Submitted:
24 August 2024
Posted:
27 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
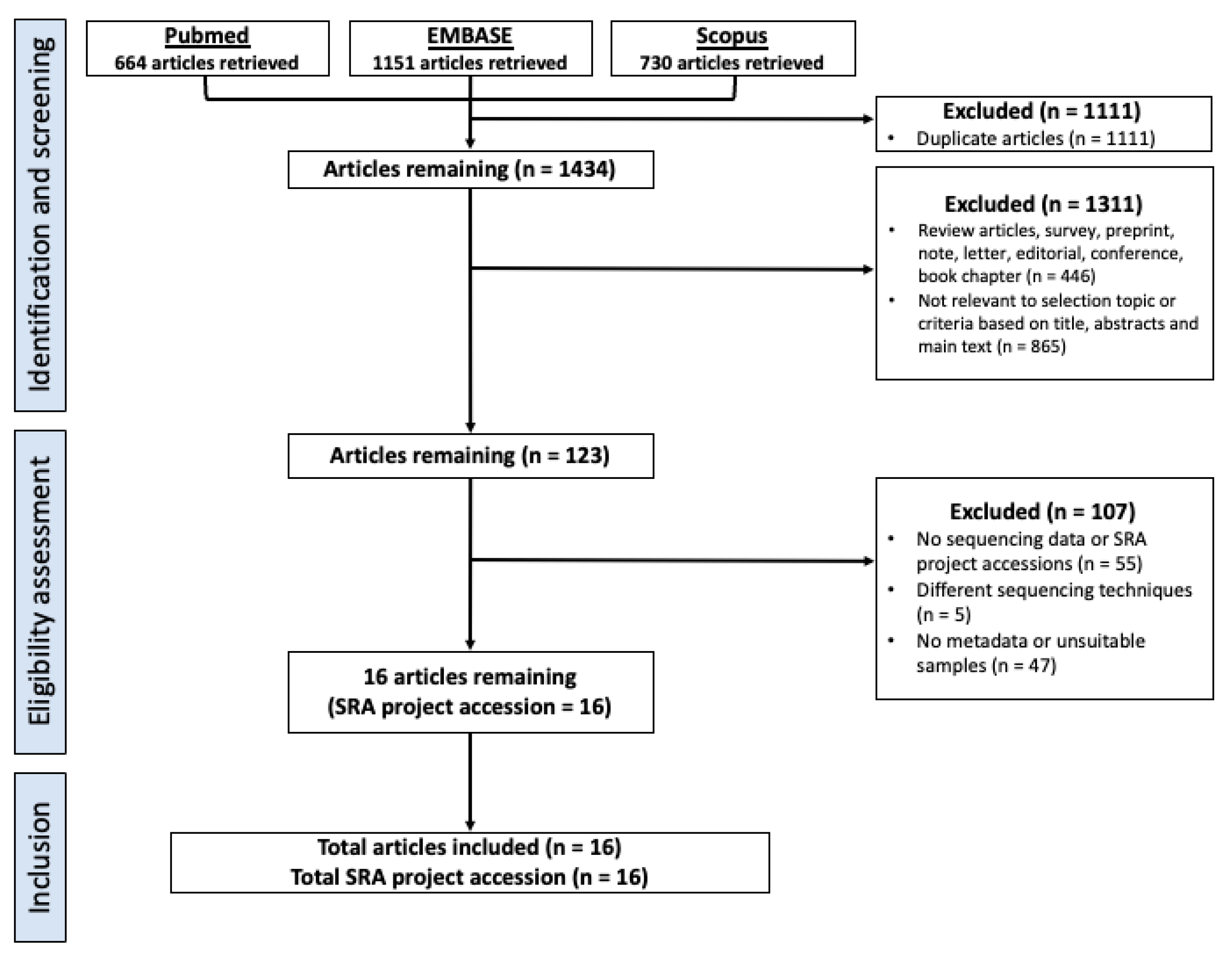
2.1. Selection Criteria, Database Search and Study design
2.2. Downloading, Pre-Processing and Metadata Collection of 16S rRNA Datasets
2.3. Evaluation and Correcting for Batch Effects
2.4. Alpha and Beta Diversity Analysis at the Genus Level
2.5. Differential Abundance Analysis between HNC, Healthy and Premalignant Oral Samples
2.6. Multivariate Sparse Partial Least Squares Discriminant Analysis (sPLS-DA) Classification to Discriminate Oral Sampling
3. Results
3.1. Study Characteristics and Dataset
3.2. Patient Type, 16s rRNA Amplification Primer Sets, and Study Batch Affects Overall Microbial Composition of Saliva and Oral Rinse Samples
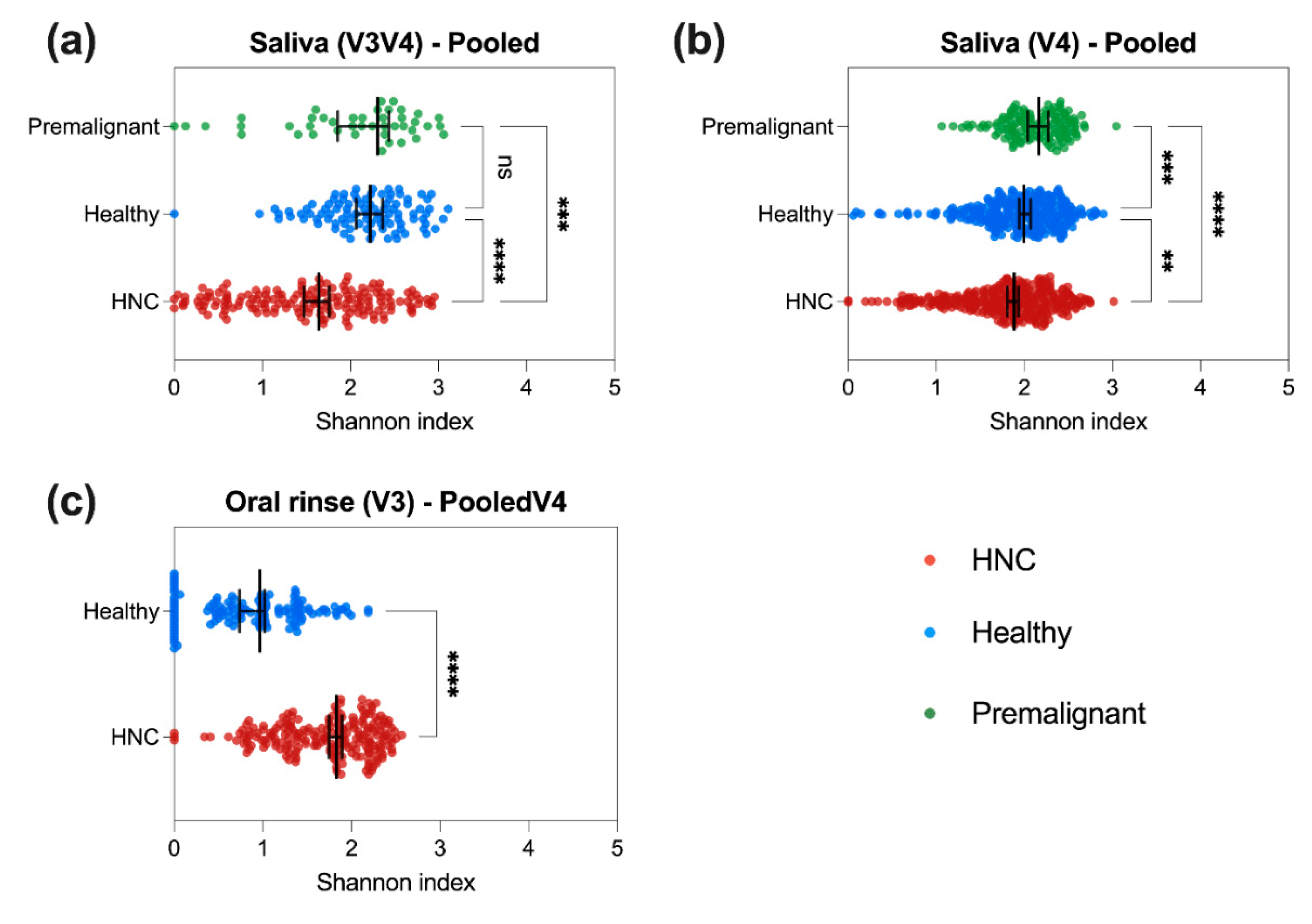
3.3. HNC Alpha Diversity Differs between Saliva and Oral Rinse
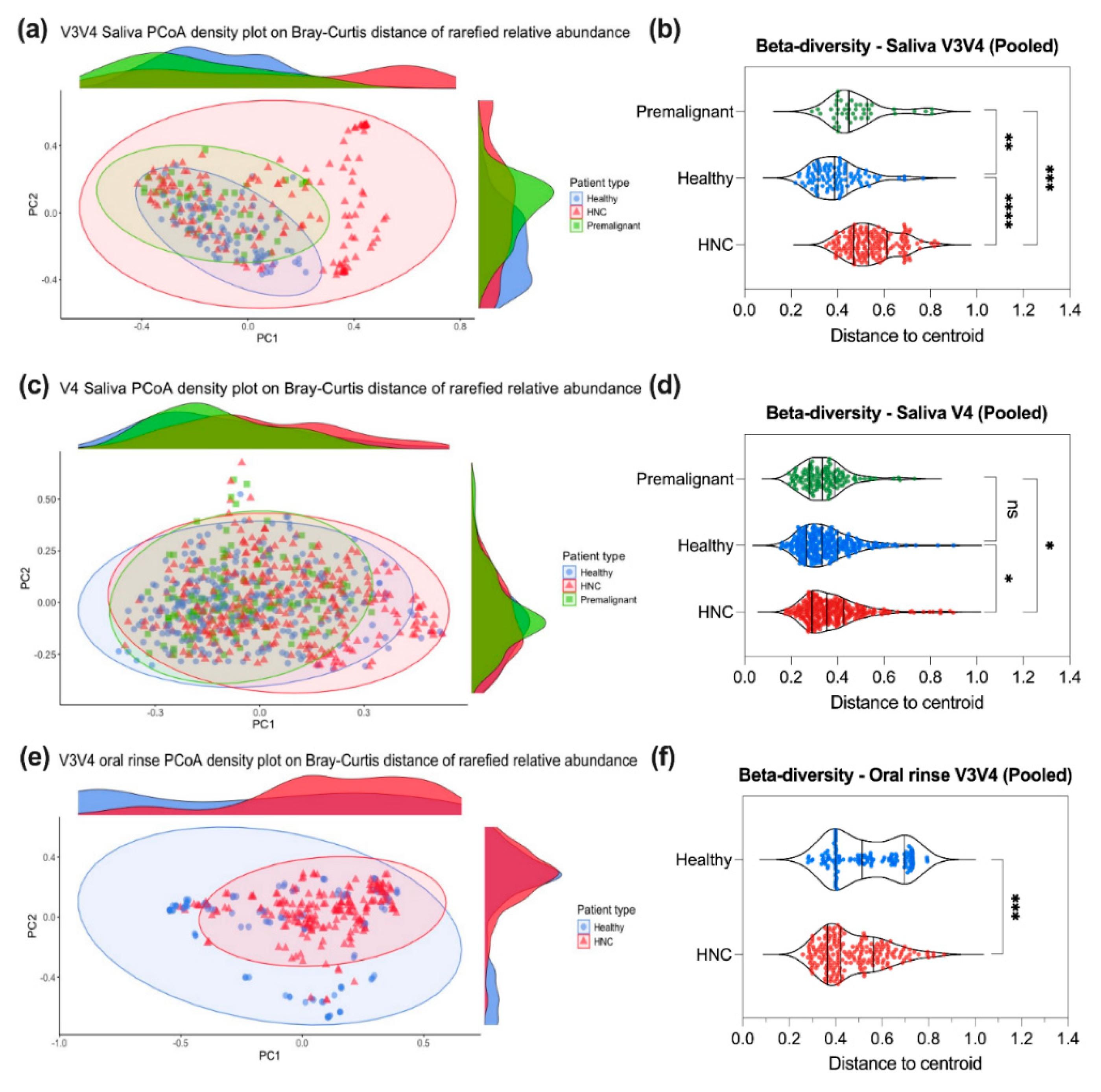
3.4. Saliva or Oral Rinse from HNC, Premalignant and Healthy Donors Have Similar Beta Diversities at the Genus Level
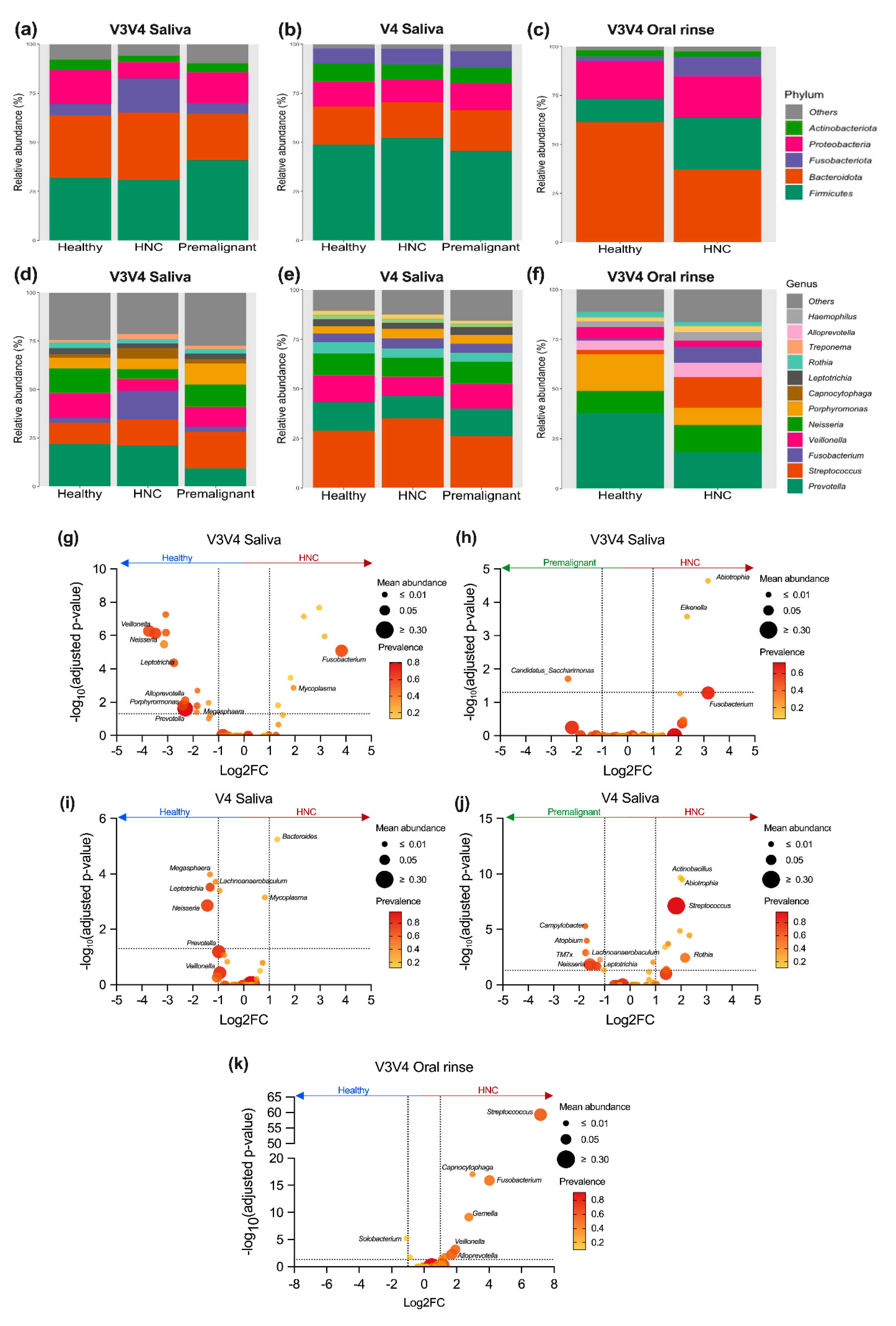
3.5. Differentially Abundant Genera between HNC and Healthy Oral Samples
| Phylum |
Saliva Relative abundance (Mean ± SD) |
Oral rinse Relative abundance (Mean ± SD) |
| Firmicutes | V3V4 = 32.4 ± 23.4 % V4 = 49.7 ± 19.0 % V4V5 = 43.2 ± 14.5 % |
V3V4 = 20.9 ± 20.4 % |
| Bacteroidota | V3V4 = 32.2 ± 27.2 % V4 = 19.1 ± 14.3 % V4V5 = 33.0 ± 14.8 % |
V3V4 = 46.5 ± 29.5 % |
| Fusobacteriota | V3V4 = 12.2 ± 21.0 % V4 = 8.00 ± 9.01 % V4V5 = 2.93 ± 2.82 % |
V3V4 = 7.06 ± 11.3 % |
| Proteobacteria | V3V4 = 12.0 ± 16.0 % V4 = 12.4 ± 11.5 % V4V5 = 16.2 ± 13.8 % |
V3V4 = 20.3 ± 24.2 % |
| Actinobacteriota | V3V4 = 4.15 ± 7.78 % V4 = 8.32 ± 7.64 % V4V5 = 3.29 ± 3.46 % |
V3V4 = 2.90 ± 5.79 % |
| Genera |
Saliva Relative abundance (Mean ± SD) |
Oral rinse Relative abundance (Mean ± SD) |
| Streptococcus | V3V4 = 13.4 ± 17.4 % V4 = 31.1 ± 18.1 % V4V5 = 29.3 ± 16.2 % |
V3V4 = 10.3 ± 15.4% |
| Neisseria | V3V4 = 8.38 ± 13.8 % V4 = 10.5 ± 10.7 % V4V5 = 9.17 ± 7.65 % |
V3V4 = 12.7 ± 19.4 % |
| Prevotella | V3V4 = 19.6 ± 21.9 % V4 = 13.0 ± 12.5 % V4V5 = 13.1 ± 9.35 % |
V3V4 = 25.8 ± 28.1 % |
| Porphyromonas | V3V4 = 6.05 ± 13.4 % V4 = 4.40 ± 7.04 % V4V5 = 12.9 ± 9.89 % |
V3V4 = 12.5 ± 21.6 % |
| Veillonella | V3V4 = 8.78 ± 10.8 % V4 = 11.6 ± 11.5 % V4V5 = 4.84 ± 4.93 % |
V3V4 = 4.1 ± 9.2 % |
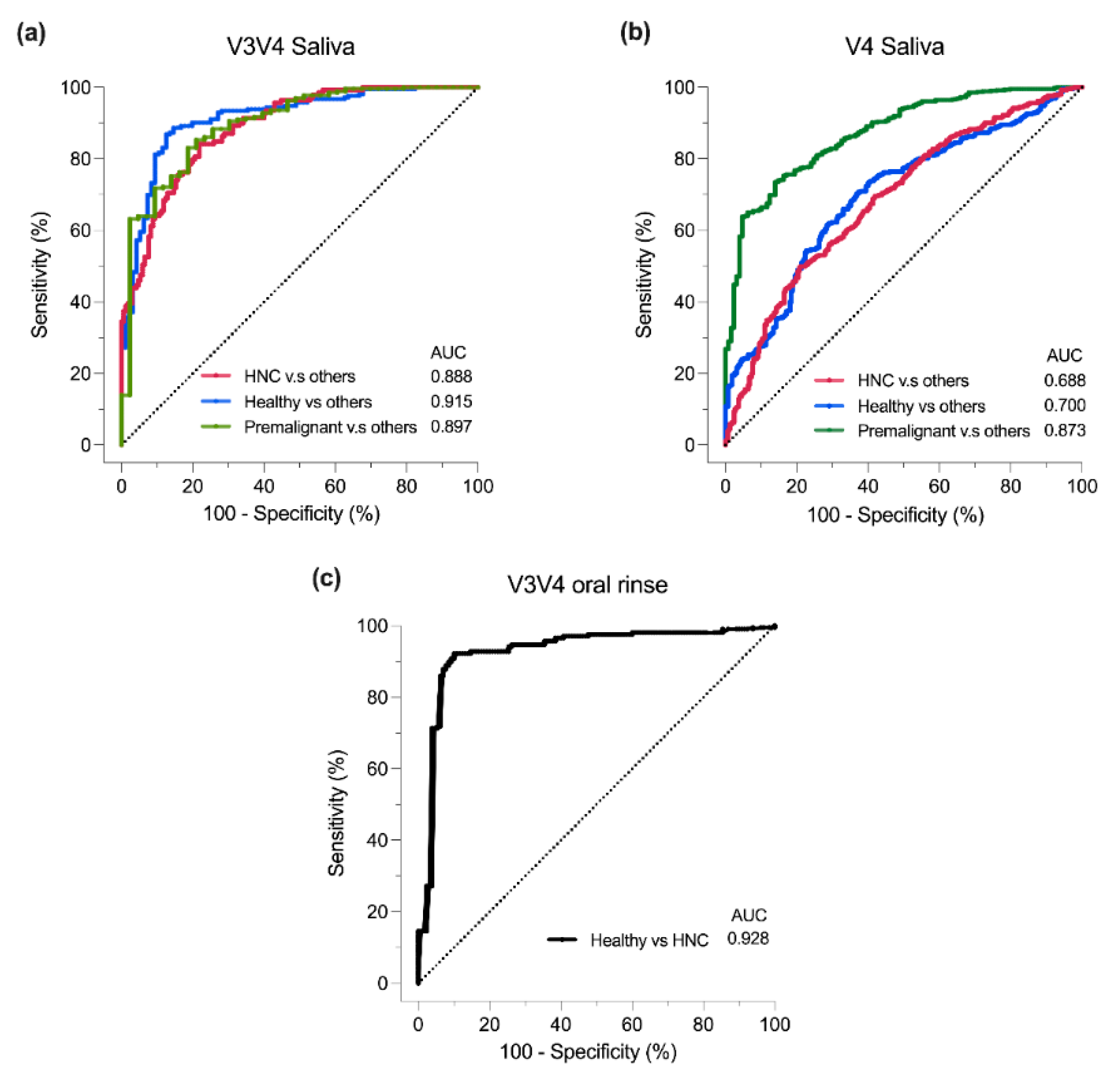
3.6. Classification of Oral Samples Based on sPLS-DA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Peng, X., L. Cheng, Y. You, C. Tang, B. Ren, Y. Li, X. Xu and X. Zhou. “Oral microbiota in human systematic diseases.” International journal of oral science 14 (2022): 14.
- Nejman, D., I. Livyatan, G. Fuks, N. Gavert, Y. Zwang, L. T. Geller, A. Rotter-Maskowitz, R. Weiser, G. Mallel, E. Gigi, et al. “The human tumor microbiome is composed of tumor type-specific intracellular bacteria.” Science 368 (2020): 973-80. [CrossRef]
