Submitted:
16 August 2024
Posted:
20 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Case study and Methods
2.1. Case Presentation
2.2. Methods
-
Prevention Genetics Hereditary Hearing Loss and Deafness Panel: https://www.preventiongenetics.com/testInfo?val=Hereditary-Hearing-Loss-and-Deafness-PanelInvitae Limb and Digital Malformations Panel: https://www.invitae.com/us/providers/test-catalog/test-55010
3. Results
4. Discussion
| Goldenhar Syndrome Phenotypes and their Prevalence [4,10] | Proband Phenotypes | 16p12.2 Deletion Phenotypes [46,47] | sSRC19 Phenotype [37] |
Possible Variant association and Genes |
|---|---|---|---|---|
| Head and Face Anomalies/Hemifacial microsomia (~83.5%) | + | - | + | + (CHUK, TCTN2) |
| Ocular Anomalies (23%) | + | - | - | + (ADGRV1, CDH23, ADAMTS10, CHUK, CSPP1) |
| Ear Anomalies/Sensorineural defects/Deafness (~55%) | + | + | - | + (MYH14, PDE1C, ADGRV1, CDH23, MYO3A) |
| Cardiac Anomalies & Congenital Heart Defects (~20.16%) | + | + | + | + (ADAMST10) |
| Skeletal Anomalies (~27%) | + | + | + | + (ADAMTS10, CHUK, TCTN2) |
| Urogenital Anomalies (~11.5%) | + | + | - | + (CHUK, TCTN2) |
| Pulmonary Anomalies (~8%) | + | - | - | + (CSPP1, ADAMTS10) |
| Gastrointestinal Anomalies (~7%) | + | - | - |
- |
| Psychiatric & Behavioral Anomalies | + | + | - | + (CSPP1, TCTN2) |
| Intellectual Disability | - | + | + | + (CSPP1, TCTN2) |
| Developmental Delay (~11.5%) | - | + | + | + (CSPP1, TCTN2) |
| Central Nervous System Anomalies (~10%) | - | - | + | + (CHUK, CSPP1, TCTN2) |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tidke SC, Vagha JD, Vagha K, Lohiya S, Hampe P. An Atypical Presentation of Goldenhar Syndrome With Seizures: A Rare Case Report. Cureus. 2023;15(10):e46627. Published 2023 Oct 7. [CrossRef]
- Tuna EB, Orino D, Ogawa K, et al. Craniofacial and dental characteristics of Goldenhar syndrome: a report of two cases. J Oral Sci. 2011;53(1):121-124. [CrossRef]
- Kulkarni V, Shah MD, Parikh A. Goldenhar syndrome (a case report). J Postgrad Med. 1985;31(3):177-179.
- Bogusiak K, Puch A, Arkuszewski P. Goldenhar syndrome: current perspectives. World J Pediatr. 2017;13(5):405-415. [CrossRef]
- Maldonado J, Revuelta Barbero JM, Rodas A, et al. Endoscopic Endonasal Odontoidectomy for Upper Cervical Spine and Brainstem Decompression in a Patient With Goldenhar Syndrome: 2-Dimensional Operative Video. Oper Neurosurg (Hagerstown). Published online November 23, 2023. [CrossRef]
- Lima Mde D, Marques YM, Alves Sde M Jr, Ortega KL, Soares MM, Magalhães MH. Distraction osteogenesis in Goldenhar Syndrome: case report and 8-year follow-up. Med Oral Patol Oral Cir Bucal. 2007;12(7):E528-E531. Published 2007 Nov 1.
- Jayaprakasan SK, Waheed MD, Batool S, Pimentel Campillo J, Nageye ME, Holder SS. Goldenhar Syndrome: An Atypical Presentation With Developmental and Speech Delay. Cureus. 2023;15(3):e36225. Published 2023 Mar 16. [CrossRef]
- Wilson GN. Cranial defects in the Goldenhar syndrome. Am J Med Genet. 1983;14(3):435-443. [CrossRef]
- Madiyal, A., Babu, S.G., Ajila, V., Madi, M., & Bhat, S. (2018). Goldenhar syndrome: Report of two cases with review of literature. CHRISMED Journal of Health and Research, 5, 67 - 71.
