Submitted:
29 August 2024
Posted:
30 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
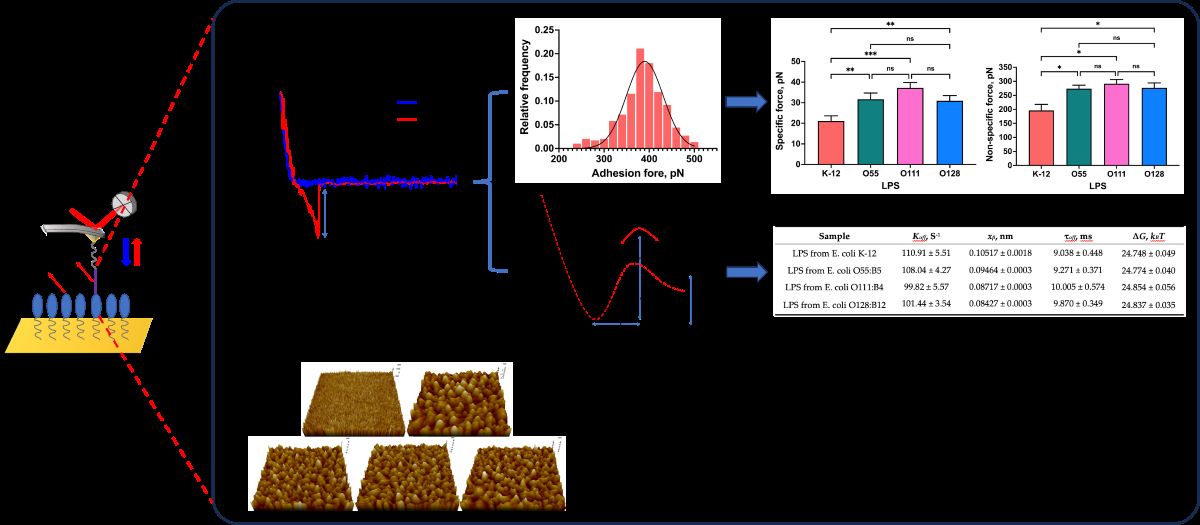
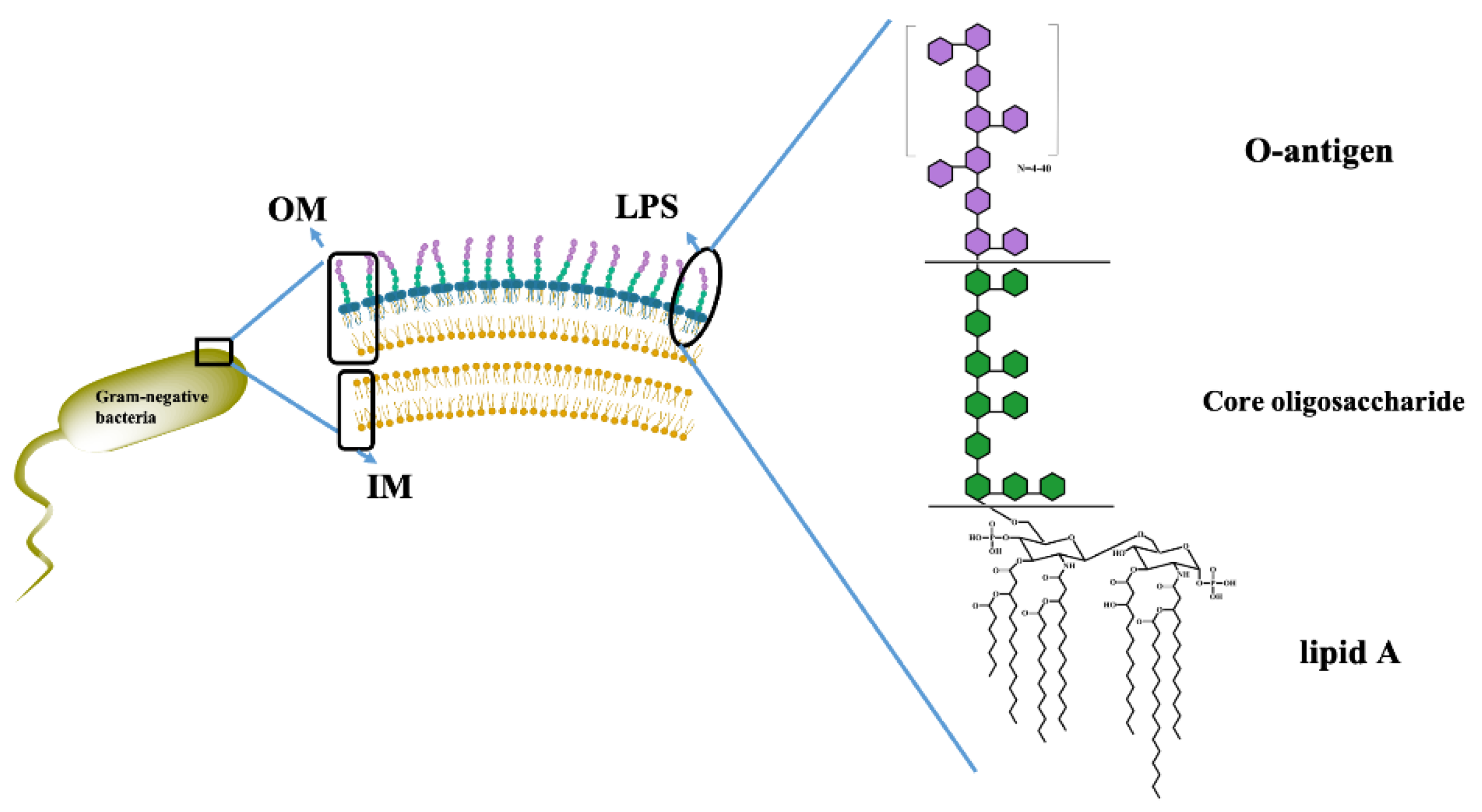
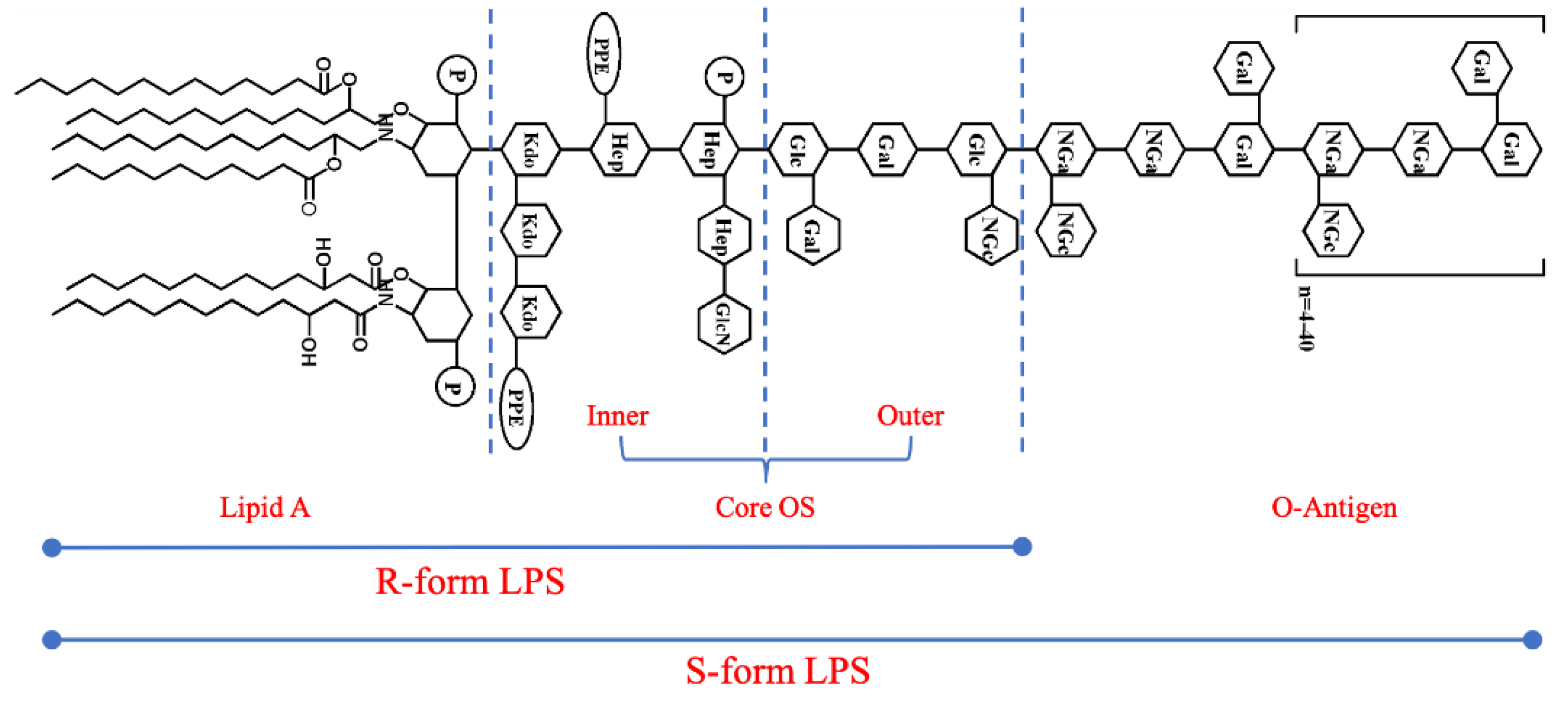
2.1. Morphological Characterization of LPSs from Different E. coli
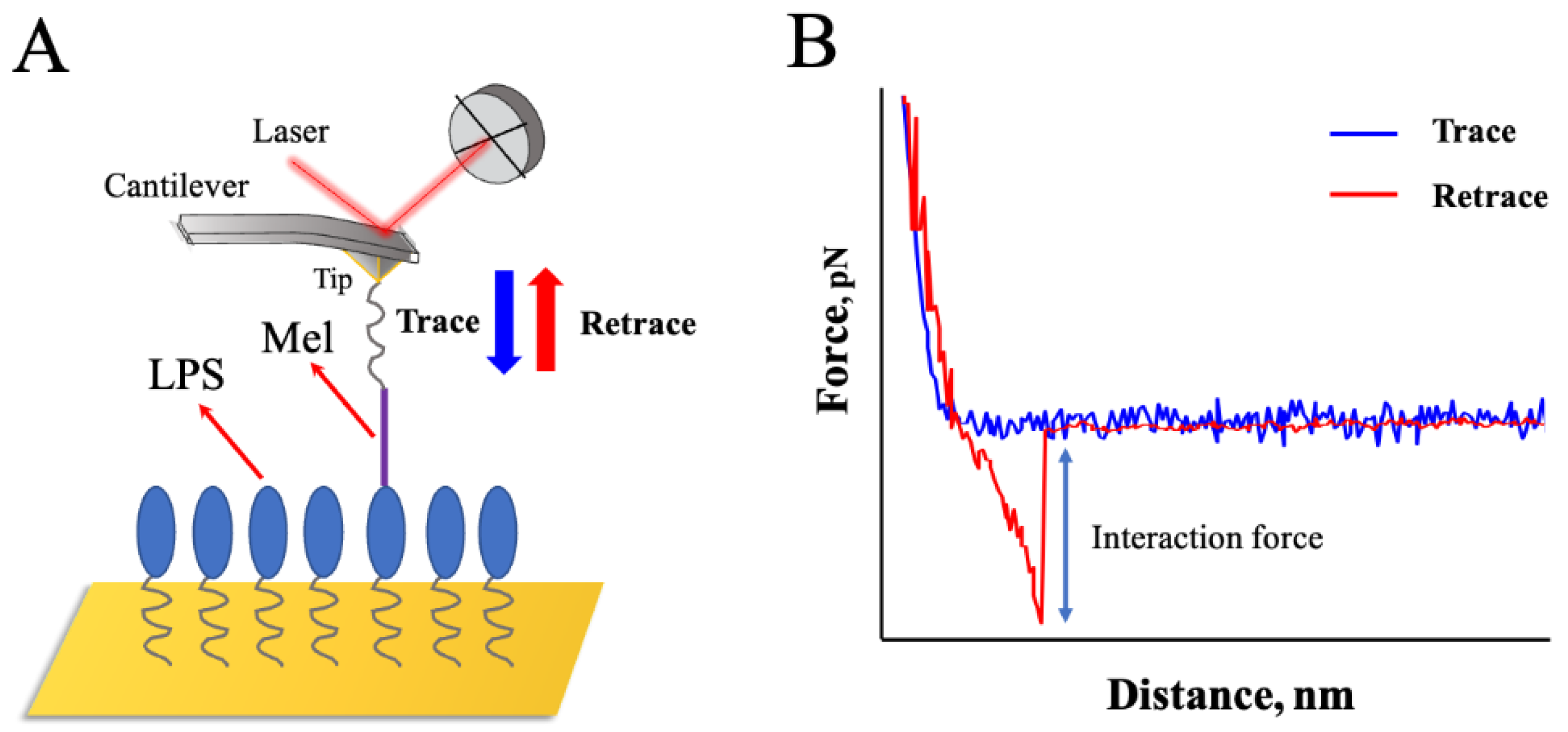
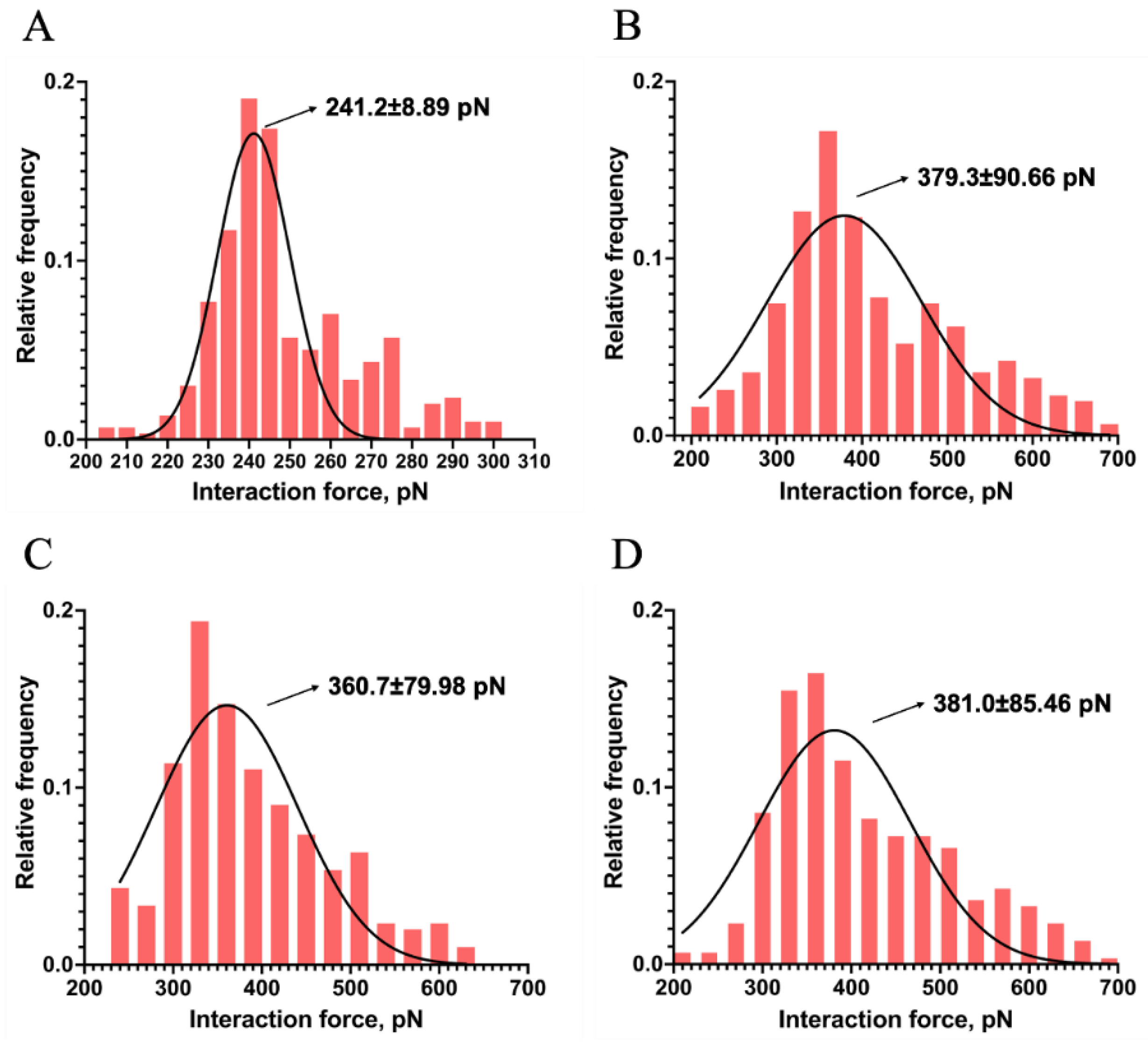
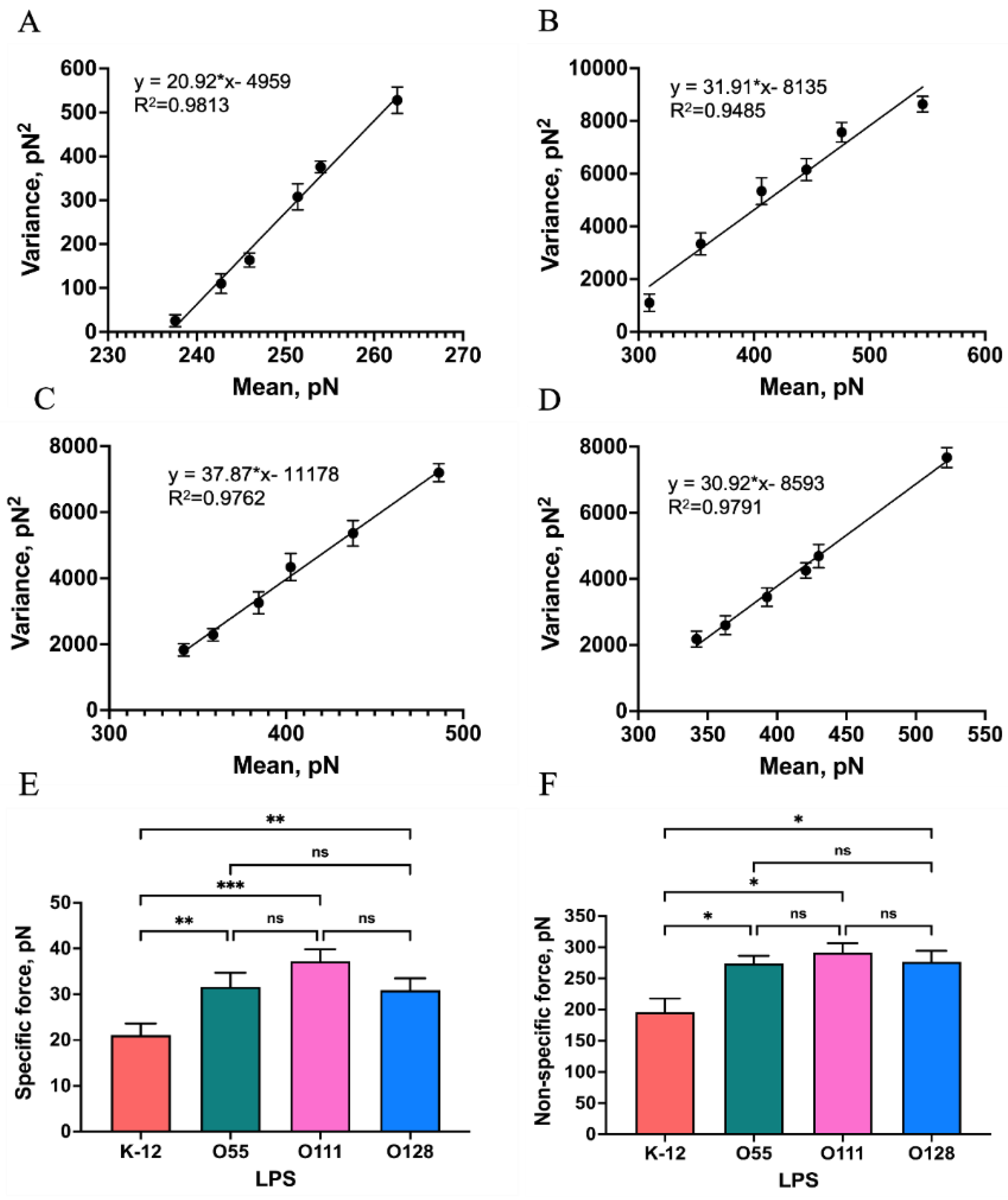
2.2. Probing the Interaction Forces between Mel and LPSs
2.3. Investigation of Single−Molecule Interaction Between Mel and LPSs
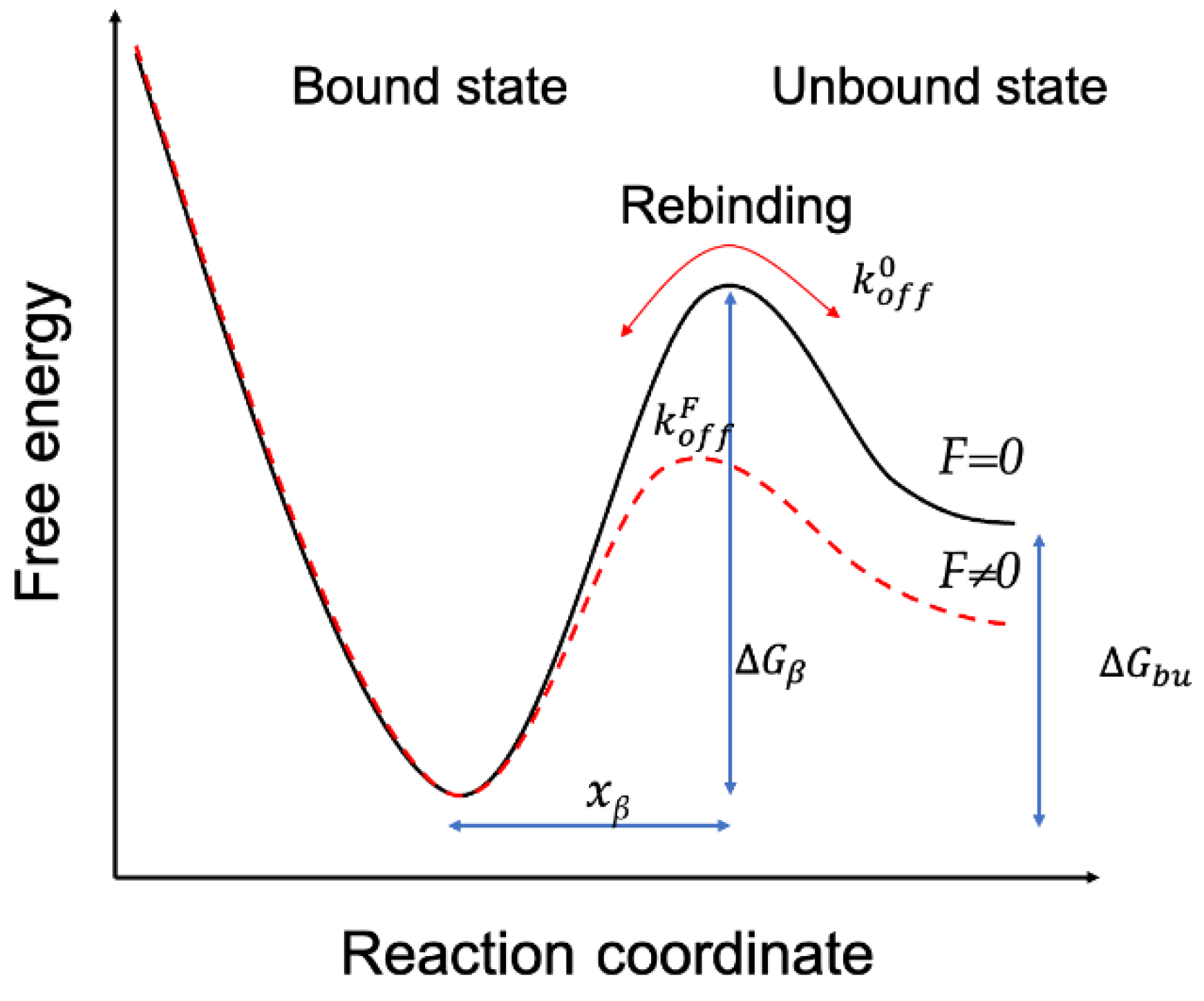
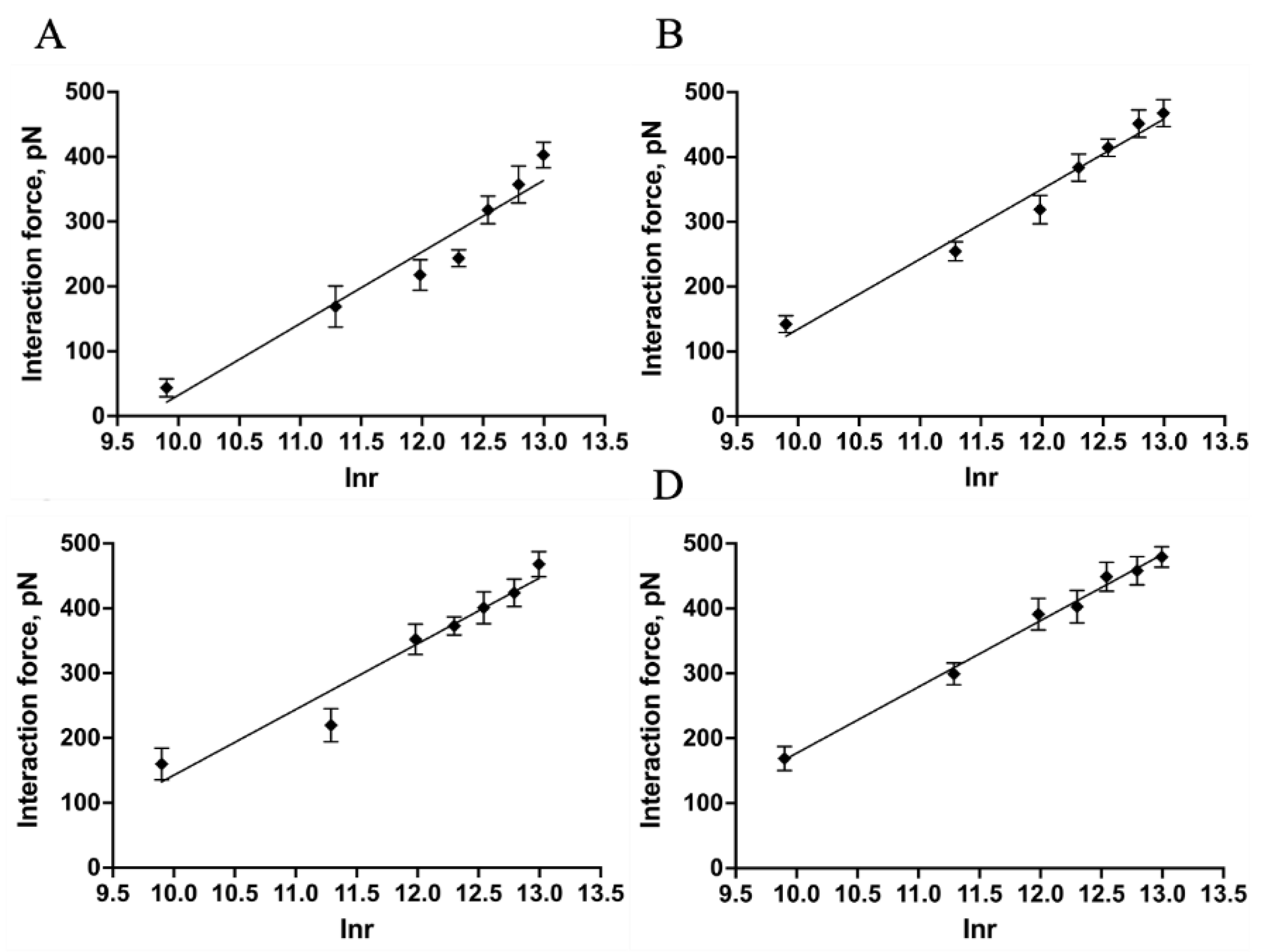
2.4. Quantitatively Probing the Kinetic Characteristics of the Mel/LPSs Dissociation
3. Materials and Methods
3.1. Materials and Reagents
3.2. Immobilization of Mel on Gold−Coated AFM Probes
3.3. Immobilization of LPSs on Mica
3.4. Interaction Force Measurements between Mel and LPSs
3.5. Dynamic Force Spectrum Measurements between Mel and LPSs
3.6. Morphological Characterization of LPSs
3.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doi, Y.; Bonomo, R.A.; Hooper, D.C.; Kaye, K.S.; Johnson, J.R.; Clancy, C.J.; Thaden, J.T.; Stryjewski, M.E.; van Duin, D. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the antibacterial resistance leadership group. Clin Infect Dis 2017, 64, S30–S35. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical properties of the bacterial outer membrane. Nat Rev Microbiol 2022, 20, 236–248. [Google Scholar] [CrossRef]
