Submitted:
25 August 2024
Posted:
26 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
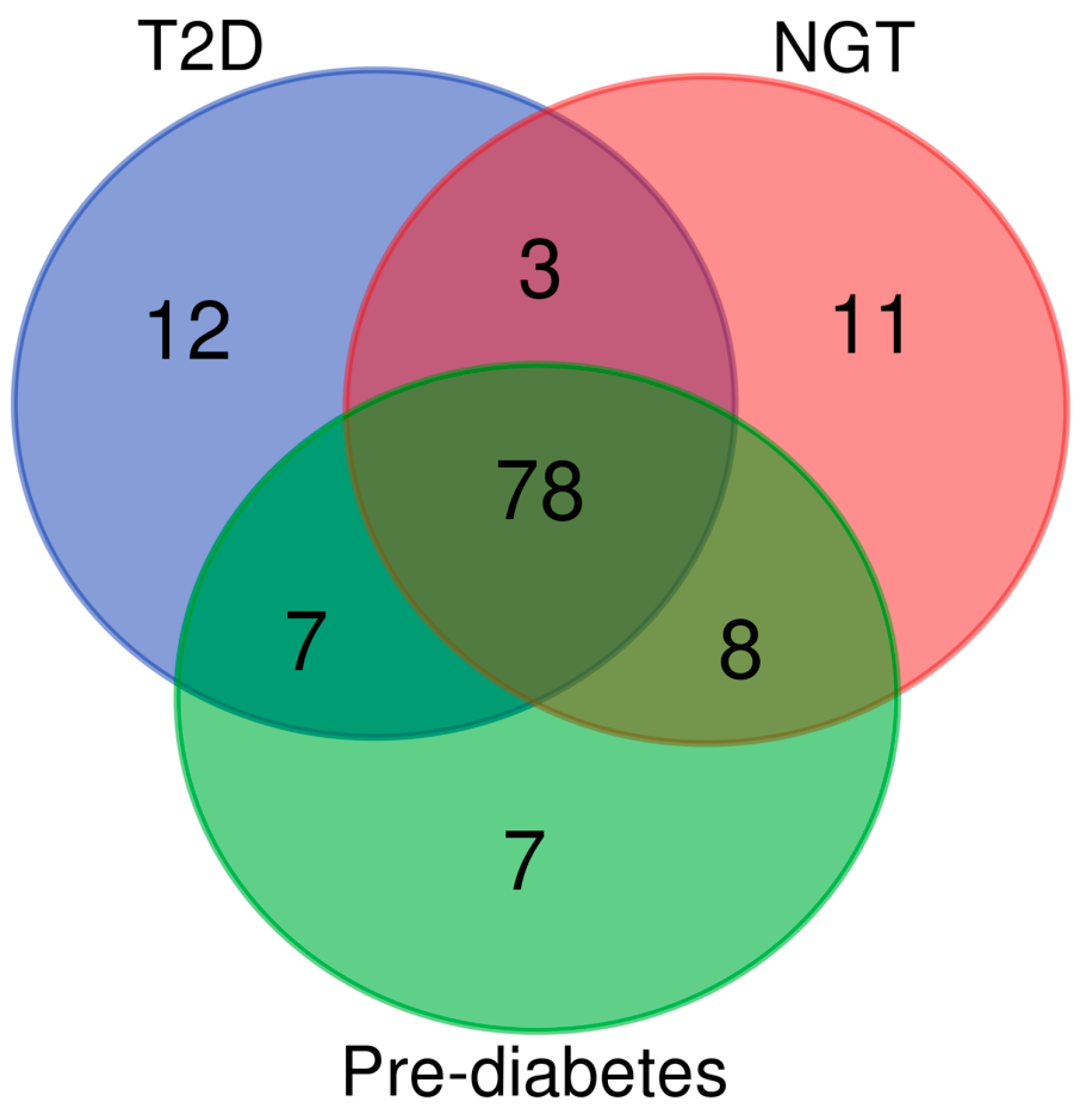
2. Results
2.1. Baseline Characteristics
2.2. Metabolites in Participants with Decreased and Normal eGFR
2.3. Effects of Glucose Tolerance on Metabolic Profile
2.4. Metabolites Associated with a Decrease in eGFR
2.5. Genetic Variants Associated with Novel Metabolites
3. Discussion
4. Materials and Methods
4.1. Study Population and Laboratory Measurements
4.2. Metabolomics
4.3. Selection of genetic variants decreasing glomerular filtration rate
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sekula, P.; Goek, O.-N.; Quaye, L.; Barrios, C.; Levey, A.S.; Römisch-Margl, W.; et al. A metabolome-wide association study of kidney function and disease in the general population. J. Am. Soc. Nephrol. 2016, 27, 1175-1188. [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.; Castro, A.F.; Feldman, H.I.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604. [CrossRef]
- Baumeister, S.E.; Böger, C.A.; Krämer, B.K.; Döring, A.; Eheberg, D.; Fischer, B.; et al. Effect of chronic kidney disease and comorbid conditions on health care costs: A 10-year observational study in a general population. Am. J. Nephrol. 2010, 31, 222-229. [CrossRef]
- Kazancioğlu, R. Risk factors for chronic kidney disease: an update. Kidney Int. Suppl. 2013, 3, 368-371. [CrossRef]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019, 51, 957-972.
