan. 1991. [CrossRef]
- D. Fournier, P. Mercier, P. Menei, F. Pouplard, T. Rizk, and G. Guy, ‘Recurrent intrinsic brain stem epidermoid cyst’, Child’s Nervous System, vol. 8, no. 8, pp. 471–474, Dec. 1992. [CrossRef]
- E. N. Weaver and R. A. Coulon, ‘Excision of a brain-stem epidermoid cyst’, J Neurosurg, vol. 51, no. 2, pp. 254–257, Aug. 1979. [CrossRef]
- R. G. Pereira, B. N. de F. Ribeiro, R. T. de L. Hollanda, L. B. de Almeida, T. B. Simeão, and E. Marchiori, ‘Non-neoplastic intracranial cystic lesions: not everything is an arachnoid cyst’, Radiol Bras, vol. 54, no. 1, pp. 49–55, Feb. 2021. [CrossRef]
- I. M. Ziyal, B. Bilginer, G. Bozkurt, O. Çataltepe, G. G. Tezel, and N. Akalan, ‘Epidermoid cyst of the brain stem symptomatic in childhood’, Child’s Nervous System, vol. 21, no. 12, pp. 1025–1029, Dec. 2005. [CrossRef]
- J. Ulrich, ‘Intracranial Epidermoids’, J Neurosurg, vol. 21, no. 12, pp. 1051–1058, Dec. 1964. [CrossRef]
- E. C. Alvord, ‘Growth rates of epidermoid tumors’, Ann Neurol, vol. 2, no. 5, pp. 367–370, Nov. 1977. [CrossRef]
- C. Bayindir, N. Balak, and A. Karasu, ‘Micro-invasive squamous cell carcinoma arising in a pre-existing intraventricular epidermoid cyst’, Acta Neurochir (Wien), vol. 138, no. 8, pp. 1008–1012, Aug. 1996. [CrossRef]
- P. Michenet, C. Vital, J. Rivel, B. Lebail, and V. Riemens, ‘[Malignant transformation of an intracranial epidermoid cyst].’, Ann Pathol, vol. 9, no. 5, pp. 360–2, 1989.
- A. Uchino et al., ‘Intracranial epidermoid carcinoma: CT and MRI’, Neuroradiology, vol. 37, no. 2, pp. 155–158, Feb. 1995. [CrossRef]
- Vajtai, D. Tassi, Z. Varga, J. Tarjányi, and E. Vörös, ‘[Malignant melanoma evolving inside a cerebral epidermoid cyst].’, Orv Hetil, vol. 136, no. 22, pp. 1171–4, May 1995.
- J. A. Cuoco, C. M. Rogers, C. M. Busch, L. S. Apfel, J. J. Entwistle, and E. A. Marvin, ‘Intracranial Squamous Cell Carcinoma Arising From a Cerebellopontine Angle Epidermoid Cyst Remnant Four Decades After Partial Resection.’, Front Oncol, vol. 9, p. 694, 2019. [CrossRef]
- H. Hasegawa et al., ‘Long-term surgical outcomes of intracranial epidermoid tumors: impact of extent of resection on recurrence and functional outcomes in 63 patients’, J Neurosurg, vol. 136, no. 6, pp. 1592–1600, Jun. 2022. [CrossRef]
- C. V. Gopalakrishnan, K. A. Ansari, S. Nair, and G. Menon, ‘Long term outcome in surgically treated posterior fossa epidermoids’, Clin Neurol Neurosurg, vol. 117, pp. 93–99, Feb. 2014. [CrossRef]
- R. A. Morshed, S. Y. Wu, P. K. Sneed, and M. W. McDermott, ‘Radiotherapy for recurrent intracranial epidermoid cysts without malignant transformation: a single-institution case series.’, J Neurooncol, vol. 144, no. 1, pp. 89–96, Aug. 2019. [CrossRef]
- S. Negrini, V. G. Gorgoulis, and T. D. Halazonetis, ‘Genomic instability an evolving hallmark of cancer’, Mar. 2010. [CrossRef]
- Y. Y. Syed, ‘Amivantamab: First Approval’, Jul. 01, 2021, Adis. [CrossRef]