- Impey, R.E.; Hawkins, D.A.; Sutton, J.M.; Soares da Costa, T.P. Overcoming intrinsic and acquired resistance mechanisms associated with the cell wall of gram-negative bacteria. Antibiotics 2020, 9, 623. [Google Scholar] [CrossRef]
- Saxena, D.; Maitra, R.; Bormon, R.; Czekanska, M.; Meiers, J.; Titz, A.; Verma, S.; Chopra, S. Tackling the outer membrane: facilitating compound entry into Gram−negative bacterial pathogens. npj Antimicrob Resist 2023, 1, 17. [Google Scholar] [CrossRef]
- Sperandeo, P.; Martorana, A.M.; Zaccaria, M.; Polissi, A. Targeting the LPS export pathway for the development of novel therapeutics. BBA−Mol Cell Res 2023, 1870, 119406. [Google Scholar] [CrossRef] [PubMed]
- Romano, K.; Hung, D. Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi−drug resistance. BBA−Mol Cell Res 2023, 1870, 119407. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Jiang, C. Antimicrobial peptides: Structure, mechanism, and modification. Eur J Med Chem 2023, 255, 115377. [Google Scholar] [CrossRef] [PubMed]
- Espeche, J.C.; Varas, R.; Maturana, P.; Cutro, A.C.; Maffía, P.C.; Hollmann, A. Membrane permeability and antimicrobial peptides: Much more than just making a hole. Peptide Sci 2024, 116, e24305. [Google Scholar] [CrossRef]