- Wuttke, M.; König, E.; Katsara, M.-A.; Kirsten, H.; Farahani, S.K.; Teumer, A.; et al. Imputation-powered whole-exome analysis identifies genes associated with kidney function and disease in the UK Biobank. Nat. Commun. 2023, 14, 1287. [CrossRef]
- Gorski, M.; Rasheed, H.; Teumer, A.; Thomas, L.F.; Graham, S.E.; Sveinbjornsson, G.; et al. Genetic loci and prioritization of genes for kidney function decline derived from a meta-analysis of 62 longitudinal genome-wide association studies. Kidney Int. 2022, 102, 624-639. [CrossRef]
- Winkler, T.W.; Rasheed, H.; Teumer, A.; Gorski, M.; Rowan, B.X.; Stanzick, K.J.; et al. Differential and shared genetic effects on kidney function between diabetic and non-diabetic individuals. Commun. Biol. 2022, 5, 580. [CrossRef]
- Titan, S.M.; Venturini, G.; Padilha, K.; Tavares, G.; Zatz, R.; Bensenor, I.; et al. Metabolites related to eGFR: Evaluation of candidate molecules for GFR estimation using untargeted metabolomics. Clin. Chim. Acta 2019, 489, 242-248. [CrossRef]
- Lee, H.; Jang, H.B.; Yoo, M.-G.; Park, S.I.; Lee, H.-J. Amino acid metabolites associated with chronic kidney disease: An eight-year follow-up Korean epidemiology study. Biomedicines 2020, 8, 222. [CrossRef]
- Lee, S.; Han, M.; Moon, S.; Kim, K.; An, W.J.; Ryu, H.; et al. Identifying genetic variants and metabolites associated with rapid estimated glomerular filtration rate decline in Korea based on genome-metabolomic integrative analysis. Metabolites 2022, 12, 1139. [CrossRef]
- Peng, H.; Liu, X.; Aoieong, C.; Tou, T.; Tsai, T.; Ngai, K.; et al. Identification of metabolite markers associated with kidney function. J. Immunol. Res. 2022, 2022, 1-9. [CrossRef]
- Peng, H.; Liu, X.; Ieong, C.A.; Tou, T.; Tsai, T.; Zhu, H.; et al. A metabolomics study of metabolites associated with the glomerular filtration rate. BMC Nephrol. 2023, 24, 105. [CrossRef]
- Grams, M.E.; Tin, A.; Rebholz, C.M.; Shafi, T.; Köttgen, A.; Perrone, R.D.; et al. Metabolomic alterations associated with cause of CKD. Clin. J. Am. Soc. Nephrol. 2017, 12, 1787-1794. [CrossRef]
- Lin, B.M.; Zhang, Y.; Yu, B.; Boerwinkle, E.; Thygarajan, B.; Yunes, M.; et al. Metabolome-wide association study of estimated glomerular filtration rates in Hispanics. Kidney Int. 2022, 101, 144-151. [CrossRef]
- Schlosser, P.; Scherer, N.; Grundner-Culemann, F.; Monteiro-Martins, S.; Haug, S.; Steinbrenner, I.; et al. Genetic studies of paired metabolomes reveal enzymatic and transport processes at the interface of plasma and urine. Nat. Genet. 2023, 55, 995-1008. [CrossRef]
- Nierenberg, J.L.; He, J.; Li, C.; Gu, X.; Shi, M.; Razavi, A.C. Novel associations between blood metabolites and kidney function among Bogalusa Heart Study and Multi-Ethnic Study of Atherosclerosis participants. Metabolomics 2019, 15, 149. [CrossRef]
- Wang, F.; Sun, L.; Sun, Q.; Liang, L.; Gao, X.; Li, R.; et al. Associations of plasma amino acid and acylcarnitine profiles with incident reduced glomerular filtration rate. Clin. J. Am. Soc. Nephrol. 2018, 13, 560-568. [CrossRef]