- Y. Y. Li and S. J. Jones, ‘Drug repositioning for personalized medicine.’, Genome Med, vol. 4, no. 3, p. 27, 2012. [CrossRef]
- M. Kubo, M. Yamane, K. Miyatani, T. Udaka, M. Mizuta, and K. Shirakawa, ‘Familial Epidermoid Cysts of the Spleen: Report of Two Cases’, Surg Today, vol. 36, no. 9, pp. 853–856, Sep. 2006. [CrossRef]
- N. N. Kulkarni et al., ‘IL-1 Receptor–Knockout Mice Develop Epidermal Cysts and Show an Altered Innate Immune Response after Exposure to UVB Radiation’, Journal of Investigative Dermatology, vol. 137, no. 11, pp. 2417–2426, Nov. 2017. [CrossRef]
- A. M. Bolger, M. Lohse, and B. Usadel, ‘Trimmomatic: a flexible trimmer for Illumina sequence data’, Bioinformatics, vol. 30, no. 15, pp. 2114–2120, Aug. 2014. [CrossRef]
- H. Li and R. Durbin, ‘Fast and accurate short read alignment with Burrows–Wheeler transform’, Bioinformatics, vol. 25, no. 14, pp. 1754–1760, Jul. 2009. [CrossRef]
- D. C. Koboldt et al., ‘VarScan: Variant detection in massively parallel sequencing of individual and pooled samples’, Bioinformatics, vol. 25, no. 17, pp. 2283–2285, 2009. [CrossRef]
- Z. Lai et al., ‘VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research’, Nucleic Acids Res, vol. 44, no. 11, Jun. 2016. [CrossRef]
- S. Kim et al., ‘Strelka2: fast and accurate calling of germline and somatic variants’, Nat Methods, vol. 15, no. 8, pp. 591–594, Aug. 2018. [CrossRef]
- Z.-K. Liu, Y.-K. Shang, Z.-C. Chen, and H. Bian, ‘A three-caller pipeline for variant analysis of cancer whole-exome sequencing data’. [CrossRef]
- K. Wang, M. Li, and H. Hakonarson, ‘ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data.’, Nucleic Acids Res, vol. 38, no. 16, p. e164, Sep. 2010. [CrossRef]
- E. Talevich, A. H. Shain, T. Botton, and B. C. Bastian, ‘CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing’, PLoS Comput Biol, vol. 12, no. 4, Apr. 2016. [CrossRef]
- C. H. Mermel, S. E. Schumacher, B. Hill, M. L. Meyerson, R. Beroukhim, and G. Getz, ‘GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers’, Genome Biol, vol. 12, no. 4, Apr. 2011. [CrossRef]
- M. V. Kuleshov et al., ‘Enrichr: a comprehensive gene set enrichment analysis web server 2016 update’, Nucleic Acids Res, vol. 44, no. 1, pp. W90–W97, Jul. 2016. [CrossRef]
- Gene Ontology Consortium et al., ‘The Gene Ontology knowledgebase in 2023.’, Genetics, vol. 224, no. 1, May 2023. [CrossRef]
- M. Kanehisa, M. Furumichi, Y. Sato, M. Kawashima, and M. Ishiguro-Watanabe, ‘KEGG for taxonomy-based analysis of pathways and genomes’, Nucleic Acids Res, vol. 51, no. D1, pp. D587–D592, Jan. 2023. [CrossRef]
- S. Wang et al., ‘Copy number signature analysis tool and its application in prostate cancer reveals distinct mutational processes and clinical outcomes’, PLoS Genet, vol. 17, no. 5, p. e1009557, May 2021. [CrossRef]
- T. Wu et al., ‘clusterProfiler 4.0: A universal enrichment tool for interpreting omics data’, The Innovation, vol. 2, no. 3, p. 100141, Aug. 2021. [CrossRef]