- Beleza-Meireles A, Clayton-Smith J, Saraiva JM, Tassabehji M. Oculo-auriculo-vertebral spectrum: a review of the literature and genetic update. J Med Genet. 2014;51(10):635-645. [CrossRef]
- Strömland K, Miller M, Sjögreen L, et al. Oculo-auriculo-vertebral spectrum: associated anomalies, functional deficits and possible developmental risk factors. Am J Med Genet A. 2007;143A(12):1317-1325. [CrossRef]
- Tingaud-Sequeira A, Trimouille A, Sagardoy T, Lacombe D, Rooryck C. Oculo-auriculo-vertebral spectrum: new genes and literature review on a complex disease. J Med Genet. 2022 May;59(5):417-427. Epub 2022 Feb 2. [CrossRef] [PubMed]
- Li T, Sang H, Chu G, et al. Genotype-phenotype correlation in 75 patients with small supernumerary marker chromosomes. Mol Cytogenet. 2020;13:30. Published 2020 Jul 14. [CrossRef]
- Jafari-Ghahfarokhi H, Moradi-Chaleshtori M, Liehr T, Hashemzadeh-Chaleshtori M, Teimori H, Ghasemi-Dehkordi P. Small supernumerary marker chromosomes and their correlation with specific syndromes. Adv Biomed Res. 2015;4:140. Published 2015 Jul 27. [CrossRef]
- Paoloni-Giacobino A, Morris MA, Dahoun SP. Prenatal supernumerary r(16) chromosome characterized by multiprobe FISH with normal pregnancy outcome. Prenat Diagn. 1998 Jul;18(7):751-2. [PubMed]
- Tasse C, Majewski F, Böhringer S, et al. A family with autosomal dominant oculo-auriculo-vertebral spectrum. Clin Dysmorphol. 2007;16(1):1-7. [CrossRef]
- Vendramini-Pittoli S, Kokitsu-Nakata NM. Oculoauriculovertebral spectrum: report of nine familial cases with evidence of autosomal dominant inheritance and review of the literature. Clin Dysmorphol. 2009;18(2):67-77. [CrossRef]
- Tsai FJ, Tsai CH. Autosomal dominant inherited oculo-auriculo-vertebral spectrum: report of one family. Zhonghua Min Guo Xiao Er Ke Yi Xue Hui Za Zhi. 1993;34(1):27-31.
- Goodin K, Prucka S, Woolley AL, et al. Familial transmission of oculoauriculovertebral spectrum (Goldenhar syndrome) is not due to mutations in either EYA1 or SALL1. Am J Med Genet A. 2009;149A(3):535-538. [CrossRef]
- Kaye CI, Martin AO, Rollnick BR, et al. Oculoauriculovertebral anomaly: segregation analysis. Am J Med Genet. 1992;43(6):913-917. [CrossRef]
- Rollnick BR. Oculoauriculovertebral anomaly: variability and causal heterogeneity. Am J Med Genet Suppl. 1988;4:41-53. [CrossRef]
- Rollnick BR, Kaye CI, Nagatoshi K, Hauck W, Martin AO. Oculoauriculovertebral dysplasia and variants: phenotypic charac-teristics of 294 patients. Am J Med Genet. 1987;26(2):361-375. [CrossRef]
- Descartes M. Oculoauriculovertebral spectrum with 5p15.33-pter deletion. Clin Dysmorphol. 2006;15(3):153-154. [CrossRef]
- Josifova DJ, Patton MA, Marks K. Oculoauriculovertebral spectrum phenotype caused by an unbalanced t(5;8)(p15.31;p23.1) rearrangement. Clin Dysmorphol. 2004;13(3):151-153. [CrossRef]
- Ladekarl S. Combination of Goldenhar’s syndrome with the Cri-Du-Chat syndrome. Acta Ophthalmol (Copenh). 1968;46(3):605-610. [CrossRef]
- Ballesta-Martínez MJ, López-González V, Dulcet LA, Rodríguez-Santiago B, Garcia-Miñaúr S, Guillen-Navarro E. Autosomal dominant oculoauriculovertebral spectrum and 14q23.1 microduplication. Am J Med Genet A. 2013;161A(8):2030-2035. [CrossRef]
- Ou Z, Martin DM, Bedoyan JK, et al. Branchiootorenal syndrome and oculoauriculovertebral spectrum features associated with duplication of SIX1, SIX6, and OTX2 resulting from a complex chromosomal rearrangement. Am J Med Genet A. 2008;146A(19):2480-2489. [CrossRef]