- Kwan, B.; Fuhrer, T.; Zhang, J.; Darshi, M.; Van Espen, B.; Montemayor, D.; et al. Metabolomic markers of kidney function decline in patients with diabetes: evidence from the Chronic Renal Insufficiency Cohort (CRIC) Study. Am. J. Kidney Dis. 2020, 76, 511-520. [CrossRef]
- Lee, S.; Han, M.; Moon, S.; Kim, K.; An, W.J.; Ryu, H.; et al. Identifying genetic variants and metabolites associated with rapid estimated glomerular filtration rate decline in Korea based on genome-metabolomic integrative analysis. Metabolites 2022, 12, 1139. [CrossRef]
- Wen, D.; Zheng, Z.; Surapaneni, A.; Yu, B.; Zhou, L.; Zhou, W.; et al. Metabolite profiling of CKD progression in the chronic renal insufficiency cohort study. JCI Insight 2022, 7, e161696. [CrossRef]
- Bernard, L.; Zhou, L.; Surapaneni, A.; Chen, J.; Rebholz, C.M.; Coresh, J.; et al. Serum metabolites and kidney outcomes: The Atherosclerosis Risk in Communities Study. Kidney Med. 2022, 4, 100522. [CrossRef]
- Guo, X.; Peng, H.; Liu, P.; Tang, L.; Fang, J.; Aoieong, C.; et al. Novel metabolites to improve glomerular filtration rate estimation. Kidney Blood Press Res. 2023, 48, 287-296. [CrossRef]
- Au, A.Y.M.; Mantik, K.; Bahadory, F.; Stathakis, P.; Guiney, H.; Erlich, J.E.; et al. Plasma arginine metabolites in health and chronic kidney disease. Nephrol. Dial. Transplant. 2023, 38, 2767-2775. [CrossRef]
- Liu, J.-J.; Ching, J.; Wee, H.N.; Liu, S.; Gurung, R.L.; Lee, J.; et al. Plasma tryptophan-kynurenine pathway metabolites and risk for progression to end-stage kidney disease in patients with type 2 diabetes. Diabetes Care 2023, 46, 2223-2231. [CrossRef]
- Hou, Y.; Xiao, Z.; Zhu, Y.; Li, Y.; Liu, Q. Blood metabolites and chronic kidney disease: a Mendelian randomization study. BMC Med. Genomics 2024, 17, 147. [CrossRef]
- Luo, Y.; Zhang, W.; Qin, G. Metabolomics in diabetic nephropathy: Unveiling novel biomarkers for diagnosis. Mol. Med. Rep. 2024, 30, 156. [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622-D631. [CrossRef]
- Stelzer, G.; Rosen, R.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; et al. The GeneCards Suite: From gene data mining to disease genome sequence analysis. Curr. Protoc. Bioinform. 2016, 54, 1.30.1-1.30.33.
- Jellum, E.; Horn, L.; Thoresen, O.; Kvittingen, E.A.; Stokke, O. Urinary excretion of N-acetyl amino acids in patients with some inborn errors of amino acid metabolism. Scand. J. Clin. Lab. Investig. 1986, 184, 21-26.
- Engelke, U.F.; Sass, O.; Van Coster, R.N.; Gerlo, E.; Olbrich, H.; Krywawych, S.; et al. NMR spectroscopy of aminoacylase 1 deficiency, a novel inborn error of metabolism. NMR Biomed. 2008, 21, 138-147. [CrossRef]
- Okajima, K.; Inoue, M.; Morino, Y. Studies on the mechanism for renal elimination of N-acetylphenylalanine: Its pathophysiologic significance in phenylketonuria. J. Lab. Clin. Med. 1985, 105, 132-138.