- A. Mayakonda, D.-C. Lin, Y. Assenov, C. Plass, and H. P. Koeffler, ‘Maftools: efficient and comprehensive analysis of somatic variants in cancer’, Genome Res, vol. 28, no. 11, pp. 1747–1756, Nov. 2018. [CrossRef]
- M. S. Lawrence et al., ‘Discovery and saturation analysis of cancer genes across 21 tumour types’, Nature, vol. 505, no. 7484, pp. 495–501, Jan. 2014. [CrossRef]
- I. Gkouveris, N. G. Nikitakis, J. Aseervatham, and K. U. E. Ogbureke, ‘The tumorigenic role of DSPP and its potential regulation of the unfolded protein response and ER stress in oral cancer cells.’, Int J Oncol, vol. 53, no. 4, pp. 1743–1751, Oct. 2018. [CrossRef]
- J. Aseervatham and K. U. E. Ogbureke, ‘Effects of DSPP and MMP20 Silencing on Adhesion, Metastasis, Angiogenesis, and Epithelial-Mesenchymal Transition Proteins in Oral Squamous Cell Carcinoma Cells.’, Int J Mol Sci, vol. 21, no. 13, Jul. 2020. [CrossRef]
- Q. Zhuang et al., ‘Prognostic and immunological roles of Fc fragment of IgG binding protein in colorectal cancer.’, Oncol Lett, vol. 22, no. 1, p. 526, Jul. 2021. [CrossRef]
- T. Yan et al., ‘FCGBP Is a Prognostic Biomarker and Associated With Immune Infiltration in Glioma.’, Front Oncol, vol. 11, p. 769033, 2021. [CrossRef]
- Q. Ding et al., ‘Non-coding RNA-related FCGBP downregulation in head and neck squamous cell carcinoma: a novel biomarker for predicting paclitaxel resistance and immunosuppressive microenvironment.’, Sci Rep, vol. 14, no. 1, p. 4426, Feb. 2024. [CrossRef]
- K. Yamada, M. Ono, N. D. Perkins, S. Rocha, and A. I. Lamond, ‘Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21.’, Mol Cell, vol. 49, no. 5, pp. 922–33, Mar. 2013. [CrossRef]
- X. Zhang, J. Wu, S. Luo, T. Lechler, and J. Y. Zhang, ‘FRA1 promotes squamous cell carcinoma growth and metastasis through distinct AKT and c-Jun dependent mechanisms.’, Oncotarget, vol. 7, no. 23, pp. 34371–83, Jun. 2016. [CrossRef]
- I. S. Fentiman, ‘Gamma-glutamyl transferase: risk and prognosis of cancer.’, Br J Cancer, vol. 106, no. 9, pp. 1467–8, Apr. 2012. [CrossRef]
- S. W. Hong et al., ‘Risk of gastrointestinal cancer in patients with an elevated level of gamma-glutamyltransferase: A nationwide population-based study.’, PLoS One, vol. 16, no. 2, p. e0245052, 2021. [CrossRef]
- J. Winter et al., ‘Prognostic role of gamma-glutamyl transferase in metastatic melanoma patients treated with immune checkpoint inhibitors’, Cancer Immunology, Immunotherapy, vol. 70, no. 4, pp. 1089–1099, Apr. 2021. [CrossRef]
- N. Allocati, M. Masulli, C. Di Ilio, and L. Federici, ‘Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases.’, Oncogenesis, vol. 7, no. 1, p. 8, Jan. 2018. [CrossRef]
- Z. Ye and H. Song, ‘Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis.’, Eur J Cancer, vol. 41, no. 7, pp. 980–9, May 2005. [CrossRef]
- K. J. Cullen, K. A. Newkirk, L. M. Schumaker, N. Aldosari, J. D. Rone, and B. R. Haddad, ‘Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors.’, Cancer Res, vol. 63, no. 23, pp. 8097–102, Dec. 2003.