- Irfan, M., R. Z. R. Delgado and J. Frias-Lopez. “The oral microbiome and cancer.” Frontiers in immunology 11 (2020): 591088. [CrossRef]
- Sykes, E. A., N. Weisbrod, E. Rival, A. Haque, R. Fu and A. Eskander. “Methods, detection rates, and survival outcomes of screening for head and neck cancers: A systematic review.” JAMA Otolaryngology–Head & Neck Surgery 149 (2023): 1047-56. [CrossRef]
- Li, Q., Y. Tie, A. Alu, X. Ma and H. Shi. “Targeted therapy for head and neck cancer: Signaling pathways and clinical studies.” Signal Transduction and Targeted Therapy 8 (2023): 31. [CrossRef]
- Johnson, D. E., B. Burtness, C. R. Leemans, V. W. Y. Lui, J. E. Bauman and J. R. Grandis. “Head and neck squamous cell carcinoma.” Nature Reviews Disease Primers 6 (2020): 92. [CrossRef]
- Ting, H. S. L., Z. Chen and J. Y. K. Chan. “Systematic review on oral microbial dysbiosis and its clinical associations with head and neck squamous cell carcinoma.” Head Neck 45 (2023): 2120-35. [CrossRef]
- Wolf, A., C. Moissl-Eichinger, A. Perras, K. Koskinen, P. V. Tomazic and D. Thurnher. “The salivary microbiome as an indicator of carcinogenesis in patients with oropharyngeal squamous cell carcinoma: A pilot study.” Scientific Reports 7 (2017): 5867. [CrossRef]
- Kumpitsch, C., C. Moissl-Eichinger, J. Pock, D. Thurnher and A. Wolf. “Preliminary insights into the impact of primary radiochemotherapy on the salivary microbiome in head and neck squamous cell carcinoma.” Scientific Reports 10 (2020): 16582. [CrossRef]
- Du, Y., R. Feng, E. T. Chang, J. W. Debelius, L. Yin, M. Xu, T. Huang, X. Zhou, X. Xiao and Y. Li. “Influence of pre-treatment saliva microbial diversity and composition on nasopharyngeal carcinoma prognosis.” Frontiers in Cellular and Infection Microbiology 12 (2022): 831409. [CrossRef]
- Chen, M. Y., J. W. Chen, L. W. Wu, K. C. Huang, J. Y. Chen, W. S. Wu, W. F. Chiang, C. J. Shih, K. N. Tsai, W. T. Hsieh, et al. “Carcinogenesis of male oral submucous fibrosis alters salivary microbiomes.” J Dent Res 100 (2021): 397-405. [CrossRef]
- Chen, J.-W., J.-H. Wu, W.-F. Chiang, Y.-L. Chen, W.-S. Wu and L.-W. Wu. “Taxonomic and functional dysregulation in salivary microbiomes during oral carcinogenesis.” Frontiers in Cellular and Infection Microbiology 11 (2021): 663068. [CrossRef]
- Lee, W. H., H. M. Chen, S. F. Yang, C. Liang, C. Y. Peng, F. M. Lin, L. L. Tsai, B. C. Wu, C. H. Hsin, C. Y. Chuang, et al. “Bacterial alterations in salivary microbiota and their association in oral cancer.” Scientific Reports 7 (2017): 16540. [CrossRef]
- Zhou, X., Y. Hao, X. Peng, B. Li, Q. Han, B. Ren, M. Li, L. Li, Y. Li, G. Cheng, et al. “The clinical potential of oral microbiota as a screening tool for oral squamous cell carcinomas.” Front Cell Infect Microbiol 11 (2021): 728933. [CrossRef]