- Rooryck C, Stef M, Burgelin I, et al. 2.3 Mb terminal deletion in 12p13.33 associated with oculoauriculovertebral spectrum and evaluation of WNT5B as a candidate gene. Eur J Med Genet. 2009;52(6):446-449. [CrossRef]
- Abdelmoity AT, Hall JJ, Bittel DC, Yu S. 1.39 Mb inherited interstitial deletion in 12p13.33 associated with developmental delay. Eur J Med Genet. 2011;54(2):198-203. [CrossRef]
- Herman GE, Greenberg F, Ledbetter DH. Multiple congenital anomaly/mental retardation (MCA/MR) syndrome with Goldenhar complex due to a terminal del(22q). Am J Med Genet. 1988;29(4):909-915. [CrossRef]
- Xu J, Fan YS, Siu VM. A child with features of Goldenhar syndrome and a novel 1.12 Mb deletion in 22q11.2 by cytogenetics and oligonucleotide array CGH: is this a candidate region for the syndrome?. Am J Med Genet A. 2008;146A(14):1886-1889. [CrossRef]
- Digilio MC, McDonald-McGinn DM, Heike C, et al. Three patients with oculo-auriculo-vertebral spectrum and microdeletion 22q11.2. Am J Med Genet A. 2009;149A(12):2860-2864. [CrossRef]
- Tan TY, Collins A, James PA, et al. Phenotypic variability of distal 22q11.2 copy number abnormalities. Am J Med Genet A. 2011;155A(7):1623-1633. [CrossRef]
- Quintero-Rivera F, Martinez-Agosto JA. Hemifacial microsomia in cat-eye syndrome: 22q11.1-q11.21 as candidate loci for facial symmetry. Am J Med Genet A. 2013;161A(8):1985-1991. [CrossRef]
- Torti EE, Braddock SR, Bernreuter K, Batanian JR. Oculo-auriculo-vertebral spectrum, cat eye, and distal 22q11 microdeletion syndromes: a unique double rearrangement. Am J Med Genet A. 2013;161A(8):1992-1998. [CrossRef]
- de Ravel TJ, Legius E, Brems H, Van Hoestenberghe R, Gillis PH, Fryns JP. Hemifacial microsomia in two patients further sup-porting chromosomal mosaicism as a causative factor. Clin Dysmorphol. 2001;10(4):263-267. [CrossRef]
- Melis D, Genesio R, Del Giudice E, et al. Selective cognitive impairment and tall stature due to chromosome 19 supernumerary ring. Clin Dysmorphol. 2012;21(1):27-32. [CrossRef]
- Novelli A, Ceccarini C, Bernardini L, et al. Pure trisomy 19p syndrome in an infant with an extra ring chromosome. Cytogenet Genome Res. 2005;111(2):182-185. [CrossRef]
- Vraneković J, Brajenović-Milić B, Modrusan-Mozetić Z, Babić I, Kapović M. Severe psychomotor retardation in a boy with a small supernumerary marker chromosome 19p. Cytogenet Genome Res. 2008;121(3-4):298-301. [CrossRef]
- Tercanli S, Hösli I, Berlinger A, Beyer R, Achermann J, Holzgreve W. Prenatal diagnosis of a partial trisomy 19q. Prenat Diagn. 2000;20(8):663-665. [CrossRef]
- Eichler EE, Hoffman SM, Adamson AA, et al. Complex beta-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 1998;8(8):791-808. [CrossRef]
- Hoovers JM, Mannens M, John R, et al. High-resolution localization of 69 potential human zinc finger protein genes: a number are clustered. Genomics. 1992;12(2):254-263. [CrossRef]
- Klug, A., Schwabe, J. W. R. Zinc fingers. FASEB J. 9, 597-604 (1995) . [CrossRef]
- Bu S, Lv Y, Liu Y, Qiao S, Wang H. Zinc Finger Proteins in Neuro-Related Diseases Progression. Front Neurosci. 2021;15:760567. Published 2021 Nov 18. [CrossRef]
- Gusic M, Schottmann G, Feichtinger RG, et al. Bi-Allelic UQCRFS1 Variants Are Associated with Mitochondrial Complex III De-ficiency, Cardiomyopathy, and Alopecia Totalis. Am J Hum Genet. 2020;106(1):102-111. [CrossRef]
- Girirajan S, Pizzo L, Moeschler J, et al. 16p12.2 Recurrent Deletion. 2015 Feb 26 [Updated 2018 Sep 13]. In: Adam MP, Feldman J, Mirzaa GM, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2024. Available from: https://www.ncbi.nlm.nih.gov/books/NBK274565/.