- Sass, J.O.; Mohr, V.; Olbrich, H.; Engelke, U.; Horvath, J.; Fliegauf, M.; et al. Mutations in ACY1, the gene encoding aminoacylase 1, cause a novel inborn error of metabolism. Am. J. Hum. Genet. 2006, 78, 401-409. [CrossRef]
- Van Coster, R.N.; Gerlo, E.A.; Giardina, T.G.; Engelke, U.F.; Smet, J.E.; De Praeter, C.M.; et al. Aminoacylase I deficiency: A novel inborn error of metabolism. Biochem. Biophys. Res. Commun. 2005, 338, 1322-1326. [CrossRef]
- Luo, G.; Ducy, P.; McKee, M.D.; et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 1997, 386, 78-81. [CrossRef]
- Chatrou, M.L.; Winckers, K.; Hackeng, T.M.; Reutelingsperger, C.P.; Schurgers, L.J. Vascular calcification: the price to pay for anticoagulation therapy with vitamin K-antagonists. Blood Rev. 2012, 26, 155-166. [CrossRef]
- Budoff, M.J.; Rader, D.J.; Reilly, M.P.; Mohler, E.R., 3rd; Lash, J.; Yang, W.; et al. Relationship of estimated GFR and coronary artery calcification in the CRIC (Chronic Renal Insufficiency Cohort) Study. Am. J. Kidney Dis. 2011, 58, 519-526.
- Jansen, R.S.; Addie, R.; Merkx, R.; et al. N-lactoyl-amino acids are ubiquitous metabolites that originate from CNDP2-mediated reverse proteolysis of lactate and amino acids. Proc. Natl. Acad. Sci. USA 2015, 112, 6601-6606. [CrossRef]
- Hennermann, J.B.; Roloff, S.; Gellermann, J.; Vollmer, I.; Windt, E.; Vetter, B.; et al. Chronic kidney disease in adolescent and adult patients with phenylketonuria. J. Inherit. Metab. Dis. 2013, 36, 747-756. [CrossRef]
- Burton, B.K.; Bradford Jones, K.; Cederbaum, S.; Rohr, F. Prevalence of comorbid conditions among adult patients diagnosed with phenylketonuria. Mol. Genet. Metab. 2018, 125, 228-234. [CrossRef]
- Hagenfeldt, L.; Naglo, A.S. New conjugated urinary metabolites in intermediate type maple syrup urine disease. Clin. Chim. Acta 1987, 169, 77-83. [CrossRef]
- Maceda, E.B.G.; Abadingo, M.E.; Magbanua-Calalo, C.J.; Dator, M.A.; Resontoc, L.P.R.; Castro-Hamoy, L.; et al. Maple syrup urine disease associated with nephrotic syndrome in a Filipino child. BMJ Case Rep. 2021, 14, e242689. [CrossRef]
- Zhang, Z.H.; He, J.Q.; Qin, W.W.; Zhao, Y.Y.; Tan, N.H. Biomarkers of obstructive nephropathy using a metabolomics approach in rat. Chem. Biol. Interact. 2018, 296, 229-239. [CrossRef]
- Rysz, J.; Franczyk, B.; Rysz-Górzyńska, M.; Gluba-Brzózka, A. Are alterations in DNA methylation related to CKD development? Int. J. Mol. Sci. 2022, 23, 7108.
- Ganesan, L.L.; O'Brien, F.J.; Sirich, T.L.; Plummer, N.S.; Sheth, R.; Fajardo, C.; et al. Association of plasma uremic solute levels with residual kidney function in children on peritoneal dialysis. Clin. J. Am. Soc. Nephrol. 2021, 16, 1531-1538. [CrossRef]
- He, W.J.; Chen, J.; Razavi, A.C.; Hu, E.A.; Grams, M.E.; Yu, B.; et al. Metabolites associated with coffee consumption and incident chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2021, 16, 1620-1629. [CrossRef]
- Laakso, M.; Kuusisto, J.; Stančáková, A.; Kuulasmaa, T.; Pajukanta, P.; Lusis, A.J.; et al. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 2017, 58, 481-493. [CrossRef]
- Yin, X.; Chan, L.S.; Bose, D.; Jackson, A.U.; VandeHaar, P.; Locke, A.E.; et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 2022, 13, 1644. [CrossRef]
- Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2010, 33, S62-S69.