- Vesty, A., K. Gear, K. Biswas, F. J. Radcliff, M. W. Taylor and R. G. Douglas. “Microbial and inflammatory-based salivary biomarkers of head and neck squamous cell carcinoma.” Clin Exp Dent Res 4 (2018): 255-62. [CrossRef]
- Oyeyemi, B. F., U. S. Kaur, A. Paramraj, R. Tandon, A. Kumar and N. S. Bhavesh. “Microbiome analysis of saliva from oral squamous cell carcinoma (oscc) patients and tobacco abusers with potential biomarkers for oral cancer screening.” Heliyon 9 (2023). [CrossRef]
- Torralba, M. G., G. Aleti, W. Li, K. J. Moncera, Y. H. Lin, Y. Yu, M. M. Masternak, W. Golusinski, P. Golusinski, K. Lamperska, et al. “Oral microbial species and virulence factors associated with oral squamous cell carcinoma.” Microb Ecol 82 (2021): 1030-46. [CrossRef]
- Zakrzewski, M., O. M. Gannon, B. J. Panizza, N. A. Saunders and A. Antonsson. “Human papillomavirus infection and tumor microenvironment are associated with the microbiota in patients with oropharyngeal cancers-pilot study.” Head Neck 43 (2021): 3324-30. [CrossRef]
- Granato, D. C., L. X. Neves, L. D. Trino, C. M. Carnielli, A. F. Lopes, S. Yokoo, B. A. Pauletti, R. R. Domingues, J. O. Sa and G. Persinoti. “Meta-omics analysis indicates the saliva microbiome and its proteins associated with the prognosis of oral cancer patients.” Biochimica et Biophysica Acta (BBA)-proteins and proteomics 1869 (2021): 140659. [CrossRef]
- Hao, Y., Z. Zeng, X. Peng, P. Ai, Q. Han, B. Ren, M. Li, H. Wang, X. Zhou and X. Zhou. “The human oral–nasopharynx microbiome as a risk screening tool for nasopharyngeal carcinoma.” Frontiers in Cellular and Infection Microbiology 12 (2022): 1013920. [CrossRef]
- Zeng, B., J. Tan, G. Guo, Z. Li, L. Yang, X. Lao, D. Wang, J. Ma, S. Zhang and G. Liao. “The oral cancer microbiome contains tumor space–specific and clinicopathology-specific bacteria.” Frontiers in Cellular and Infection Microbiology 12 (2022): 942328. [CrossRef]
- Nie, F., L. Wang, Y. Huang, P. Yang, P. Gong, Q. Feng and C. Yang. “Characteristics of microbial distribution in different oral niches of oral squamous cell carcinoma.” Frontiers in Cellular and Infection Microbiology 12 (2022): 905653. [CrossRef]
- Pandey, D., M. Szczesniak, J. Maclean, H. C. H. Yim, F. Zhang, P. Graham, E. M. El-Omar and P. Wu. “Dysbiosis in head and neck cancer: Determining optimal sampling site for oral microbiome collection.” Pathogens 11 (2022): 1550. [CrossRef]
- Medeiros, M. C. d., S. The, E. Bellile, N. Russo, L. Schmitd, E. Danella, P. Singh, R. Banerjee, C. Bassis and G. R. Murphy III. “Salivary microbiome changes distinguish response to chemoradiotherapy in patients with oral cancer.” Microbiome 11 (2023): 268. [CrossRef]
- Mäkinen, A. I., V. Y. Pappalardo, M. J. Buijs, B. W. Brandt, A. A. Mäkitie, J. H. Meurman and E. Zaura. “Salivary microbiome profiles of oral cancer patients analyzed before and after treatment.” Microbiome 11 (2023): 171. [CrossRef]
