1. Introduction
Nowadays, Pharmacology plays a primary role, studying drugs and the interactions that take place between them and living organisms. Several online servers are available, in which it is possible to predict the different chemical-physical and pharmacological characteristics of drugs with excellent reliability, with the aim of lowering production costs and focusing in a targeted way in therapy research. One of the methods currently used by the scientific community to support medical and drug therapy is the use of various predictive software of biological properties and Molecular Docking. It predicts the preferred orientation of a molecule towards a second one when these bind together to form a stable complex. The latter, through well-known various score functions, for instance Autodock Vina and Autodock 4 , allows to accurately predict and estimate of the Binding Energy (kcal mol -1) and the constant inhibition Ki of small molecules complexed in the active site of protein targets, often over-expressed in different neoplastic processes. In fact, the Virtual Screening and Molecular Docking methods are often effective methods that allow time to obtain preliminary results, before carrying out the in Vitro or in Vivo tests in the Laboratories. Our work aims to investigate Dual specificity tyrosine-phosphorylation-regulated kinase 2 which it is an enzyme that in humans is encoded by the
DYRK2 gene [
1]. DYRK2 belongs to a family of protein kinases, a positive regulator of the 26S proteasome, whose members are presumed to be involved in cellular growth and development [
2,
3]. From scientific studies, this kinase is involved as a key protein in cancer development [
4,
5]. Some researchers report that this protein is involved in p53 through the phosphorylation of Ser46 in the apoptotic response to DNA damage [
6]. This short communication lead to focus on possible role of Hypericin by Docking and MD Analysis
. From several studies reported in Literature, Hypericin is an anthraquinone derivative that is naturally found in the yellow flower of Hypericum perforatum (St. John’s wort) with antidepressant, potential antiviral, antineoplastic and immunostimulating activities [