- Stephani, J.C.; Gerhards, L.; Khairalla, B.; Solov’yov, I.A.; Brand, I. How do antimicrobial peptides interact with the outer membrane of Gram-negative bacteria? role of lipopolysaccharides in peptide binding, anchoring, and penetration. ACS Infect. Dis. 2024, 10, 763–778. [Google Scholar] [CrossRef]
- Lima, W.G.; de Lima, M.E. Therapeutic Prospection of animal venoms-derived antimicrobial peptides against infections by multidrug−resistant acinetobacter baumannii: A systematic review of pre−clinical studies. Toxins 2023, 15, 268. [Google Scholar] [CrossRef]
- Gong, H.; Hu, X.; Zhang, L.; Fa, K.; Liao, M.; Liu, H.; Fragneto, G.; Campana, M.; Lu, J.R. How do antimicrobial peptides disrupt the lipopolysaccharide membrane leaflet of Gram-negative bacteria? J Colloid Interf Sci 2023, 637, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Su, G.; Jiang, S.; Chen, L.; Huang, J.; Yang, F. New N−terminal fatty−acid−modified melittin analogs with potent biological activity. Int J Mol Sci 2024, 25, 867. [Google Scholar] [CrossRef] [PubMed]
- Brand, I.; Khairalla, B. Structural changes in the model of the outer cell membrane of Gram−negative bacteria interacting with melittin: an in situ spectroelectrochemical study. Faraday Discuss 2021, 232, 68–85. [Google Scholar] [CrossRef]