- Chen, Z., P. Y. Wong, C. W. K. Ng, L. Lan, S. Fung, J. W. Li, L. Cai, P. Lei, Q. Mou, S. H. Wong, et al. “The intersection between oral microbiota, host gene methylation and patient outcomes in head and neck squamous cell carcinoma.” Cancers (Basel) 12 (2020). [CrossRef]
- Chan, J. Y. K., C. W. K. Ng, L. Lan, S. Fung, J. W. Li, L. Cai, P. Lei, Q. Mou, K. Meehan, E. H. L. Lau, et al. “Restoration of the oral microbiota after surgery for head and neck squamous cell carcinoma is associated with patient outcomes.” Front Oncol 11 (2021): 737843. [CrossRef]
- Sawant, S., J. Dugad, D. Parikh, S. Srinivasan and H. Singh. “Identification & correlation of bacterial diversity in oral cancer and long-term tobacco chewers- a case-control pilot study.” J Med Microbiol 70 (2021). [CrossRef]
- Zhu, H., H. C. Yip, M. K. Cheung, H. C. Chan, C. Ng, E. H. Lau, Z. W. Yeung, E. W. Wong, L. Leung and X. Qu. “Convergent dysbiosis of upper aerodigestive microbiota between patients with esophageal and oral cavity squamous cell carcinoma.” International Journal of Cancer 152 (2023): 1903-15. [CrossRef]
- Chen, Z., P. Y. Wong, C. W. K. Ng, L. Lan, S. Fung, J. W. Li, L. Cai, P. Lei, Q. Mou, S. H. Wong, et al. “The intersection between oral microbiota, host gene methylation and patient outcomes in head and neck squamous cell carcinoma.” Cancers 12 (2020): 3425. [CrossRef]
- Benjamin, W., K. Wang, K. Zarins, E. Bellile, F. Blostein and I. Argirion. Oral microbiome community composition in head and neck squamous cell carcinoma. Cancers (basel). 2023; 15 (9): 2549. [CrossRef]
- Yang, C.-Y., Y.-M. Yeh, H.-Y. Yu, C.-Y. Chin, C.-W. Hsu, H. Liu, P.-J. Huang, S.-N. Hu, C.-T. Liao and K.-P. Chang. “Oral microbiota community dynamics associated with oral squamous cell carcinoma staging.” Frontiers in microbiology 9 (2018): 862. [CrossRef]
- Lim, Y., N. Fukuma, M. Totsika, L. Kenny, M. Morrison and C. Punyadeera. “The performance of an oral microbiome biomarker panel in predicting oral cavity and oropharyngeal cancers.” Frontiers in Cellular and Infection Microbiology 8 (2018): 267. [CrossRef]
- Srivastava, A., S. Mishra, P. K. Garg, A. K. Dubey, S. Deo and D. Verma. “Comparative and analytical characterization of the oral bacteriome of smokeless tobacco users with oral squamous cell carcinoma.” Applied Microbiology and Biotechnology 106 (2022): 4115-28. [CrossRef]
- Shitozawa, Y., K. Haro, M. Ogawa, A. Miyawaki, M. Saito and K. Fukuda. “Differences in the microbiota of oral rinse, lesion, and normal site samples from patients with mucosal abnormalities on the tongue.” Scientific Reports 12 (2022): 16839. [CrossRef]
- Yu, X., Y. Shi, R. Yuan, Z. Chen, Q. Dong, L. Han, L. Wang and J. Zhou. “Microbial dysbiosis in oral squamous cell carcinoma: A systematic review and meta-analysis.” Heliyon 9 (2023). [CrossRef]