- D. Mock, B. Whitestone, and J. Freeman, ‘Gamma-glutamyl transpeptidase activity in human oral squamous cell carcinoma’, Oral Surgery, Oral Medicine, Oral Pathology, vol. 64, no. 2, pp. 197–201, Aug. 1987. [CrossRef]
- M. D. M. Noblejas-López et al., ‘Expression of MHC class I, HLA-A and HLA-B identifies immune-activated breast tumors with favorable outcome.’, Oncoimmunology, vol. 8, no. 10, p. e1629780, 2019. [CrossRef]
- T. Michelakos et al., ‘Differential role of HLA-A and HLA-B, C expression levels as prognostic markers in colon and rectal cancer.’, J Immunother Cancer, vol. 10, no. 3, Mar. 2022. [CrossRef]
- G. Dyckhoff, C. Herold-Mende, S. Scherer, P. K. Plinkert, and R. Warta, ‘Human Leucocyte Antigens as Prognostic Markers in Head and Neck Squamous Cell Carcinoma.’, Cancers (Basel), vol. 14, no. 15, Aug. 2022. [CrossRef]
- M. Griffioen, I. J. M. Ouwerkerk, V. Harten, and P. I. Schrier, ‘HLA-B down-regulation in human melanoma is mediated by sequences located downstream of the transcription-initiation site’, Int J Cancer, vol. 80, no. 4, pp. 573–580, Feb. 1999. [CrossRef]
- H. Deng, Y. Chen, J. Wang, and R. An, ‘HLA-DRB1: A new potential prognostic factor and therapeutic target of cutaneous melanoma and an indicator of tumor microenvironment remodeling.’, PLoS One, vol. 17, no. 9, p. e0274897, 2022. [CrossRef]
- D. K. Lester et al., ‘Fucosylation of HLA-DRB1 regulates CD4+ T cell-mediated anti-melanoma immunity and enhances immunotherapy efficacy.’, Nat Cancer, vol. 4, no. 2, pp. 222–239, Feb. 2023. [CrossRef]
- M.-F. Senosain et al., ‘HLA-DR cancer cells expression correlates with T cell infiltration and is enriched in lung adenocarcinoma with indolent behavior.’, Sci Rep, vol. 11, no. 1, p. 14424, Jul. 2021. [CrossRef]
- K. Amrane et al., ‘HLA-DR expression in melanoma: from misleading therapeutic target to potential immunotherapy biomarker.’, Front Immunol, vol. 14, p. 1285895, 2023. [CrossRef]
- Y. Heng et al., ‘High Expression of Tumor HLA-DR Predicts Better Prognosis and Response to Anti-PD-1 Therapy in Laryngeal Squamous Cell Carcinoma.’, Transl Oncol, vol. 33, p. 101678, Jul. 2023. [CrossRef]
- Q. Ma, K. Geng, P. Xiao, and L. Zeng, ‘Identification and Prognostic Value Exploration of Radiotherapy Sensitivity-Associated Genes in Non-Small-Cell Lung Cancer.’, Biomed Res Int, vol. 2021, p. 5963868, 2021. [CrossRef]
- S. P. Verma and P. Das, ‘Novel splicing in IGFN1 intron 15 and role of stable G-quadruplex in the regulation of splicing in renal cell carcinoma.’, PLoS One, vol. 13, no. 10, p. e0205660, 2018. [CrossRef]
- M. Hoffman et al., ‘Intratumoral Genetic and Functional Heterogeneity in Pediatric Glioblastoma.’, Cancer Res, vol. 79, no. 9, pp. 2111–2123, May 2019. [CrossRef]
- I. Lee, S.-J. Lee, T. M. Kang, W. K. Kang, and C. Park, ‘Unconventional role of the inwardly rectifying potassium channel Kir2.2 as a constitutive activator of RelA in cancer.’, Cancer Res, vol. 73, no. 3, pp. 1056–62, Feb. 2013. [CrossRef]
- N. Khalilipour, A. Baranova, A. Jebelli, A. Heravi-Moussavi, S. Bruskin, and M. R. Abbaszadegan, ‘Familial Esophageal Squamous Cell Carcinoma with damaging rare/germline mutations in KCNJ12/KCNJ18 and GPRIN2 genes.’, Cancer Genet, vol. 221, pp. 46–52, Feb. 2018. [CrossRef]