- Inker, L.A.; Eneanya, N.D.; Coresh, J.; Tighiouart, H.; Wang, D.; Sang, Y.; et al. New creatinine- and cystatin C-based equations to estimate GFR without race. N. Engl. J. Med. 2021, 385, 1737-1749. [CrossRef]
- Fernandes Silva, L.; Vangipurapu, J.; Kuulasmaa, T.; Laakso, M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019, 9, 10240. [CrossRef]
| Measurements | NGT (n=3034) |
Prediabetes (n=5715) |
T2D (n=1410) |
p |
|---|---|---|---|---|
| Age (years) | 56.8 ± 6.9 | 57.4 ± 7.2 | 60.6 ± 6.7 | 1.1E-63 |
| Systolic blood pressure (mmHg) | 134.3 ± 15.9 | 138.7 ± 16.2 | 145.2 ± 18.1 | 2.1E-93 |
| Body mass index (kg/m2) | 25.8 ± 3.38 | 27.4 ± 3.9 | 30.2 ± 5.2 | 1.1E-247 |
| Current smoking (%) | 18.0 | 18.4 | 17.2 | 0.606 |
| Total triglycerides (mmol/l) | 1.22 ± 0.65 | 1.49 ± 1.08 | 1.90 ± 1.21 | 1.2E-143 |
| Fasting glucose (mmol/l) | 5.24 ± 0.24 | 5.97 ± 0.37 | 7.51 ± 2.01 | < 1E-250 |
| HbA1C (%) | 5.59 ± 0.31 | 5.71 ± 0.34 | 6.58 ± 1.13 | < 1E-250 |
| Fasting plasma insulin (mU/l) | 6.25 ± 4.11 | 9.32 ± 6.4 | 19.6 ± 28.5 | < 1E-250 |
| Creatinine (umol/l) | 84.6 ± 15.9 | 83.4 ± 12.8 | 84. 6 ± 22.3 | 0.0003 |
| eGFR (ml/min/1.73 m2) | 87.9 ± 12.3 | 88.6 ± 12.2 | 86.1 ± 14.5 | 4.5E-10 |
| Urine albumin (mg/l) | 18.4 ± 110.9 | 20.6 ± 82.5 | 93.5 ± 380.1 | 7.2E-181 |
| hs-CRP (mg/l) | 1.82 ± 2.96 | 2.13 ± 4.5 | 3.22 ± 6.07 | 3.4E-40 |
|
Abbreviations: NGT, normal glucose tolerance; T2D, type 2 diabetes; HbA1C, hemoglobin A1C; eGFR, estimated glomerular filtration rate; hs-CRP, high sensitivity C-reactive protein | ||||
| Metabolite | Sub-class | N | Beta | p * | Beta | p ** |
|---|---|---|---|---|---|---|
| Amino acids | ||||||
| N-acetylmethionine |
Methionine, cysteine, taurine metab. | 7080 | -0.334 | 1.4E-183 | -0.087 | 5.5E-24 |
| N-acetylvaline | Leucine, isoleucine, valine metab. | 7082 | -0.343 | 1.0E-194 | -0.082 | 2.6E-21 |
| γ-carboxyglutamate | Glutamate metab. | 6929 | -0.295 | 1.1E-138 | -0.065 | 2.6E-14 |
| 3-methylglutaryl- carnitine (2) |
Leucine, isoleucine, valine metab. | 7001 | -0.257 | 1.1E-105 | -0.058 | 5.8E-12 |
| Proline | Urea cycle; arginine proline metab. | 7081 | -0.107 | 1.3E-19 | -0.048 | 3.9E-9 |
| Pro-hydroxy-pro | Urea cycle; arginine proline metab. | 7079 | -0.155 | 1.9E-39 | -0.047 | 5.2E-9 |
| 4-guanidinobutanoate | Guanidino acetamido metab. | 7049 | -0.158 | 1.7E-40 | -0.049 | 2.3E-9 |
| N-acetyltaurine | Methionine, cysteine, taurine metab. | 7048 | -0.208 | 1.4E-69 | -0.041 | 7.6E-7 |
| Hydantoin-5-propionate | Histidine metab. | 6154 | -0.211 | 3.6E-63 | -0.043 | 1.1E-6 |