- Gopinath, D., R. K. Menon, M. Banerjee, R. S. Yuxiong, M. G. Botelho and N. W. Johnson. “Culture-independent studies on bacterial dysbiosis in oral and oropharyngeal squamous cell carcinoma: A systematic review.” Critical reviews in oncology/hematology 139 (2019): 31-40. [CrossRef]
- Bronzato, J. D., R. A. Bomfim, D. H. Edwards, D. Crouch, M. P. Hector and B. P. Gomes. “Detection of fusobacterium in oral and head and neck cancer samples: A systematic review and meta-analysis.” Archives of oral biology 112 (2020): 104669. [CrossRef]
- Baker, J. L., J. L. Mark Welch, K. M. Kauffman, J. S. McLean and X. He. “The oral microbiome: Diversity, biogeography and human health.” Nature Reviews Microbiology 22 (2024): 89-104.
- Caselli, E., C. Fabbri, M. D’Accolti, I. Soffritti, C. Bassi, S. Mazzacane and M. Franchi. “Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture.” BMC microbiology 20 (2020): 1-19. [CrossRef]
- Ruan, X., J. Luo, P. Zhang and K. Howell. “The salivary microbiome shows a high prevalence of core bacterial members yet variability across human populations.” npj Biofilms and Microbiomes 8 (2022): 85. [CrossRef]
- Takahashi, Y., J. Park, K. Hosomi, T. Yamada, A. Kobayashi, Y. Yamaguchi, S. Iketani, J. Kunisawa, K. Mizuguchi and N. Maeda. “Analysis of oral microbiota in japanese oral cancer patients using 16s rrna sequencing.” Journal of Oral Biosciences 61 (2019): 120-28. [CrossRef]
- Yeo, K., R. Li, F. Wu, G. Bouras, L. T. H. Mai, E. Smith, P. J. Wormald, R. Valentine, A. J. Psaltis, S. Vreugde, et al. “Identification of consensus head and neck cancer-associated microbiota signatures: A systematic review and meta-analysis of 16s rrna and the cancer microbiome atlas datasets.” J Med Microbiol 73 (2024). [CrossRef]
- Chan, J. Y. K., M. K. Cheung, L. Lan, C. Ng, E. H. L. Lau, Z. W. C. Yeung, E. W. Y. Wong, L. Leung, X. Qu, L. Cai, et al. “Characterization of oral microbiota in hpv and non-hpv head and neck squamous cell carcinoma and its association with patient outcomes.” Oral Oncol 135 (2022): 106245. [CrossRef]
- Neuzillet, C., M. Marchais, S. Vacher, M. Hilmi, A. Schnitzler, D. Meseure, R. Leclere, C. Lecerf, C. Dubot, E. Jeannot, et al. “Prognostic value of intratumoral fusobacterium nucleatum and association with immune-related gene expression in oral squamous cell carcinoma patients.” Scientific Reports 11 (2021): 7870. [CrossRef]
- Hayes, R. B., J. Ahn, X. Fan, B. A. Peters, Y. Ma, L. Yang, I. Agalliu, R. D. Burk, I. Ganly and M. P. Purdue. “Association of oral microbiome with risk for incident head and neck squamous cell cancer.” JAMA oncology 4 (2018): 358-65. [CrossRef]
- Shay, E., N. Sangwan, R. Padmanabhan, S. Lundy, B. Burkey and C. Eng. “Bacteriome and mycobiome and bacteriome-mycobiome interactions in head and neck squamous cell carcinoma.” Oncotarget 11 (2020): 2375. [CrossRef]
- Heng, W., W. Wang, T. Dai, P. Jiang, Y. Lu, R. Li, M. Zhang, R. Xie, Y. Zhou and M. Zhao. “Oral bacteriome and mycobiome across stages of oral carcinogenesis.” Microbiology spectrum 10 (2022): e02737-22. [CrossRef]