- H. Choi et al., ‘Polymorphisms of Killer Ig-like Receptors and the Risk of Glioblastoma.’, J Clin Med, vol. 12, no. 14, Jul. 2023. [CrossRef]
- X. Liu et al., ‘Immune checkpoints HLA-E:CD94-NKG2A and HLA-C:KIR2DL1 complementarily shield circulating tumor cells from NK-mediated immune surveillance.’, Cell Discov, vol. 10, no. 1, p. 16, Feb. 2024. [CrossRef]
- R. Zou et al., ‘Elevated LILRB1 expression predicts poor prognosis and is associated with tumor immune infiltration in patients with glioma.’, BMC Cancer, vol. 23, no. 1, p. 403, May 2023. [CrossRef]
- T. Zeller et al., ‘Perspectives of targeting LILRB1 in innate and adaptive immune checkpoint therapy of cancer.’, Front Immunol, vol. 14, p. 1240275, 2023. [CrossRef]
- M. Xian et al., ‘Leukocyte immunoglobulin-like receptor B1 (LILRB1) protects human multiple myeloma cells from ferroptosis by maintaining cholesterol homeostasis.’, Nat Commun, vol. 15, no. 1, p. 5767, Jul. 2024. [CrossRef]
- T. Utsunomiya et al., ‘Expression of cancer-testis antigen (CTA) genes in intrahepatic cholangiocarcinoma.’, Ann Surg Oncol, vol. 11, no. 10, pp. 934–40, Oct. 2004. [CrossRef]
- J. Peng et al., ‘Expression of cancer/testis (CT) antigens in Chinese hepatocellular carcinoma and its correlation with clinical parameters.’, Cancer Lett, vol. 219, no. 2, pp. 223–32, Mar. 2005. [CrossRef]
- X. Jin et al., ‘MST1 inhibits the progression of breast cancer by regulating the Hippo signaling pathway and may serve as a prognostic biomarker.’, Mol Med Rep, vol. 23, no. 5, May 2021. [CrossRef]
- Y. Hayashi, M. Osanai, and G.-H. Lee, ‘NOTCH2 signaling confers immature morphology and aggressiveness in human hepatocellular carcinoma cells.’, Oncol Rep, vol. 34, no. 4, pp. 1650–8, Oct. 2015. [CrossRef]
- K. Dai, L. Huang, Y.-B. Huang, Z.-B. Chen, L.-H. Yang, and Y.-A. Jiang, ‘1810011o10 Rik Inhibits the Antitumor Effect of Intratumoral CD8+ T Cells through Suppression of Notch2 Pathway in a Murine Hepatocellular Carcinoma Model.’, Front Immunol, vol. 8, p. 320, 2017. [CrossRef]
- E. S. Demitrack and L. C. Samuelson, ‘Notch as a Driver of Gastric Epithelial Cell Proliferation.’, Cell Mol Gastroenterol Hepatol, vol. 3, no. 3, pp. 323–330, May 2017. [CrossRef]
- R.-H. Gan et al., ‘High expression of Notch2 drives tongue squamous cell carcinoma carcinogenesis.’, Exp Cell Res, vol. 399, no. 1, p. 112452, Feb. 2021. [CrossRef]
- D. Mikheil et al., ‘Notch Signaling Suppresses Melanoma Tumor Development in BRAF/Pten Mice.’, Cancers (Basel), vol. 15, no. 2, Jan. 2023. [CrossRef]
- N. P. Kren, I. S. Zagon, and P. J. McLaughlin, ‘Mutations in the opioid growth factor receptor in human cancers alter receptor function.’, Int J Mol Med, vol. 36, no. 1, pp. 289–93, Jul. 2015. [CrossRef]
- N. Qu, X. Wang, Y. Meng, and F. Shan, ‘Prospective oncotarget for gynecological cancer: Opioid growth factor (OGF) - opioid growth factor receptor (OGFr) axis.’, Int Immunopharmacol, vol. 75, p. 105723, Oct. 2019. [CrossRef]
- P. J. McLaughlin and I. S. Zagon, ‘Progression of squamous cell carcinoma of the head and neck is associated with down-regulation of the opioid growth factor receptor.’, Int J Oncol, vol. 28, no. 6, pp. 1577–83, Jun. 2006.