7,
8,
9]. In addition it useful in case of panic attacks, anxiety, stress, nervousness and depression [
7,
8,
9].
This natural compound has a chemical structure similar to Nogalaviketone, Myricetin, Quercetin, Tricetin, Gossypol, Pholoxine B and Rose Bengal. In fact, we have performed, through Swiss similarly prediction (
http://swisssimilarity.ch/) to confirm what has been said previously.
Results and Discussion
The goal of this short computational analysis evaluated by Autodock Vina [
12] with PyRx program (
https://pyrx.sourceforge.io) 6and Autodock 4 [
12] with Autodock Tools (
http//:AutoDock.scripps.edu) was to find out one natural molecule that could have the effective biological role in the key protein of Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2). This protein, according to several studies reported in the literature, is involved in different biological processes mainly in the cancer field [
4,
5] . The first step, before to perform docking analysis we focused on Ligand Binding site of this protein and we investigate the crystal ligand, through Validation Method in order to obtain good estimation values in terms of Binding Energy onto Active site of DYRK2 protein. Regarding
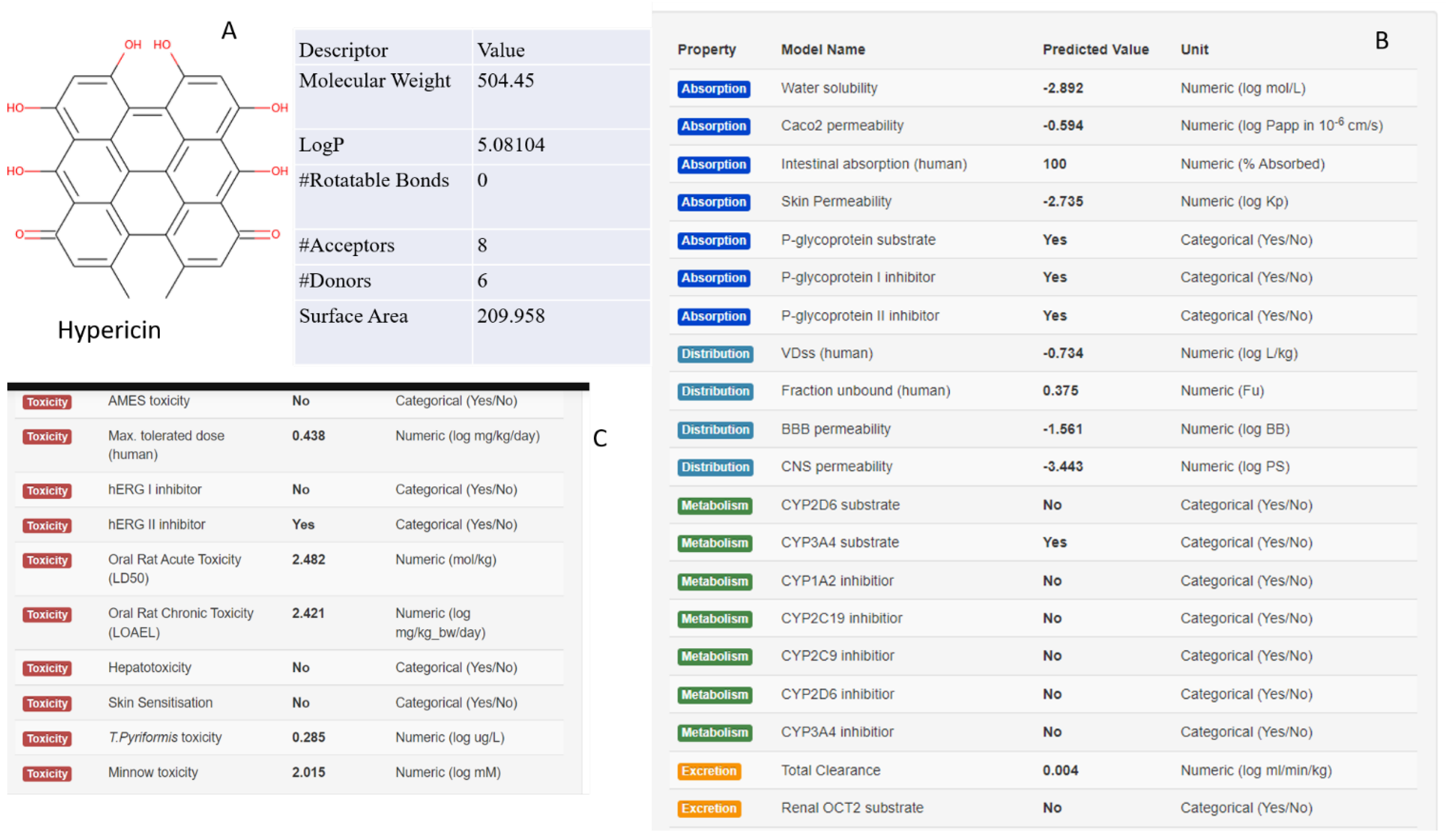
Figure 2 we focused on Drug-likeness evaluation, according to Lipinski
’s rule states Toxicity prediction and ADMET ( Absorption, Distribution, Metabolism, Excretion and Toxicity) parameters of natural compound Hypericin, evaluated by PkCSM Server [
13]. while in
Table 3 we report the chemical-physical characteristics of proposed best ZINC 95480894, according to Phamit Server 14 and Zinc 15 Database (
Http://zinc15.docking.org). In addition, we highlights which it could be the biological function of Hypericin, a carbopolycyclic compound. It has a role as an antidepressant. Moreover, it derives from a hydride of a bisanthene, through PASS Online Server (
http://www.way2drug.com) [
15]. This analysis aims to predict biological activity profile , according to over screened 4000 kinds of biological activity. In short it estimates the probability that your compound