- Li, Y., X. Tan, X. Zhao, Z. Xu, W. Dai, W. Duan, S. Huang, E. Zhang, J. Liu and S. Zhang. “Composition and function of oral microbiota between gingival squamous cell carcinoma and periodontitis.” Oral Oncology 107 (2020): 104710. [CrossRef]
- Zuo, H. J., M. R. Fu, H. L. Zhao, X. W. Du, Z. Y. Hu, X. Y. Zhao, X. Q. Ji, X. Q. Feng, W. Zhumajiang, T. H. Zhou, et al. “Study on the salivary microbial alteration of men with head and neck cancer and its relationship with symptoms in southwest china.” Frontiers in Cellular and Infection Microbiology 10 (2020): 514943. [CrossRef]
- Peter, T. K., M. H. H. Withanage, C. L. Comnick, C. Pendleton, S. Dabdoub, S. Ganesan, D. Drake, J. Banas, X. J. Xie and E. Zeng. “Systematic review and meta-analysis of oral squamous cell carcinoma associated oral microbiome.” Front Microbiol 13 (2022): 968304. [CrossRef]
- Metsäniitty, M., S. Hasnat, T. Salo and A. Salem. “Oral microbiota-a new frontier in the pathogenesis and management of head and neck cancers.” Cancers (Basel) 14 (2021). [CrossRef]
- Su Mun, L., S. Wye Lum, G. Kong Yuiin Sze, C. Hock Yoong, K. Ching Yung, L. Kah Lok and D. Gopinath. “Association of microbiome with oral squamous cell carcinoma: A systematic review of the metagenomic studies.” Int J Environ Res Public Health 18 (2021). [CrossRef]
- Ramos, R. T., C. S. Sodré, P. M. G. R. de Sousa Rodrigues, A. M. P. da Silva, M. S. Fuly, H. F. Dos Santos, L. S. Gonçalves, D. de Carvalho Ferreira and M. G. Ribeiro. “High-throughput nucleotide sequencing for bacteriome studies in oral squamous cell carcinoma: A systematic review.” Oral and Maxillofacial Surgery 24 (2020): 387-401. [CrossRef]
- Delaney, C., C. L. R. Veena, M. C. Butcher, W. McLean, S. M. A. Shaban, C. J. Nile and G. Ramage. “Limitations of using 16s rrna microbiome sequencing to predict oral squamous cell carcinoma.” APMIS (2023). [CrossRef]
- Gloor, G. B., J. M. Macklaim, V. Pawlowsky-Glahn and J. J. Egozcue. “Microbiome datasets are compositional: And this is not optional.” Front Microbiol 8 (2017): 2224. [CrossRef]
- Nearing, J. T., A. M. Comeau and M. G. I. Langille. “Identifying biases and their potential solutions in human microbiome studies.” Microbiome 9 (2021): 113. [CrossRef]
- Johnson, J. S., D. J. Spakowicz, B. Y. Hong, L. M. Petersen, P. Demkowicz, L. Chen, S. R. Leopold, B. M. Hanson, H. O. Agresta, M. Gerstein, et al. “Evaluation of 16s rrna gene sequencing for species and strain-level microbiome analysis.” Nat Commun 10 (2019): 5029. [CrossRef]
- Wang, S., F. Song, H. Gu, X. Wei, K. Zhang, Y. Zhou and H. Luo. “Comparative evaluation of the salivary and buccal mucosal microbiota by 16s rrna sequencing for forensic investigations.” Frontiers in microbiology 13 (2022): 777882. [CrossRef]