- D. Maßberg et al., ‘Monoterpene (-)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor.’, Arch Biochem Biophys, vol. 566, pp. 100–9, Jan. 2015. [CrossRef]
- L. Weber et al., ‘Olfactory Receptors as Biomarkers in Human Breast Carcinoma Tissues.’, Front Oncol, vol. 8, p. 33, 2018. [CrossRef]
- Y. Cao, S. Wu, Y. Gu, Y. H. Wong, Y. Shi, and L. Zhang, ‘Disulfidptosis-related PABPC3 promotes tumor progression and inhibits immune activity in osteosarcoma.’, J Gene Med, vol. 26, no. 1, p. e3641, Jan. 2024. [CrossRef]
- W. Yao et al., ‘PABPC1 promotes cell proliferation and metastasis in pancreatic adenocarcinoma by regulating COL12A1 expression.’, Immun Inflamm Dis, vol. 11, no. 7, p. e919, Jul. 2023. [CrossRef]
- P. Brock et al., ‘PDPR Gene Variants Predisposing to Papillary Thyroid Cancer.’, Thyroid, vol. 34, no. 5, pp. 575–582, May 2024. [CrossRef]
- L. Sui et al., ‘PRSS2 remodels the tumor microenvironment via repression of Tsp1 to stimulate tumor growth and progression.’, Nat Commun, vol. 13, no. 1, p. 7959, Dec. 2022. [CrossRef]
- Y. Chen, B. Wang, Z. Zhao, M. Li, and F. Wang, ‘PRSS2 overexpression relates to poor prognosis and promotes proliferation, migration and invasion in gastric cancer.’, Tissue Cell, vol. 79, p. 101949, Dec. 2022. [CrossRef]
- Q. Yan et al., ‘RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing.’, Oncogene, vol. 40, no. 15, pp. 2635–2650, Apr. 2021. [CrossRef]
- H. Tuersun, L. Liu, J. Zhang, R. Maimaitizunong, X. Tang, and H. Li, ‘m6A reading protein RBMX as a biomarker for prognosis and tumor progression in esophageal cancer.’, Transl Cancer Res, vol. 12, no. 9, pp. 2319–2335, Sep. 2023. [CrossRef]
- M. Xie, F. Wang, B. Chen, Z. Wu, C. Chen, and J. Xu, ‘Systematic pan-cancer analysis identifies SLC35C1 as an immunological and prognostic biomarker.’, Sci Rep, vol. 13, no. 1, p. 5331, Apr. 2023. [CrossRef]
- H. D. K. Ta et al., ‘Novel Insights into the Prognosis and Immunological Value of the SLC35A (Solute Carrier 35A) Family Genes in Human Breast Cancer.’, Biomedicines, vol. 9, no. 12, Nov. 2021. [CrossRef]
- A. Kuemmel et al., ‘Humoral immune responses of lung cancer patients against the Transmembrane Phosphatase with TEnsin homology (TPTE).’, Lung Cancer, vol. 90, no. 2, pp. 334–41, Nov. 2015. [CrossRef]
- N. Zainodini, M. Abolhasani, M. Mohsenzadegan, M. M. Farajollahi, and E. Rismani, ‘Overexpression of Transmembrane Phosphatase with Tensin homology (TPTE) in prostate cancer is clinically significant, suggesting its potential as a valuable biomarker.’, J Cancer Res Clin Oncol, vol. 150, no. 3, p. 165, Mar. 2024. [CrossRef]