| N-lactoyl valine | Lactoyl amino acid | 6781 | -0.182 | 2.5E-51 | -0.043 | 3.1E-6 |
| N-lactoylisoleucine | Lactoyl amino acid | 5437 | -0.189 | 4.4E-45 | -0.043 | 1.6E-5 |
| N-lactoyl phenylalanine | Lactoyl amino acid | 7033 | -0.233 | 2.7E-87 | -0.037 | 4.4E-5 |
| Lipids | ||||||
| 11beta-hydroxy etiocholanolone glucuronide |
Androgenic steroids | 4891 | -0.204 | 2.9E-47 | -0.050 | 4.0E-7 |
| 3-decenoylcarnitine | Fatty acid metab. | 5395 | -0.217 | 2.9E-58 | -0.042 | 9.2E-6 |
| Cis-3,4-methylene heptanoylglycine | Fatty acid metab. | 6825 | -0.161 | 5.2E-41 | -0.038 | 4.8E-6 |
| 2-methylmalonyl carnitine (C4-DC) |
Fatty acid metab. | 5827 | -0.235 | 8.0E-74 | -0.042 | 3.1E-6 |
| Propionylglycine | Fatty acid metab | 3960 | -0.119 | 4.9E-14 | -0.049 | 1.3E-5 |
| Nucleotide | ||||||
| 5-methyluridine(ribothymidine) | Pyrimidine metab. | 7082 | -0.134 | 6.8E-30 | -0.038 | 3.1E-6 |
| Peptide | ||||||
| Pyroglutamylvaline | Modified peptides | 6398 | -0.202 | 7.7E-60 | -0.051 | 2.6E-9 |
| Xenobiotics | ||||||
| 2,3-dihydroxyisovalerate | Food component/plant | 6998 | -0.206 | 3.8E-68 | -0.048 | 6.8E-9 |
| (S)-a-amino-omega-caprolactam | Food component/plant | 7007 | -0.296 | 1.3E-141 | -0.050 | 1.0E-8 |
| 3-methoxycatechol sulfate (2) | Benzoate metab. | 5379 | -0.185 | 2.0E-42 | -0.044 | 1.9E-6 |
| 3-methyl catechol sulfate (1) | Benzoate metab. | 7065 | -0.209 | 3.0E-70 | -0.040 | 2.1E-6 |
| 3-methoxycatechol sulfate (1) | Benzoate metab. | 6318 | -0.174 | 4.0E-44 | -0.039 | 5.5E-6 |
| 2-acetamidophenol sulfate | Food component/plant | 5939 | -0.153 | 2.9E-32 | -0.042 | 3.6E-6 |
| N-(2-furoyl)glycine | Food component/plant | 5025 | -0.235 | 5.0E-64 | -0.042 | 2.4E-5 |
| 2-aminophenol sulfate | Food component/plant | 7066 | -0.147 | 2.8E-35 | -0.036 | 1.1E-5 |
| Other metabolite | ||||||
| Glutamine_degradant | Partially characterized molecules | 7060 | -0.222 | 7.3E-80 | -0.071 | 2.2E-17 |
| p*: non-adjusted; p**: adjusted for eGFR at baseline, age, BMI, smoking, fasting glucose, total triglycerides and systolic blood pressure. | ||||||
| Gene-variant | Metabolite | p |
|---|---|---|
| KLHDC7B-rs470118 | 5-methyluridine | 9.9E-199 |
| CPS1-rs715 | Glycine | 8.1E-90 |
| AC007326.4-rs5992344 | Proline | 2.0E -63 |
| DOCK3- rs138144932 | N-acetylmethionine | 1.3E -44 |
| AOX1-rs7562507 | Hydantoin-5-propionate | 1.4E-17 |
| COLEC10-rs13264172 | Pro-hydroxy-pro | 3.5E-10 |
| MAGI1-rs264676 | 2.3-dihydroxy-5-methylthio-4-penenoate | 2.9E-8 |
| DCBLD2- rs192423025 | Pyroglutamylvaline | 3.4E-8 |
| CNTNAP2-rs533473709 | γ-carboxyglutamate | 5.3 E-8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