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010 Mar;42(3):203-9. Epub 2010 Feb 14. [CrossRef] [PubMed] [PubMed Central]
- Zamariolli M, Colovati M, Moysés-Oliveira M, Nunes N, Caires Dos Santos L, Alvarez Perez AB, Bragagnolo S, Melaragno MI. Rare single-nucleotide variants in oculo-auriculo-vertebral spectrum (OAVS). Mol Genet Genomic Med 2019;7:e00959. [CrossRef]
- Choi BO, Kang SH, Hyun YS, Kanwal S, Park SW, Koo H, Kim SB, Choi YC, Yoo JH, Kim JW, Park KD, Choi KG, Kim SJ, Züchner S, Chung KW. A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum Mutat. 2011 Jun;32(6):669-77. Epub 2011 Apr 7. [CrossRef] [PubMed] [PubMed Central]
- Kim, B. J., Kim, A. R., Han, J. H., Lee, C., Oh, D. Y., & Choi, B. Y. (2017). Discovery of MYH14 as an important and unique deafness gene causing prelingually severe autosomal dominant nonsyndromic hearing loss. The Journal of Gene Medicine, 19(4). [CrossRef]
- Wang, L., Feng, Y., Yan, D., Qin, L., Grati, M., Mittal, R., Li, T., Sundhari, A. K., Liu, Y., Chapagain, P., Blanton, S. H., Liao, S., & Liu, X. (2018). A dominant variant in the PDE1C gene is associated with nonsyndromic hearing loss. Human Genetics, 137(6–7), 437–446. [CrossRef]
- Yoon, P. , Sumalde, A. , Ray, D. , Newton, S. , Cass, S. , Chan, K. & Santos-Cortez, R. (2020). Novel Variants in Hearing Loss Genes and Associations With Audiometric Thresholds in a Multi-ethnic Cohort of US Patients With Cochlear Implants. Otology & Neurotology, 41 (7), 978-985. [CrossRef]