- Page, M. J., J. E. McKenzie, P. M. Bossuyt, I. Boutron, T. C. Hoffmann, C. D. Mulrow, L. Shamseer, J. M. Tetzlaff, E. A. Akl, S. E. Brennan, et al. “The prisma 2020 statement: An updated guideline for reporting systematic reviews.” Rev Esp Cardiol (Engl Ed) 74 (2021): 790-99. Declaración PRISMA 2020: una guía actualizada para la publicación de revisiones sistemáticas. [CrossRef]
- Sterne, J. A., J. Savović, M. J. Page, R. G. Elbers, N. S. Blencowe, I. Boutron, C. J. Cates, H.-Y. Cheng, M. S. Corbett and S. M. Eldridge. “Rob 2: A revised tool for assessing risk of bias in randomised trials.” bmj 366 (2019).
- Team, S. T. D. “Sra toolkit.” (2020).
- Callahan, B. J., P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. Johnson and S. P. Holmes. “Dada2: High-resolution sample inference from illumina amplicon data.” Nat Methods 13 (2016): 581-3. [CrossRef]
- Bolyen, E., J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, et al. “Reproducible, interactive, scalable and extensible microbiome data science using qiime 2.” Nat Biotechnol 37 (2019): 852-57. [CrossRef]
- Martin, M. “Cutadapt removes adapter sequences from high-throughput sequencing reads.” EMBnet. journal 17 (2011): 10-12. [CrossRef]
- Wang, Y. and L. C. KA. “Plsda-batch: A multivariate framework to correct for batch effects in microbiome data.” Brief Bioinform 24 (2023). [CrossRef]
- McMurdie, P. J. and S. Holmes. “Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data.” PLoS ONE 8 (2013): e61217. [CrossRef]
- Ma, S., D. Shungin, H. Mallick, M. Schirmer, L. H. Nguyen, R. Kolde, E. Franzosa, H. Vlamakis, R. Xavier and C. Huttenhower. “Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using mmuphin.” Genome biology 23 (2022): 208. [CrossRef]
- Curry, K. D., Q. Wang, M. G. Nute, A. Tyshaieva, E. Reeves, S. Soriano, Q. Wu, E. Graeber, P. Finzer, W. Mendling, et al. “Emu: Species-level microbial community profiling of full-length 16s rrna oxford nanopore sequencing data.” Nat Methods 19 (2022): 845-53. [CrossRef]
- Kodikara, S., S. Ellul and K.-A. Lê Cao. “Statistical challenges in longitudinal microbiome data analysis.” Briefings in Bioinformatics 23 (2022): bbac273. [CrossRef]
- Susin, A., Y. Wang, K.-A. Lê Cao and M. L. Calle. “Variable selection in microbiome compositional data analysis.” NAR Genomics and Bioinformatics 2 (2020): lqaa029. [CrossRef]
- Yeo, K., J. Connell, G. Bouras, E. Smith, W. Murphy, J. C. Hodge, S. Krishnan, P. J. Wormald, R. Valentine, A. J. Psaltis, et al. “A comparison between full-length 16s rrna oxford nanopore sequencing and illumina v3-v4 16s rrna sequencing in head and neck cancer tissues.” Arch Microbiol 206 (2024): 248. [CrossRef]
- Liu, C., Y. Cui, X. Li and M. Yao. “Microeco: An r package for data mining in microbial community ecology.” FEMS Microbiol Ecol 97 (2021). [CrossRef]
- Oksanen, J. “Vegan: Community ecology package-r package version 1.17-8.” http://CRAN. R-project. org/package= vegan (2011).