- Bhunia, A.; Domadia, P.N.; Bhattacharjya, S. Structural and thermodynamic analyses of the interaction between melittin and lipopolysaccharide. BBA−Biomembranes 2007, 1768, 3282–3291. [Google Scholar] [CrossRef]
- Lostao, A.; Lim, K.; Pallarés, M.C.; Ptak, A.; Marcuello, C. Recent advances in sensing the inter−biomolecular interactions at the nanoscale–A comprehensive review of AFM-based force spectroscopy. Int J Biol Macromol 2023, 124089. [Google Scholar] [CrossRef]
- Sun, H.; Tian, Y.; Fu, Y.; Lei, Y.; Wang, Y.; Yan, X.; Wang, J. Single-molecule scale quantification reveals interactions underlying protein-protein interface: from forces to non-covalent bonds. Phys Chem Chem Phys 2023, 25, 31791–31803. [Google Scholar] [CrossRef]
- Ananchenko, B.; Belozerov, V.; Byvalov, A.; Konyshev, I.; Korzhavina, A.; Dudina, L. Evaluation of intermolecular forces between lipopolysaccharides and monoclonal antibodies using atomic force microscopy. Int J Biol Macromol 2020, 156, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, J.; Sun, H.; Fu, Y.; Wang, Y. Probing changes in Ca2+-induced interaction forces between calmodulin and melittin by atomic force microscopy. Micromachines 2020, 11, 906. [Google Scholar] [CrossRef]