is belonging to the particular classes of active compounds In our case PASS predict that the Hypericin has consisted of the possible probability values for CYP2C12 ( cytochrome P450, family 2, subfamily c, polypeptide 12 ) substrate with Pa value of about 0.898 % and Pi value of 0.012%. Regarding the possible adverse and toxic effects, the same prediction based on clinical manifestations, Hypericin could be negative effect on Reproductive Dysfunction with Pa value of ca 0.874 and Pi value 0.011 % score value, estimated by PASS analysis. Moreover, we have studied other predicted available methods to confirm the its biological activity. Indeed, we have investigate Swiss Target Prediction (
http//:SwissTargetPrediction.ch/)[
16] , developed by Swiss Istitute of Bioinformatics. From this computed analysis Hyperin could be target on with a probable score of ca 20% of Oxdorectase, ca 20% of Membrane receptor of Cytochrome 450, of ca 6,7%, Ligand-gated ion channel of ca 6,7%, of ca Surface Antigen of 6,7 % , of ca 6,7 % Protease , Primary active transporter of 13.3%, Kinase of 13.3 %. In fact, as we see in figure 3 we evaluate the quasi-perfect overlap ligand structures between Crystal ligand IRB and re-docked ligand with lower Reference RMSD, About 1 A° angstrom. The following step we have performed by Virtual screening method several repeatability tests by Docking analysis, onto Active site pocked of DYRK2 protein. Indeed, we report in
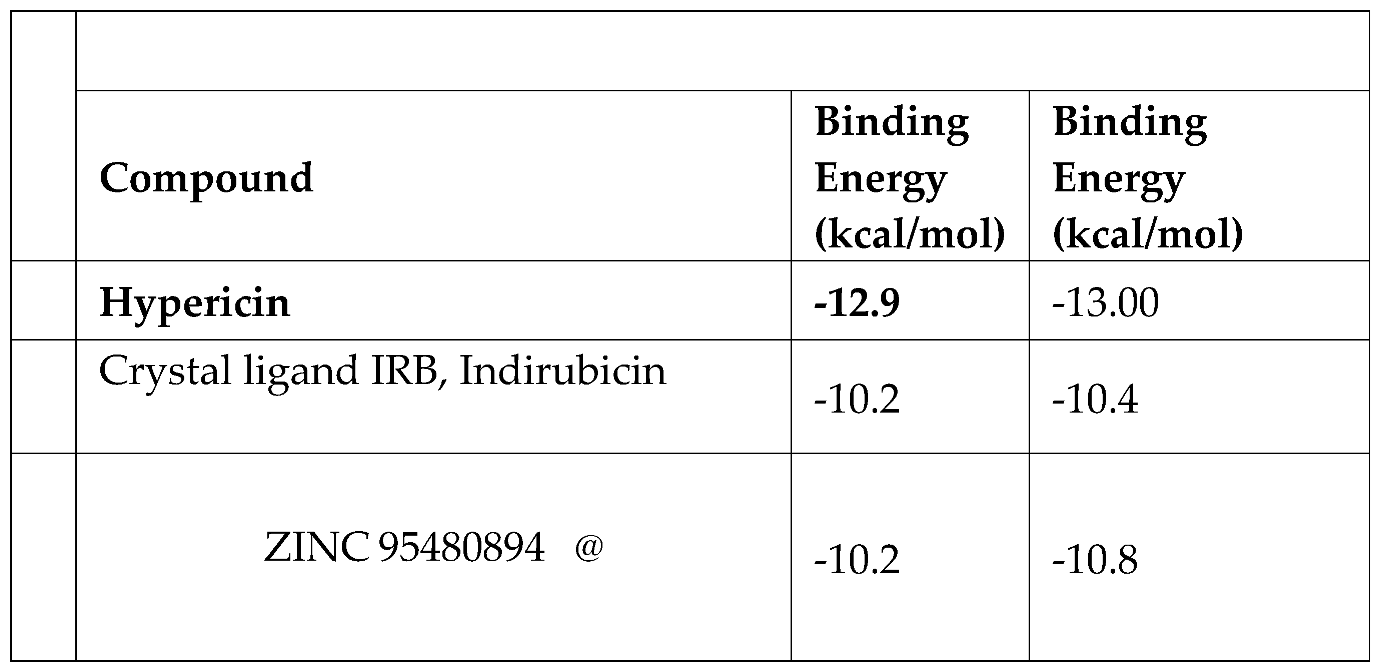
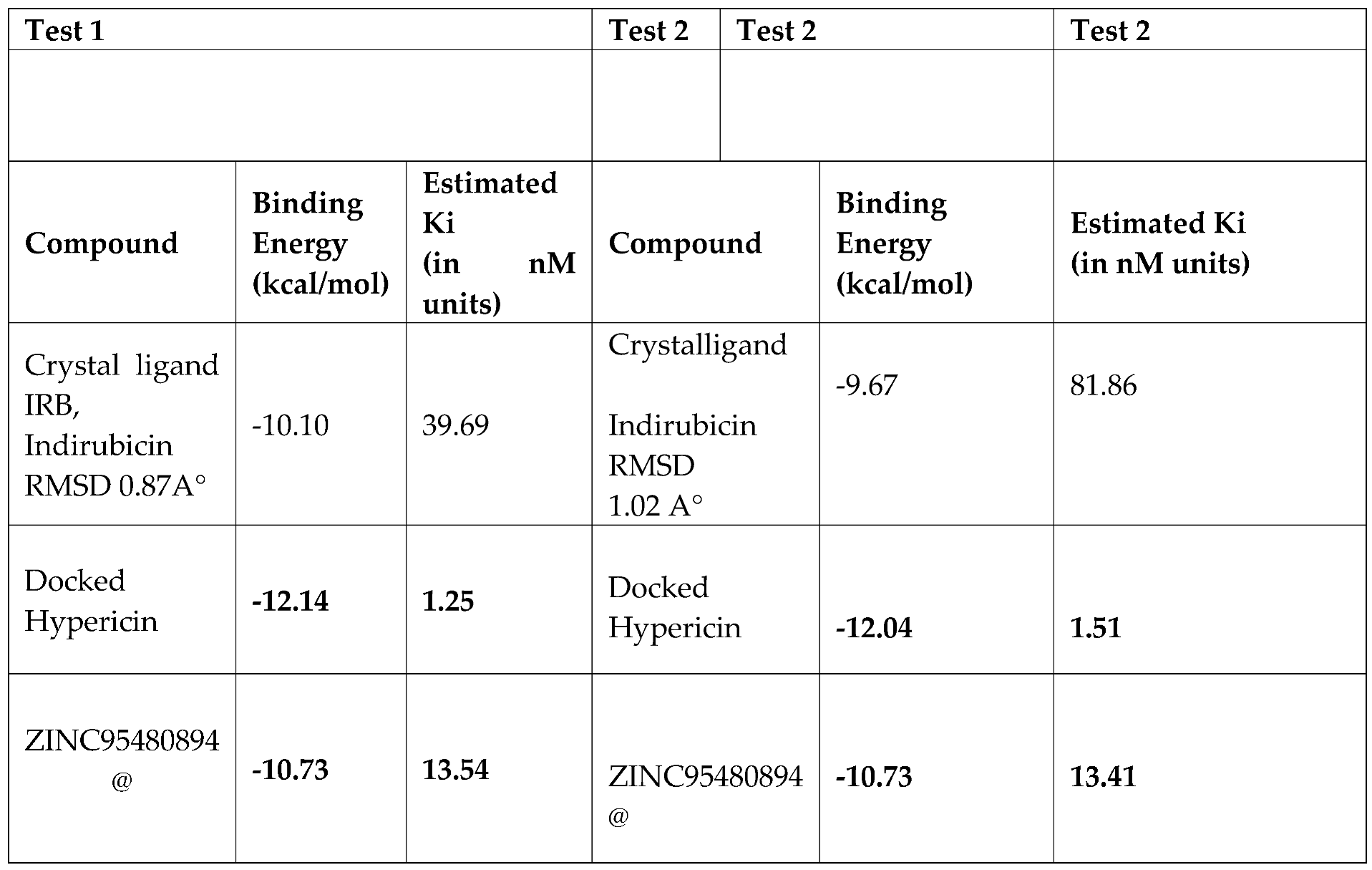
Table 1-
2 the best results in terms of binding energies poses of natural compound called Hypericn, an anthraquinone derivative product found in Hypericum olympicum and one of the best similar compound, analysed by Pharmit Server Engine and Zinc Database, complexed in Ligand Binding Site pocket of Tyrosine phosphorylation regulated kinase. Indeed, they have shown an excellent Binding energy value, of ca -12/-13 kcal/mol value and ca -10/-11 respectively in DYRK2 protein, estimated both by AutoDock Vina [
6] and by AutoDock 4[
7] , respectively ( See below
Table 1-
2). In
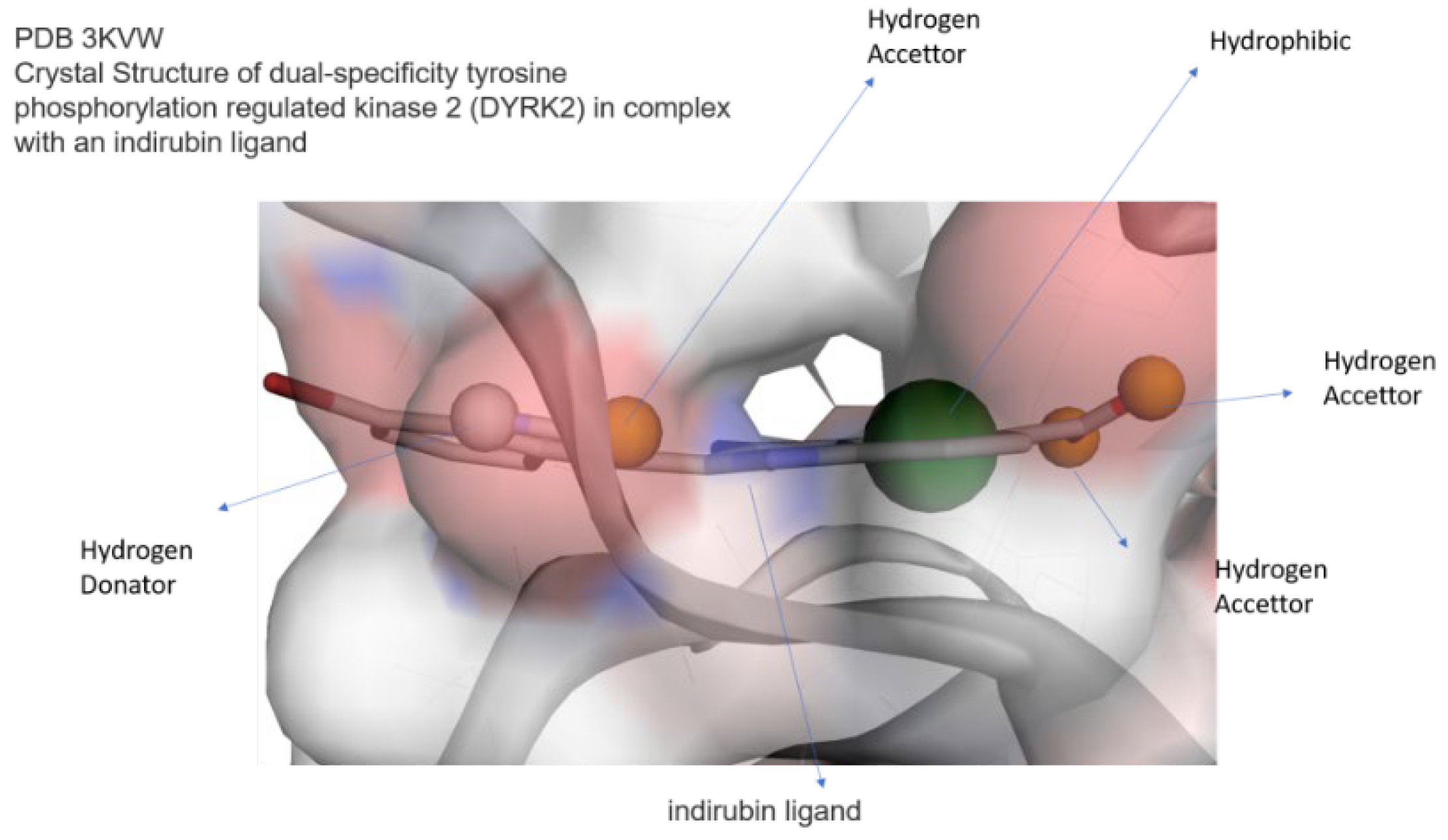