- Zhou, H., K. He, J. Chen and X. Zhang. “Linda: Linear models for differential abundance analysis of microbiome compositional data.” Genome biology 23 (2022): 95. [CrossRef]
- Rohart, F., B. Gautier, A. Singh and K.-A. Lê Cao. “Mixomics: An r package for ‘omics feature selection and multiple data integration.” PLoS Computational Biology 13 (2017): e1005752. [CrossRef]
- Abellan-Schneyder, I., M. S. Matchado, S. Reitmeier, A. Sommer, Z. Sewald, J. Baumbach, M. List and K. Neuhaus. “Primer, pipelines, parameters: Issues in 16s rrna gene sequencing.” mSphere 6 (2021). [CrossRef]
- Gibbons, S. M., C. Duvallet and E. J. Alm. “Correcting for batch effects in case-control microbiome studies.” PLoS Computational Biology 14 (2018): e1006102. [CrossRef]
- Topçuoğlu, B. D., Z. Lapp, K. L. Sovacool, E. Snitkin, J. Wiens and P. D. Schloss. “Mikropml: User-friendly r package for supervised machine learning pipelines.” Journal of open source software 6 (2021). [CrossRef]
- Sharma, A. K., W. T. DeBusk, I. Stepanov, A. Gomez and S. S. Khariwala. “Oral microbiome profiling in smokers with and without head and neck cancer reveals variations between health and disease.” Cancer Prev Res (Phila) 13 (2020): 463-74. [CrossRef]
- Guerrero-Preston, R., F. Godoy-Vitorino, A. Jedlicka, A. Rodríguez-Hilario, H. González, J. Bondy, F. Lawson, O. Folawiyo, C. Michailidi, A. Dziedzic, et al. “16s rrna amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment.” Oncotarget 7 (2016): 51320-34. [CrossRef]
- Frank, D. N., Y. Qiu, Y. Cao, S. Zhang, L. Lu, J. M. Kofonow, C. E. Robertson, Y. Liu, H. Wang, C. L. Levens, et al. “A dysbiotic microbiome promotes head and neck squamous cell carcinoma.” Oncogene 41 (2022): 1269-80. [CrossRef]
- Zhu, W., W. Shen, J. Wang, Y. Xu, R. Zhai, J. Zhang, M. Wang, M. Wang and L. Liu. “Capnocytophaga gingivalis is a potential tumor promotor in oral cancer.” Oral Dis 30 (2024): 353-62. [CrossRef]
- Liu, Q. Y., Y. Liao, Y. X. Wu, H. Diao, Y. Du, Y. W. Chen, J. R. Xie, W. Q. Xue, Y. Q. He, T. M. Wang, et al. “The oral microbiome as mediator between oral hygiene and its impact on nasopharyngeal carcinoma.” Microorganisms 11 (2023). [CrossRef]
- Banavar, G., O. Ogundijo, R. Toma, S. Rajagopal, Y. K. Lim, K. Tang, F. Camacho, P. J. Torres, S. Gline, M. Parks, et al. “The salivary metatranscriptome as an accurate diagnostic indicator of oral cancer.” npj Genomic Medicine 6 (2021): 105. [CrossRef]
- Omori, M., N. Kato-Kogoe, S. Sakaguchi, N. Fukui, K. Yamamoto, Y. Nakajima, K. Inoue, H. Nakano, D. Motooka and T. Nakano. “Comparative evaluation of microbial profiles of oral samples obtained at different collection time points and using different methods.” Clinical Oral Investigations 25 (2021): 2779-89. [CrossRef]
- Zaura, E., B. J. F. Keijser, S. M. Huse and W. Crielaard. “Defining the healthy “core microbiome” of oral microbial communities.” BMC microbiology 9 (2009): 259. [CrossRef]
- Dhakal, A., R. Upadhyay, C. Wheeler, R. Hoyd, V. Karivedu, M. E. Gamez, S. Valentin, M. Vanputten, P. Bhateja, M. Bonomi, et al. “Association between tumor microbiome and hypoxia across anatomic subsites of head and neck cancers.” Int J Mol Sci 23 (2022). [CrossRef]
- Yano, Y., X. Hua, Y. Wan, S. Suman, B. Zhu, C. L. Dagnall, A. Hutchinson, K. Jones, B. D. Hicks, J. Shi, et al. “Comparison of oral microbiota collected using multiple methods and recommendations for new epidemiologic studies.” mSystems 5 (2020). [CrossRef]
- Mougeot, J. C., M. F. Beckman, H. C. Langdon, R. V. Lalla, M. T. Brennan and F. K. Bahrani Mougeot. “Haemophilus pittmaniae and leptotrichia spp. Constitute a multi-marker signature in a cohort of human papillomavirus-positive head and neck cancer patients.” Front Microbiol 12 (2021): 794546. [CrossRef]
- Hayes, R. B., J. Ahn, X. Fan, B. A. Peters, Y. Ma, L. Yang, I. Agalliu, R. D. Burk, I. Ganly, M. P. Purdue, et al. “Association of oral microbiome with risk for incident head and neck squamous cell cancer.” JAMA Oncol 4 (2018): 358-65. [CrossRef]
- Kers, J. G. and E. Saccenti. “The power of microbiome studies: Some considerations on which alpha and beta metrics to use and how to report results.” Front Microbiol 12 (2021): 796025. [CrossRef]
- Unlu, O., M. Demirci, T. Paksoy, A. B. Eden, H. D. Tansuker, A. Dalmizrak, C. Aktan, F. Senel, A. V. Sunter, O. Yigit, et al. “Oral microbial dysbiosis in patients with oral cavity cancers.” Clinical Oral Investigations 28 (2024): 377. [CrossRef]
- Shen, X., B. Zhang, X. Hu, J. Li, M. Wu, C. Yan, Y. Yang and Y. Li. “Neisseria sicca and corynebacterium matruchotii inhibited oral squamous cell carcinomas by regulating genome stability.” Bioengineered 13 (2022): 14094-106. [CrossRef]
- Baraniya, D., V. Jain, R. Lucarelli, V. Tam, L. Vanderveer, S. Puri, M. Yang and N. N. Al-Hebshi. “Screening of health-associated oral bacteria for anticancer properties in vitro.” Front Cell Infect Microbiol 10 (2020): 575656. [CrossRef]
- Connell, J. T., K. Yeo, G. Bouras, A. Bassiouni, K. Fenix, C. Cooksley, S. Vreugde, P. J. Wormald and A. J. Psaltis. “Enhanced phylogenetic insights into the microbiome of chronic rhinosinusitis through the novel application of long read 16s rrna gene amplicon sequencing.” Rhinology (2024). [CrossRef]
- Kumar, B., E. Lorusso, B. Fosso and G. Pesole. “A comprehensive overview of microbiome data in the light of machine learning applications: Categorization, accessibility, and future directions.” Front Microbiol 15 (2024): 1343572. [CrossRef]
- “Microbiome test spots oral cancers.” Nature Biotechnology 39 (2021): 650-50. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