- Sabnis, A.; Edwards, A.M. Lipopolysaccharide as an antibiotic target. BBA−Mol Cell Res 2023, 1870, 119507. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Duda, K.A.; Lanzetta, R.; Silipo, A.; De Castro, C.; Molinaro, A. A journey from structure to function of bacterial lipopolysaccharides. Chem. Rev. 2021, 122, 15767–15821. [Google Scholar] [CrossRef]
- Golomidova, A.; Naumenko, O.; Senchenkova, S.; Knirel, Y.; Letarov, A. The O−polysaccharide of Escherichia coli F5, which is structurally related to that of E. coli O28ab, provides cells only weak protection against bacteriophage attack. Arch Virol 2019, 164, 2783–2787. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu Rev Biochem 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Lu, Q.; Wang, J.; Faghihnejad, A.; Zeng, H.; Liu, Y. Understanding the molecular interactions of lipopolysaccharides during E. coli initial adhesion with a surface forces apparatus. Soft Matter 2011, 7, 9366–9379. [Google Scholar] [CrossRef]
- Zhou, Y.; Cao, W.; Xu, Z.; Zhang, X.F.; Liu, Y. Binding kinetics of liposome conjugated E−selectin and P−selectin glycoprotein ligand-1 measured with atomic force microscopy. Colloid Surface B 2021, 207, 112002. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, C.; Ling, L.; Wan, L.; Fang, X.; Bai, C. Specific aptamer−protein interaction studied by atomic force microscopy. Anal. Chem. 2003, 75, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.S.; Huefner, N.D.; Chan, W.S.; Stevens, F.; Harris, J.M.; Beebe, T.P. Specific interactions between biotin and avidin studied by atomic force microscopy using the Poisson statistical analysis method. Langmuir 1999, 15, 1373–1382. [Google Scholar] [CrossRef]
- Jucker, B.; Harms, H.; Hug, S.; Zehnder, A. Adsorption of bacterial surface polysaccharides on mineral oxides is mediated by hydrogen bonds. Colloid Surface B 1997, 9, 331–343. [Google Scholar] [CrossRef]
- Guo, W.; Xu, S.; Reichart, T.M.; Xiao, M.; Lu, T.; Mello, C.; Chen, Z. Probing molecular interactions between surface−immobilized antimicrobial peptides and lipopolysaccharides in situ. Langmuir 2020, 36, 12383–12393. [Google Scholar] [CrossRef]
- Uzarski, J.R.; Mello, C.M. Detection and classification of related lipopolysaccharides via a small array of immobilized antimicrobial peptides. Anal. Chem. 2012, 84, 7359–7366. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Z.; Liu, H.; Nash, M.A. Next generation methods for single−molecule force spectroscopy on polyproteins and receptor-ligand complexes. Front Mol Biosci 2020, 7, 85. [Google Scholar] [CrossRef]