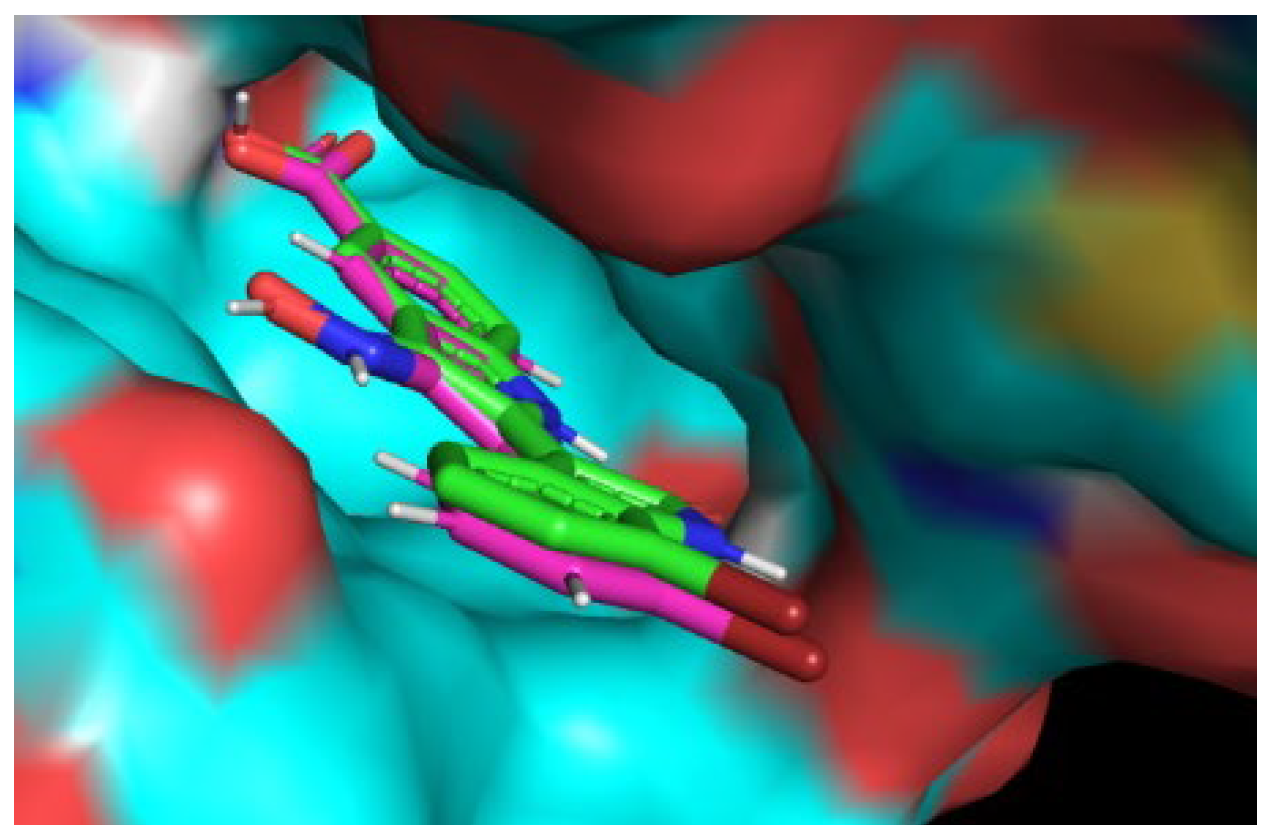
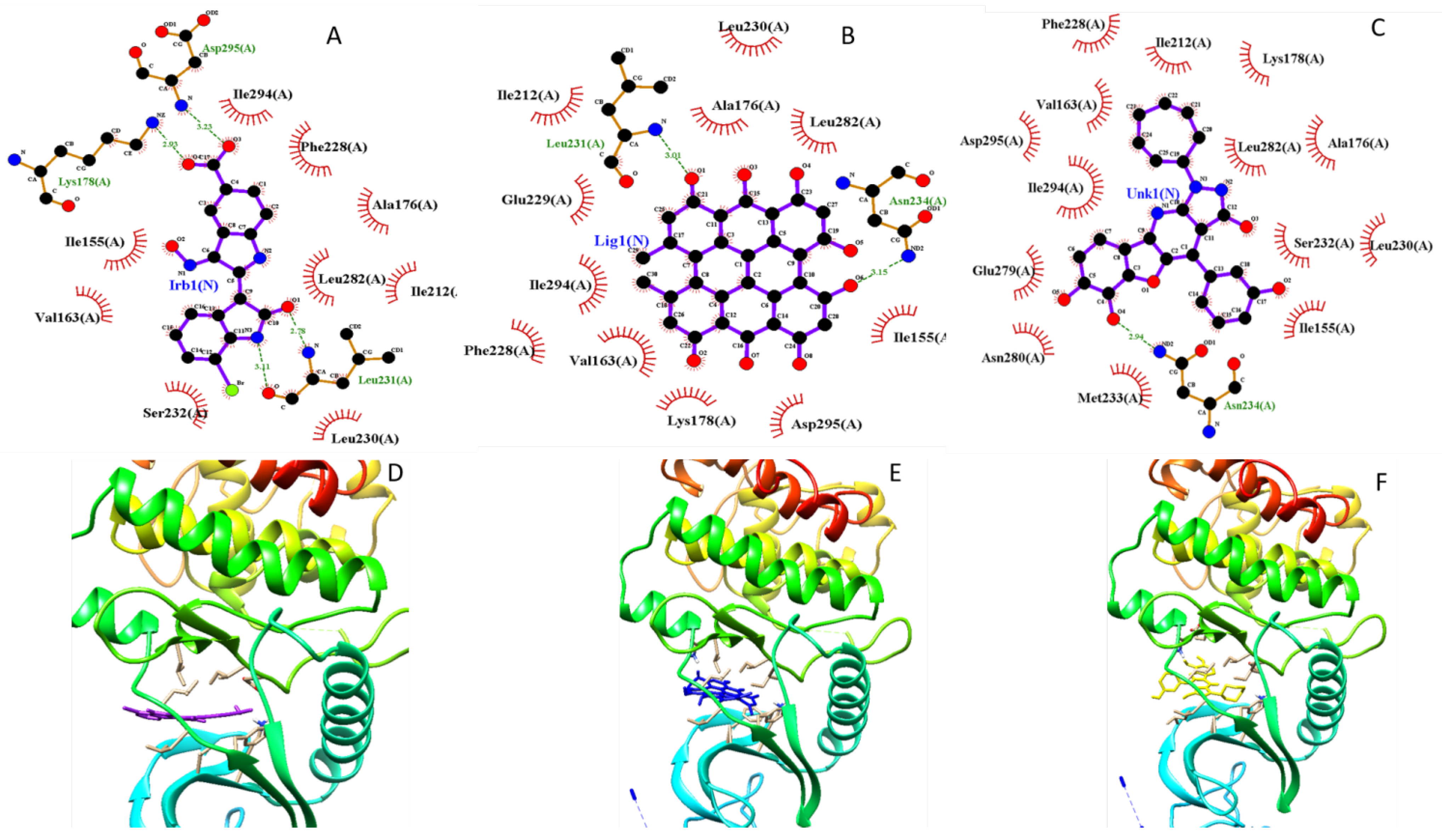
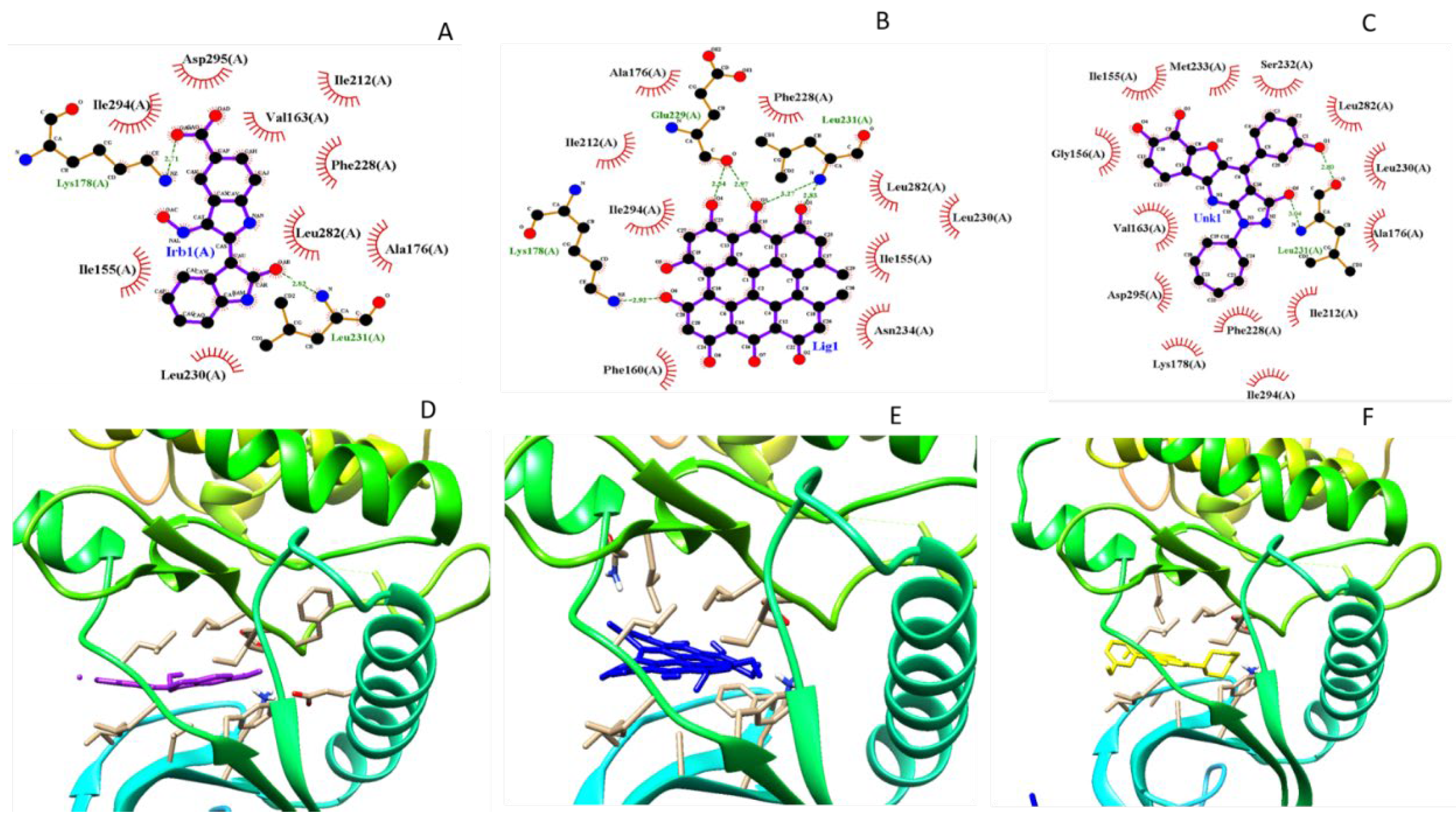
Figure 4 we report the comparison of 2D a schematic diagram of the interactions, characterized by LIGPLOT software between protein DYRK2 between crystal ligand IRB ( Indirubicin), docked Hypericin and docked Zinc 95480894 residues interactions and also their 3D structure in Ligand Binding Site pocket, evaluated by Autodock Vina with Pyrx program. In figure 5 we show the same investigation, estimated by Autodock 4 with Autodock Tool.
In
Figure 6 focused on Normal mode analysis [
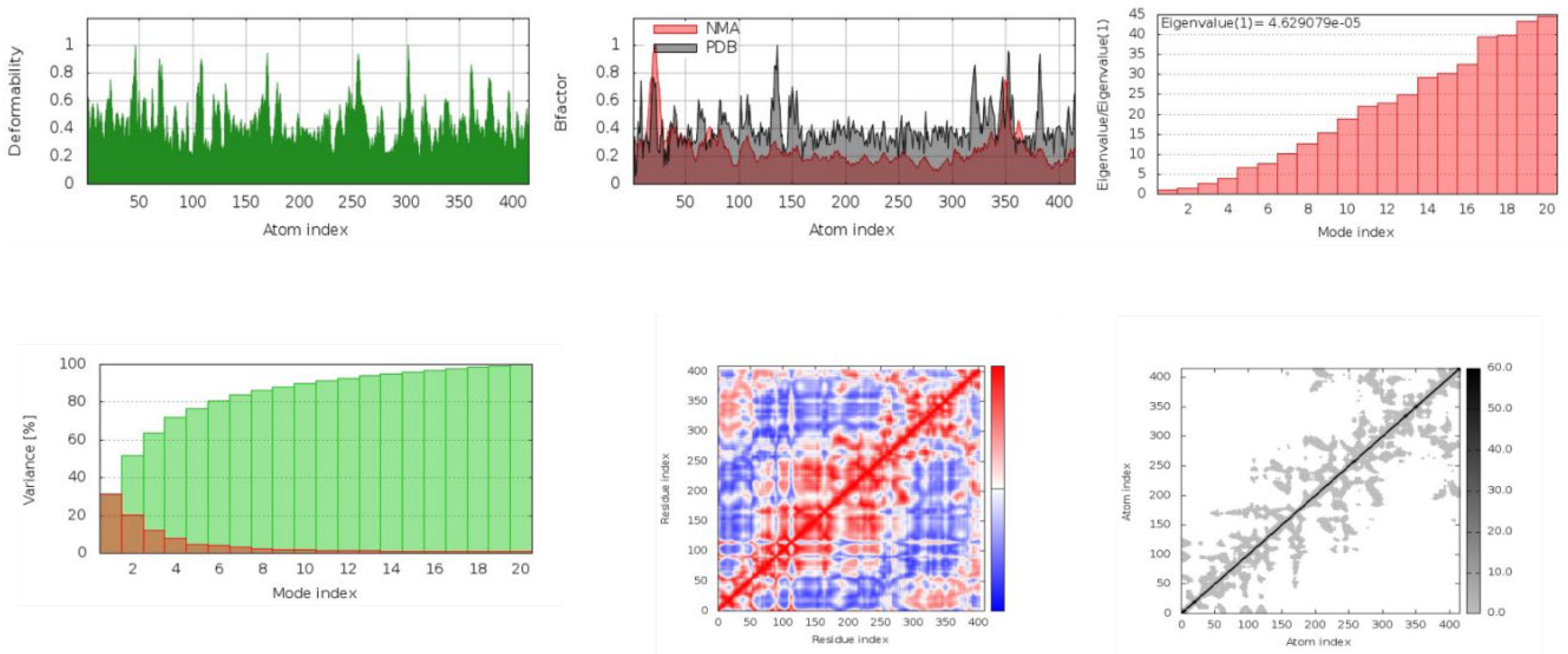
17], “NMA” of DYRK2-docked Hypericin complex, evaluated by versitile iMod Server ( iMod.chaconlab.org) to validate docking results. Generally speaking, NMA is a simulation technique used to sudy vibrational motion of a harmonic oscillating system in the vicinity of its equilibrium. In our case several parameters have been estimated of this protein-ligand complex, for instance Deformability, B-Factor, Eigenvalue, Valence %, Residue and Atom index.