- Pater, J. A., Green, J., O’Rielly, D. D., Griffin, A., Squires, J., Burt, T., Fernandez, S., Fernandez, B., Houston, J., Zhou, J., Roslin, N. M., & Young, T. L. (2019). Novel Usher syndrome pathogenic variants identified in cases with hearing and vision loss. BMC Medical Genetics, 20(1), 68–68. [CrossRef]
- Usami, S., Isaka, Y., Miyagawa, M., & Nishio, S. (2022). Variants in CDH23 cause a broad spectrum of hearing loss: from non-syndromic to syndromic hearing loss as well as from congenital to age-related hearing loss. Human Genetics, 141(3–4), 903–914. [CrossRef]
- Doll, J., Hofrichter, M. A. H., Bahena, P., Heihoff, A., Segebarth, D., Müller, T., Dittrich, M., Haaf, T., & Vona, B. (2020). A novel missense variant in MYO3A is associated with autosomal dominant high-frequency hearing loss in a German family. Molecular Genetics & Genomic Medicine, 8(8), e1343-n/a. [CrossRef]
- Bueno, A. S., Nunes, K., Dias, A. M. M., Alves, L. U., Mendes, B. C. A., Sampaio-Silva, J., Smits, J., Yntema, H. G., Meyer, D., Lezirovitz, K., & Mingroni-Netto, R. C. (2022). Frequency and origin of the c.2090T>G p.(Leu697Trp) MYO3A variant associated with autosomal dominant hearing loss. European Journal of Human Genetics: EJHG, 30(1), 13–21. [CrossRef]
- González-Del Pozo, M., Fernández-Suárez, E., Martín-Sánchez, M., Bravo-Gil, N., Méndez-Vidal, C., Rodríguez-De La Rúa, E., Borrego, S., & Antiñolo, G. (2020). Unmasking Retinitis Pigmentosa complex cases by a whole genome sequencing algorithm based on open-access tools: Hidden recessive inheritance and potential oligogenic variants. Journal of Translational Medicine, 18(1), 73–73. [CrossRef]