- Yang, J.; Petitjean, S.J.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS−CoV−2 binding to the ACE2 receptor. Nat Commun 2020, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, J.; Wang, Y.; Sun, H. Investigating the effect of tyrosine kinase inhibitors on the interaction between human serum albumin by atomic force microscopy. Biomolecules 2022, 12, 819. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, A.R.; Cannistraro, S. The application of atomic force spectroscopy to the study of biological complexes undergoing a biorecognition process. Chem Soc Rev 2010, 39, 734–749. [Google Scholar] [CrossRef]
- Akhunzada, M.J.; Yoon, H.J.; Deb, I.; Braka, A.; Wu, S. Bell-Evans model and steered molecular dynamics in uncovering the dissociation kinetics of ligands targeting G−protein−coupled receptors. Sci Rep−UK 2022, 12, 15972. [Google Scholar] [CrossRef] [PubMed]
- Rico-Pasto, M.; Zaltron, A.; Ritort, F. Force dependence of proteins’ transition state position and the Bell–Evans model. Nanomaterials 2021, 11, 3023. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jin, Y.; Desai, U.R.; Yadavalli, V.K. Investigation of the heparin–thrombin interaction by dynamic force spectroscopy. BBA-Gen Subjects 2015, 1850, 1099–1106. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, M.; Guan, Y.; Guo, X.; Li, Z.; Rankl, C.; Tang, J. Imaging and quantifying analysis the binding behavior of PD−L1 at molecular resolution by atomic force microscopy. Anal Chim Acta 2022, 1191, 339281. [Google Scholar] [CrossRef]