- X. Chen, C. Chen, R. He, Y. Huang, and Y. Wang, ‘UBXN11 Predicts as a Poor Index for Colorectal Cancer and Contributes to the Tumorigenesis by Activating NF-κB Signaling.’, Dig Dis Sci, vol. 69, no. 6, pp. 2074–2082, Jun. 2024. [CrossRef]
- F. Xie et al., ‘USP8 promotes cancer progression and extracellular vesicle-mediated CD8+ T cell exhaustion by deubiquitinating the TGF-β receptor TβRII.’, EMBO J, vol. 41, no. 16, p. e108791, Aug. 2022. [CrossRef]
- W. Xiong et al., ‘USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy.’, Nat Commun, vol. 13, no. 1, p. 1700, Mar. 2022. [CrossRef]
- B. Duan, C. Wang, Z. Liu, and X. Yang, ‘USP8 is a Novel Therapeutic Target in Melanoma Through Regulating Receptor Tyrosine Kinase Levels.’, Cancer Manag Res, vol. 13, pp. 4181–4189, 2021. [CrossRef]
- J. Cui et al., ‘Comprehensive characterization of the genomic alterations in human gastric cancer.’, Int J Cancer, vol. 137, no. 1, pp. 86–95, Jul. 2015. [CrossRef]
- Y. Liang et al., ‘Discovery of Aberrant Alteration of Genome in Colorectal Cancer by Exome Sequencing.’, Am J Med Sci, vol. 358, no. 5, pp. 340–349, Nov. 2019. [CrossRef]
- P. Rentzsch, D. Witten, G. M. Cooper, J. Shendure, and M. Kircher, ‘CADD: predicting the deleteriousness of variants throughout the human genome.’, Nucleic Acids Res, vol. 47, no. D1, pp. D886–D894, Jan. 2019. [CrossRef]
- C. Féral, G. Guellaën, and A. Pawlak, ‘Human testis expresses a specific poly(A)-binding protein.’, Nucleic Acids Res, vol. 29, no. 9, pp. 1872–83, May 2001. [CrossRef]
- M. Yan, C. Zhao, N. Wei, X. Wu, J. Cui, and Y. Xing, ‘High Expression of Ubiquitin-Specific Protease 8 (USP8) Is Associated with Poor Prognosis in Patients with Cervical Squamous Cell Carcinoma.’, Med Sci Monit, vol. 24, pp. 4934–4943, Jul. 2018. [CrossRef]
- B. Sha et al., ‘USP8 inhibitor-induced DNA damage activates cell cycle arrest, apoptosis, and autophagy in esophageal squamous cell carcinoma.’, Cell Biol Toxicol, vol. 39, no. 5, pp. 2011–2032, Oct. 2023. [CrossRef]
- M. Reincke et al., ‘Mutations in the deubiquitinase gene USP8 cause Cushing’s disease’, Nat Genet, vol. 47, no. 1, pp. 31–38, Jan. 2015. [CrossRef]
- M.-X. Xiu and Y.-M. Liu, ‘The role of oncogenic Notch2 signaling in cancer: a novel therapeutic target.’, Am J Cancer Res, vol. 9, no. 5, pp. 837–854, 2019.