- Brodie, K. D., Moore, A. T., Slavotinek, A. M., Meyer, A. K., Nadaraja, G. S., Conrad, D. E., Weinstein, J. E., & Chan, D. K. (2021). Genetic Testing Leading to Early Identification of Childhood Ocular Manifestations of Usher Syndrome. The Laryngoscope, 131(6), E2053–E2059. [CrossRef]
- Steinkellner, H., Etzler, J., Gogoll, L., Neesen, J., Stifter, E., Brandau, O., & Laccone, F. (2015). Identification and molecular characterisation of a homozygous missense mutation in the ADAMTS10 gene in a patient with Weill–Marchesani syndrome. European Journal of Human Genetics : EJHG, 23(9), 1186–1191. [CrossRef]
- Levitas, A., Aspit, L., Lowenthal, N., Shaki, D., Krymko, H., Slanovic, L., Yagev, R., & Parvari, R. (2023). A Novel Mutation in the ADAMTS10 Associated with Weil–Marchesani Syndrome with a Unique Presentation of Developed Membranes Causing Severe Stenosis of the Supra Pulmonic, Supramitral, and Subaortic Areas in the Heart. International Journal of Molecular Sciences, 24(10), 8864-. [CrossRef]
- Marzin P, Rondeau S, Alessandri J, et al. Weill-Marchesani syndrome: natural history and genotype-phenotype correlations from 18 news cases and review of literature Journal of Medical Genetics Published Online First: 21 September 2023. [CrossRef]
- Khandelwal, K. D., Ockeloen, C. W., Venselaar, H., Boulanger, C., Brichard, B., Sokal, E., Pfundt, R., Rinne, T., van Beusekom, E., Bloemen, M., Vriend, G., Revencu, N., Carels, C. E. L., van Bokhoven, H., & Zhou, H. (2017). Identification of a de novo variant in CHUK in a patient with an EEC/AEC syndrome-like phenotype and hypogammaglobulinemia. American Journal of Medical Genetics. Part A, 173(7), 1813–1820. [CrossRef]