Table 1.
Comparison repeability tests of Docking analysis by Autodock Vina of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with Hypericin .
Table 1.
Comparison repeability tests of Docking analysis by Autodock Vina of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with Hypericin .
Table 2.
Docking repeability analysis estimated by Autodock 4 with Autodock Tool of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with Hypericin .
Table 2.
Docking repeability analysis estimated by Autodock 4 with Autodock Tool of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with Hypericin .
Figure 2.
A) Drug-likeness B) ADMET ( Absorption, Distribution, Metabolism, Excretion and Toxicity) parameters and C) Toxicity investigation of natural compound of Hypericin, estimatimated by PkCSM server .
Figure 2.
A) Drug-likeness B) ADMET ( Absorption, Distribution, Metabolism, Excretion and Toxicity) parameters and C) Toxicity investigation of natural compound of Hypericin, estimatimated by PkCSM server .
Table 3.
Chemical and physical characteristics of ZINC95480894 best proposed similar compound of Crystal Ligand IRB, evaluated by Zinc Database (
https://zinc15.docking.org/) and Pharmit Server Engine.
Table 3.
Chemical and physical characteristics of ZINC95480894 best proposed similar compound of Crystal Ligand IRB, evaluated by Zinc Database (
https://zinc15.docking.org/) and Pharmit Server Engine.
Figure 3.
Comparison and overlapping between Crystal Ligand B IRB and re-docked Ligand in Ligand Binding site Pocked of DYRK2protein. The figure was reproduced by the Pymol program.
Figure 3.
Comparison and overlapping between Crystal Ligand B IRB and re-docked Ligand in Ligand Binding site Pocked of DYRK2protein. The figure was reproduced by the Pymol program.
Figure 4.
Comparison 2D residues interactions and 3D structure of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 chain A (DYRK2), in complex with best pose proposed docked compound a) Crystal Ligand, b) docked Hypericin and c) ZINC95480894 respectively Autodock Vina with Autodock tool. This figure was reproduced by LIGPLOT program.
Figure 4.
Comparison 2D residues interactions and 3D structure of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 chain A (DYRK2), in complex with best pose proposed docked compound a) Crystal Ligand, b) docked Hypericin and c) ZINC95480894 respectively Autodock Vina with Autodock tool. This figure was reproduced by LIGPLOT program.
Figure 5.
Comparison 2D residues interactions and 3D structure of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 chain A (DYRK2), in complex with best pose proposed docked compound a) Crystal Ligand, b) docked Hypericin and ZINC95480894 respectively Autodock 4 with Autodock tool. This figure was reproduced by LIGPLOT program.
Figure 5.
Comparison 2D residues interactions and 3D structure of Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 chain A (DYRK2), in complex with best pose proposed docked compound a) Crystal Ligand, b) docked Hypericin and ZINC95480894 respectively Autodock 4 with Autodock tool. This figure was reproduced by LIGPLOT program.
Figure 6.
Several predicted Parameters ( Deformability, B-Factor, Eigenvalue, Variance %, Residue index and atom-atom index) plotted of Crystal. chain A (DYRK2), in complex with docked Hypericin. This figure was reproduced by iMODS server.
Figure 6.
Several predicted Parameters ( Deformability, B-Factor, Eigenvalue, Variance %, Residue index and atom-atom index) plotted of Crystal. chain A (DYRK2), in complex with docked Hypericin. This figure was reproduced by iMODS server.
Figure 6 shows the prediction of the deformability parameter as a function of the atom index. The main-chain deformability is a measure of the capability of a given molecule to deform at each of its residues. The location of the chain ‘hinges’ can be derived from high deformability regions. The B-factor, also called the temperature factor, is one of the many parameters used to fit a potential structure to the electron density map obtained the B-factor, also called the temperature factor, from the corresponding PDB field and the calculated from NMA is obtained by multiplying the NMA mobility by (8pi^2). The eigenvalue associated with each normal mode representing the motion stiffness. Its value is directly related to the energy required to deform the structure. The lower the eigenvalue, the easier the deformation. In our case eigenvalue (1) is 4.629079 e-05. The variance associated with each normal mode is inversely related to the eigenvalue. Coloured bars show the individual (red) and cumulative (green) variances. Moreover the Reside index evaluates Covariance matrix indicates coupling between pairs of residues, i.e. whether they experience correlated (red), uncorrelated (white), or anti-correlated (blue) motions. The elastic network model defines which pairs of atoms are connected by springs. Each dot in the graph of atom-atom index represents one spring between the corresponding pair of atoms. Dots are coloured according to their stiffness, the darker greys indicate stiffer springs and vice versa.