- C.-H. Lu et al., ‘USP17 mediates macrophage-promoted inflammation and stemness in lung cancer cells by regulating TRAF2/TRAF3 complex formation’, Oncogene, vol. 37, no. 49, pp. 6327–6340, Dec. 2018. [CrossRef]
- C. Ducker and P. E. Shaw, ‘USP17-mediated de-ubiquitination and cancer: Clients cluster around the cell cycle’, Int J Biochem Cell Biol, vol. 130, p. 105886, Jan. 2021. [CrossRef]
- B. Wang, D. Chen, and H. Hua, ‘TBC1D3 family is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma’, Mol Ther Oncolytics, vol. 22, pp. 528–538, Sep. 2021. [CrossRef]
- B. Wang et al., ‘Up-regulation of OLR1 expression by TBC1D3 through activation of TNFα/NF-κB pathway promotes the migration of human breast cancer cells.’, Cancer Lett, vol. 408, pp. 60–70, Nov. 2017. [CrossRef]
- J. Tian et al., ‘TBC1D2 Promotes Ovarian Cancer Metastasis via Inducing E-Cadherin Degradation’, Front Oncol, vol. 12, Apr. 2022. [CrossRef]
- S. K. Ghosh, T. S. McCormick, and A. Weinberg, ‘Human Beta Defensins and Cancer: Contradictions and Common Ground’, Front Oncol, vol. 9, May 2019. [CrossRef]
- M. Griffith et al., ‘DGIdb: mining the druggable genome.’, Nat Methods, vol. 10, no. 12, pp. 1209–10, Dec. 2013. [CrossRef]
- M. Cannon et al., ‘DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms.’, Nucleic Acids Res, vol. 52, no. D1, pp. D1227–D1235, Jan. 2024. [CrossRef]
- R. Dienstmann, J. Rodon, J. Barretina, and J. Tabernero, ‘Genomic medicine frontier in human solid tumors: prospects and challenges.’, J Clin Oncol, vol. 31, no. 15, pp. 1874–84, May 2013. [CrossRef]
- D. Hanahan and R. A. Weinberg, ‘Hallmarks of cancer: the next generation.’, Cell, vol. 144, no. 5, pp. 646–74, Mar. 2011. [CrossRef]
- C. M. Melo, T. Vidotto, L. P. Chaves, W. Lautert-Dutra, R. B. Dos Reis, and J. A. Squire, ‘The Role of Somatic Mutations on the Immune Response of the Tumor Microenvironment in Prostate Cancer.’, Int J Mol Sci, vol. 22, no. 17, Sep. 2021. [CrossRef]
- G. P. Dunn, L. J. Old, and R. D. Schreiber, ‘The immunobiology of cancer immunosurveillance and immunoediting.’, Immunity, vol. 21, no. 2, pp. 137–48, Aug. 2004. [CrossRef]
- X. Gong and R. Karchin, ‘Pan-Cancer HLA Gene-Mediated Tumor Immunogenicity and Immune Evasion.’, Mol Cancer Res, vol. 20, no. 8, pp. 1272–1283, Aug. 2022. [CrossRef]
- C. J. Forlenza et al., ‘KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma.’, J Clin Oncol, vol. 34, no. 21, pp. 2443–51, Jul. 2016. [CrossRef]
- V. Varbanova, E. Naumova, and A. Mihaylova, ‘Killer-cell immunoglobulin-like receptor genes and ligands and their role in hematologic malignancies.’, Cancer Immunol Immunother, vol. 65, no. 4, pp. 427–40, Apr. 2016. [CrossRef]
- W. Wang et al., ‘Killer immunoglobulin-like receptor (KIR) and KIR–ligand genotype do not correlate with clinical outcome of renal cell carcinoma patients receiving high-dose IL2’, Cancer Immunology, Immunotherapy, vol. 65, no. 12, pp. 1523–1532, Dec. 2016. [CrossRef]
- I. Milo et al., ‘The immune system profoundly restricts intratumor genetic heterogeneity.’, Sci Immunol, vol. 3, no. 29, Nov. 2018. [CrossRef]
- M. Reincke et al., ‘Mutations in the deubiquitinase gene USP8 cause Cushing’s disease.’, Nat Genet, vol. 47, no. 1, pp. 31–8, Jan. 2015. [CrossRef]
- M. Gounder et al., ‘Nirogacestat, a γ-Secretase Inhibitor for Desmoid Tumors.’, N Engl J Med, vol. 388, no. 10, pp. 898–912, Mar. 2023. [CrossRef]
| Sample | Age | Sex | Symptoms | Previous Treatment | Extent of Resection | Recurrence\Progression (Months) |
|---|---|---|---|---|---|---|
| H766 | 41 | F | Syncope Headache Vision |
None | STR | 7 |
| 4151 | 57 | F | Neuralgia | None | GTR | None |
| 1777 | 30 | M | Epilepsy | None | GTR | None |
| 2939 | 34 | F | Vision Adrenal insufficiency |
None | STR | None |
| 810 | 27 | F | Headaches Emesis Hydrocephalus Ataxia Nerve palsy Cognitive |
None | nGTR | None |
| 4316 | 32 | F | Headache | 2 | GTR | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