- Cadieux-Dion, M., Safina, N. P., Engleman, K., Saunders, C., Repnikova, E., Raje, N., Canty, K., Farrow, E., Miller, N., Zellmer, L., & Thiffault, I. (2018). Novel heterozygous pathogenic variants in CHUK in a patient with AEC-like phenotype, immune deficiencies and 1q21.1 microdeletion syndrome: A case report. BMC Medical Genetics, 19(1), 41–41. [CrossRef]
- Wei, C., Zhang, H., Fu, M., Ye, J., & Yao, B. (2024). Novel compound heterozygous variants in the CSPP1 gene causes Joubert syndrome: case report and literature review of the CSPP1 gene’s pathogenic mechanism. Frontiers in Pediatrics, 12, 1305754-. [CrossRef]
- Ben-Omran, T., Alsulaiman, R., Kamel, H., Shaheen, R., & Alkuraya, F. S. (2015). Intrafamilial clinical heterogeneity of CSPP1-related ciliopathy. American Journal of Medical Genetics. Part A, 167A(10), 2478–2480. [CrossRef]
- Huppke, P., Wegener, E., Böhrer-Rabel, H., Bolz, H. J., Zoll, B., Gärtner, J., & Bergmann, C. (2015). Tectonic gene mutations in patients with Joubert syndrome. European Journal of Human Genetics : EJHG, 23(5), 616–620. [CrossRef]
- Fang, L., Wang, L., Yang, L., Xu, X., Pei, S., & Wu, D. (2023). Novel variants identified in five Chinese families with Joubert Syndrome: a case report. BMC Medical Genomics, 16(1), 1–221. [CrossRef]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010 May 14;86(5):749-64. [CrossRef] [PubMed] [PubMed Central]
- Srivastava, S., Love-Nichols, J.A., Dies, K.A. et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 21, 2413–2421 (2019). [CrossRef]