- Müller, D.J.; Dumitru, A.C.; Lo Giudice, C.; Gaub, H.E.; Hinterdorfer, P.; Hummer, G.; De Yoreo, J.J.; Dufrêne, Y.F.; Alsteens, D. Atomic force microscopy−based force spectroscopy and multiparametric imaging of biomolecular and cellular systems. Chem. Rev. 2020, 121, 11701–11725. [Google Scholar] [CrossRef]
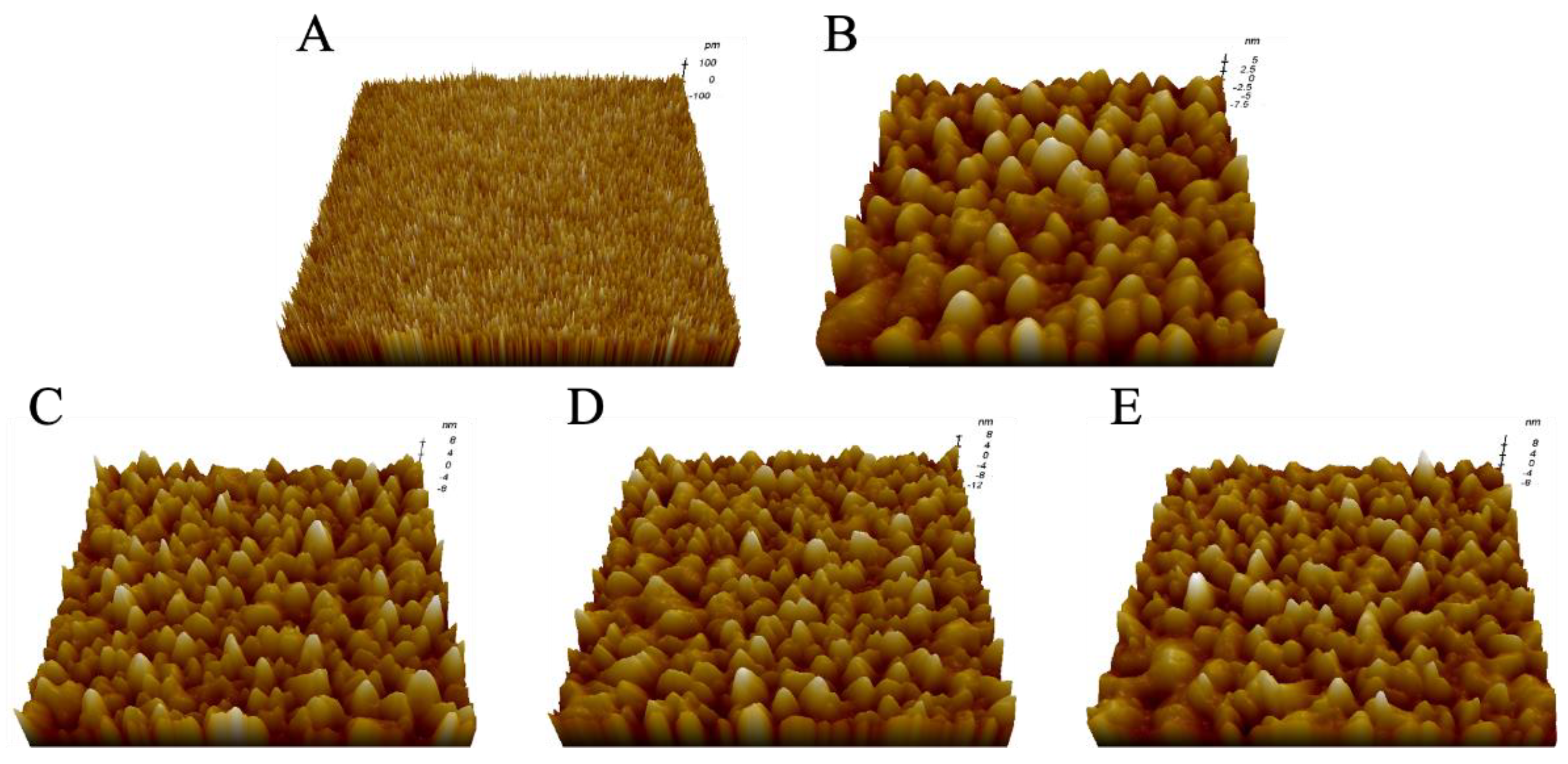
| No. | Sample | Parameter | |
|---|---|---|---|
| Ra, nm2 | μ, nm | ||
| 1 | Mica | 0.057 ± 0.003 | 0.231 ± 0.004 |
| 2 | LPS from E. coli K-12 | 0.216 ± 0.004 | 13.245 ± 0.071 |
| 3 | LPS from E. coli O55:B5 | 0.304 ± 0.002 | 17.352 ± 0.045 |
| 4 | LPS from E. coli O111:B4 | 0.335 ± 0.007 | 18.106 ± 0.062 |
| 5 | LPS from E. coli O128:B12 | 0.315 ± 0.011 | 17.566 ± 0.051 |
| Sample | Koff, S-1 | xβ, nm | τoff, ms | ∆G, kBT |
|---|---|---|---|---|
| LPS from E. coli K-12 | 110.91 ± 5.51 | 0.10517 ± 0.0018 | 9.038 ± 0.448 | 24.748 ± 0.049 |
| LPS from E. coli O55:B5 | 108.04 ± 4.27 | 0.09464 ± 0.0003 | 9.271 ± 0.371 | 24.774 ± 0.040 |
| LPS from E. coli O111:B4 | 99.82 ± 5.57 | 0.08717 ± 0.0003 | 10.005 ± 0.574 | 24.854 ± 0.056 |
| LPS from E. coli O128:B12 | 101.44 ± 3.54 | 0.08427 ± 0.0003 | 9.870 ± 0.349 | 24.837 ± 0.035 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