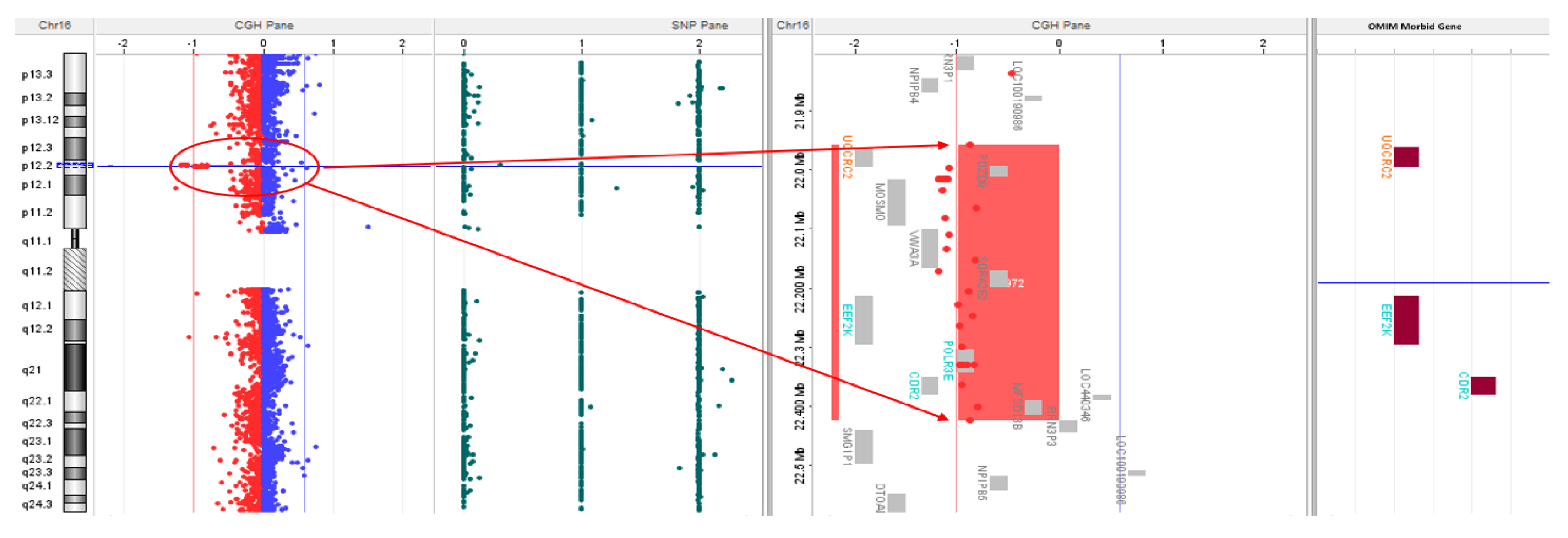
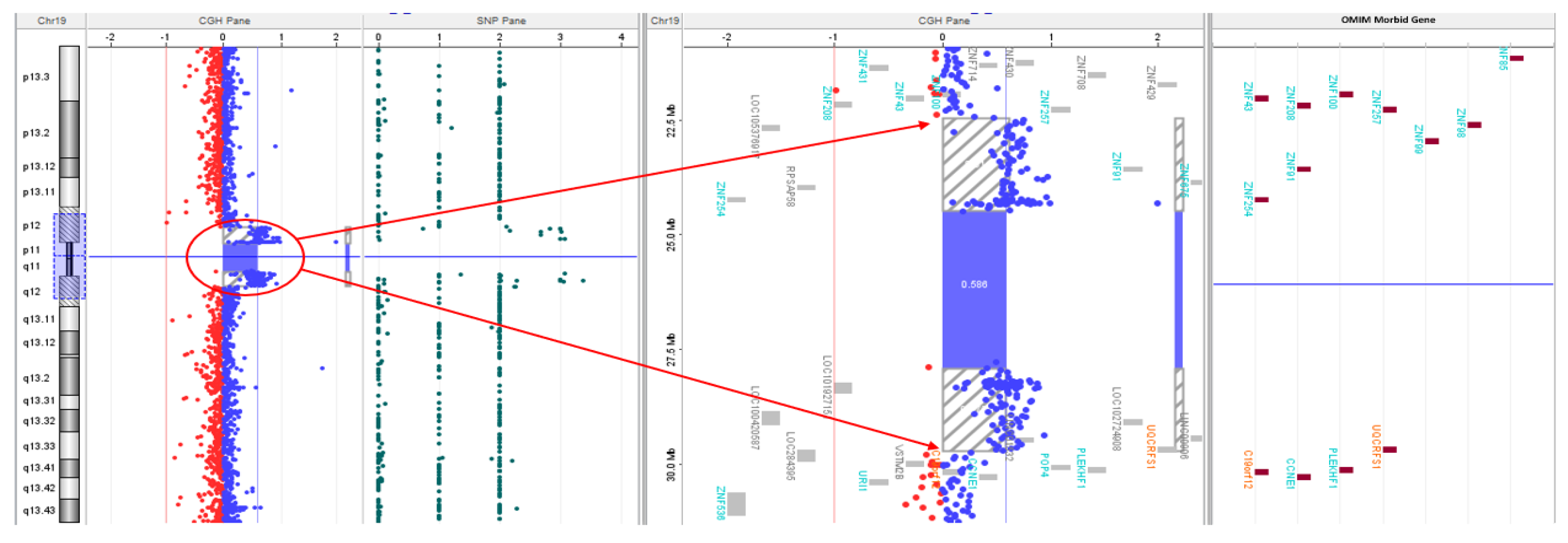
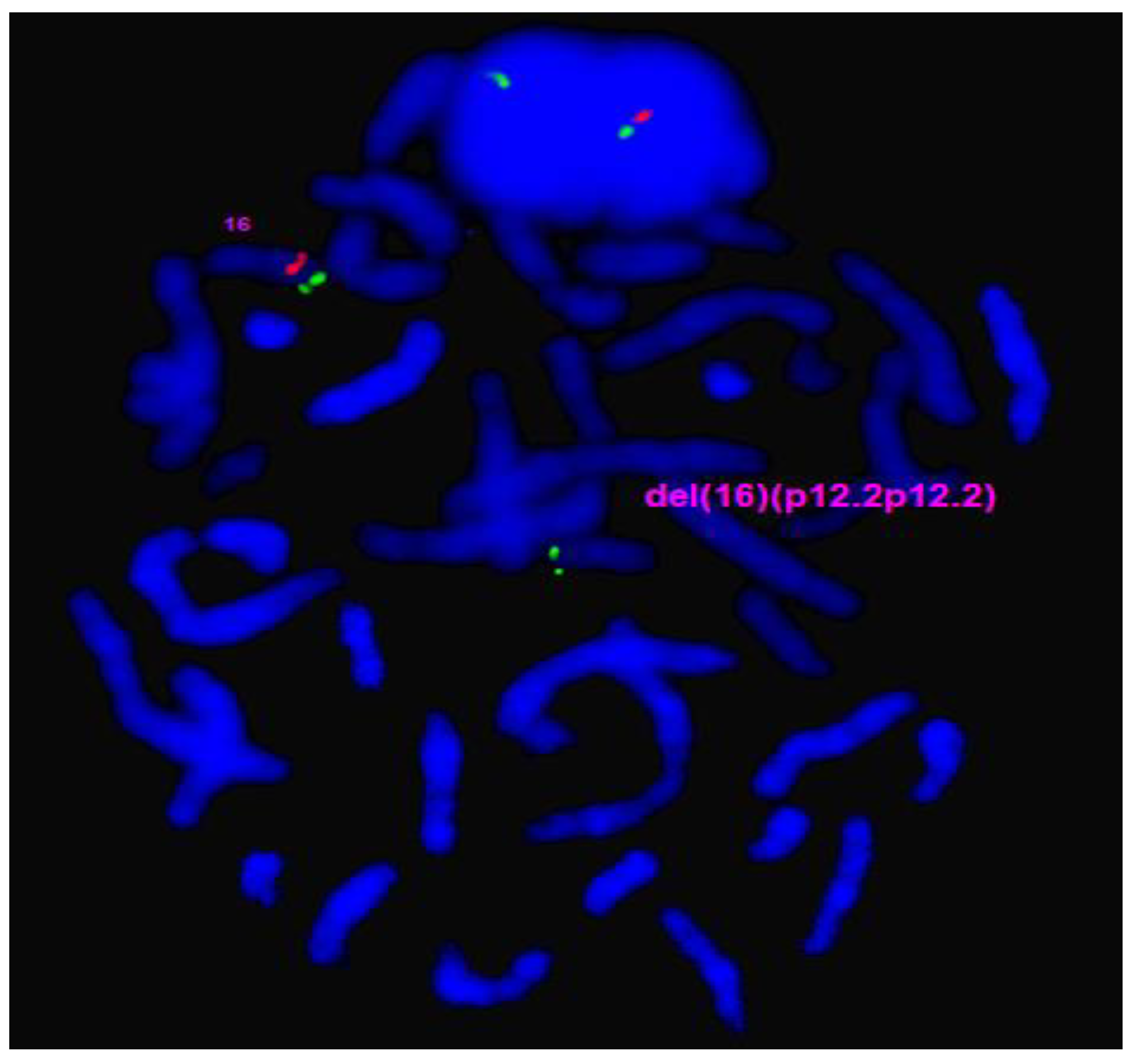
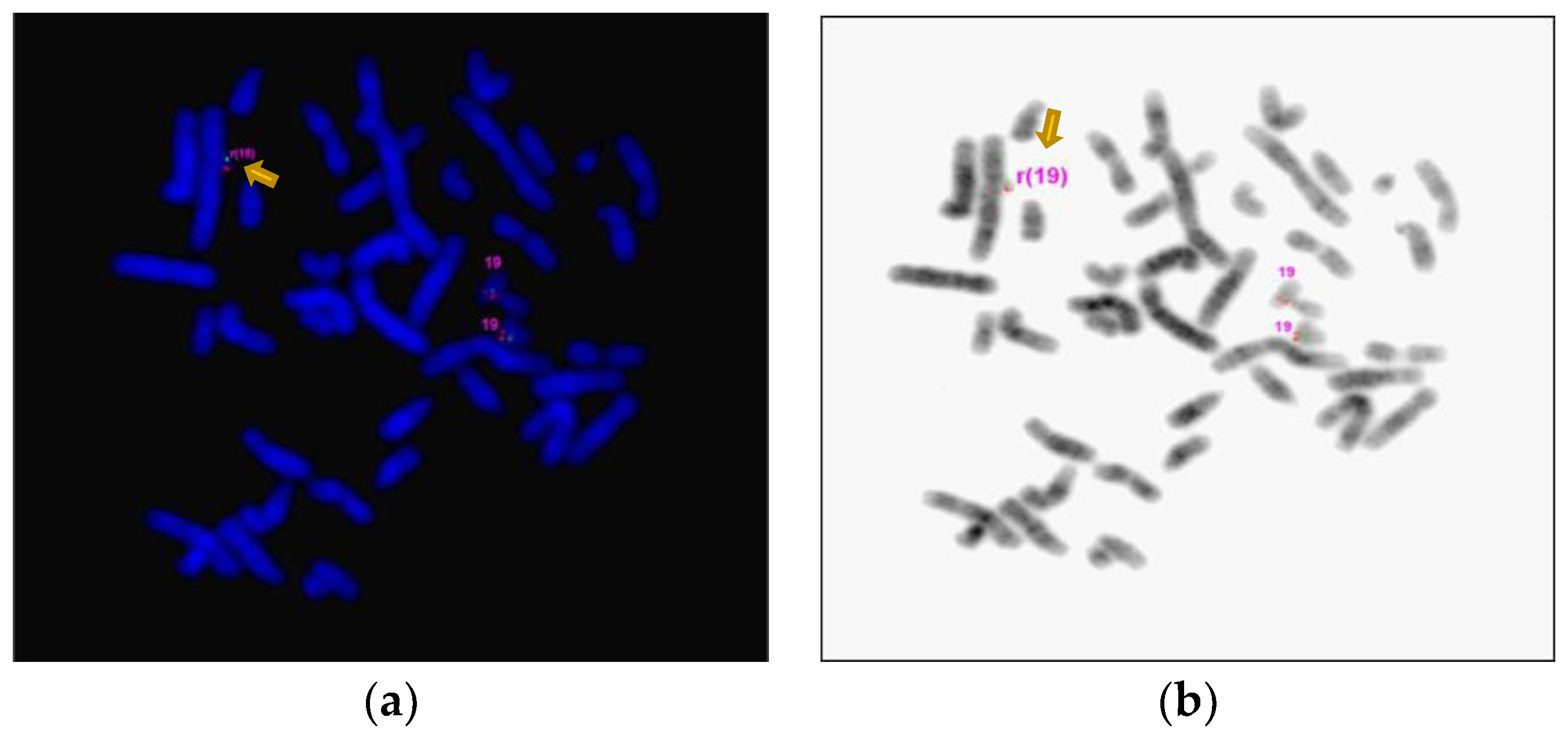
| Gene Transcript | Mode of Inheritance, OMIM ID | DNA variants, Predicted effects, Zygosity |
Highest Allele Frequency in a gnomAD Population |
|---|---|---|---|
|
MYH14, NM_001145809.1 |
Autosomal Dominant, 608568 |
c.4001C>T, p.Ala1334Val, Heterozygous |
0.0071% East Asian |
|
PDE1C, NM_001322059.1 |
Autosomal Dominant, 602987 |
c.8C>T, p.Ser3Leu, Heterozygous |
0.026% European (Non-Finnish) |
|
ADGRV1, NM_032119.3 |
Autosomal Recessive, 602851 |
c.15502A>G, p.Thr5168Ala, Heterozygous |
Not present |
|
CDH23, NM_022124.5 |
Autosomal Recessive, 605516 |
c.8772C>A, p.Ser2924Arg, Apparently Mosaic |
0.00089% European (Non-Finnish) |
|
MYO3A, NM_017433.4 |
Autosomal Recessive, 606808 |
c.484G>A, p.Gly162Ser, Heterozygous |
Not present |
| Gene Transcript |
Mode of Inheritance, OMIM ID |
DNA variants, Predicted effects, Zygosity |
Highest Allele Frequency in a gnomAD Population |
|
ADAMTS10, NM_030957.3 |
Autosomal Recessive, 608990 |
c.506G>A, p.Thr5168Ala, Heterozygous |
Not present |
|
CHUK, NM_001278.4 |
Autosomal Recessive, 600664 |
c.1294C>T, p.Ala1334Val, Heterozygous |
0.0017% European (Non-Finnish) |
|
CSPP1, NM_024790.6 |
Autosomal Recessive, 611654 |
c.1972A>G, p.Arg658Gly, Heterozygous |
0.092% (Admixed American) |
|
TCTN2, NM_024809.4 |
Autosomal Recessive, 613846 |
c.160G>A, p.Val54Met, Heterozygous |
0.0061% (African/African-American) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





