Submitted:
16 August 2024
Posted:
20 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.1.1. Search Strategy
2.1.2. Eligibility Criteria
2.1.3. Study Selection Process
2.2. Data Extraction
2.2.1. Study Characteristics
2.2.2. Quantitative Outcome Measures Of 3D Reconstruction Methods
Trajectory reconstruction errors
Volume reconstruction errors
2.2.3. Analysis of Ablation Experiments
3. Results
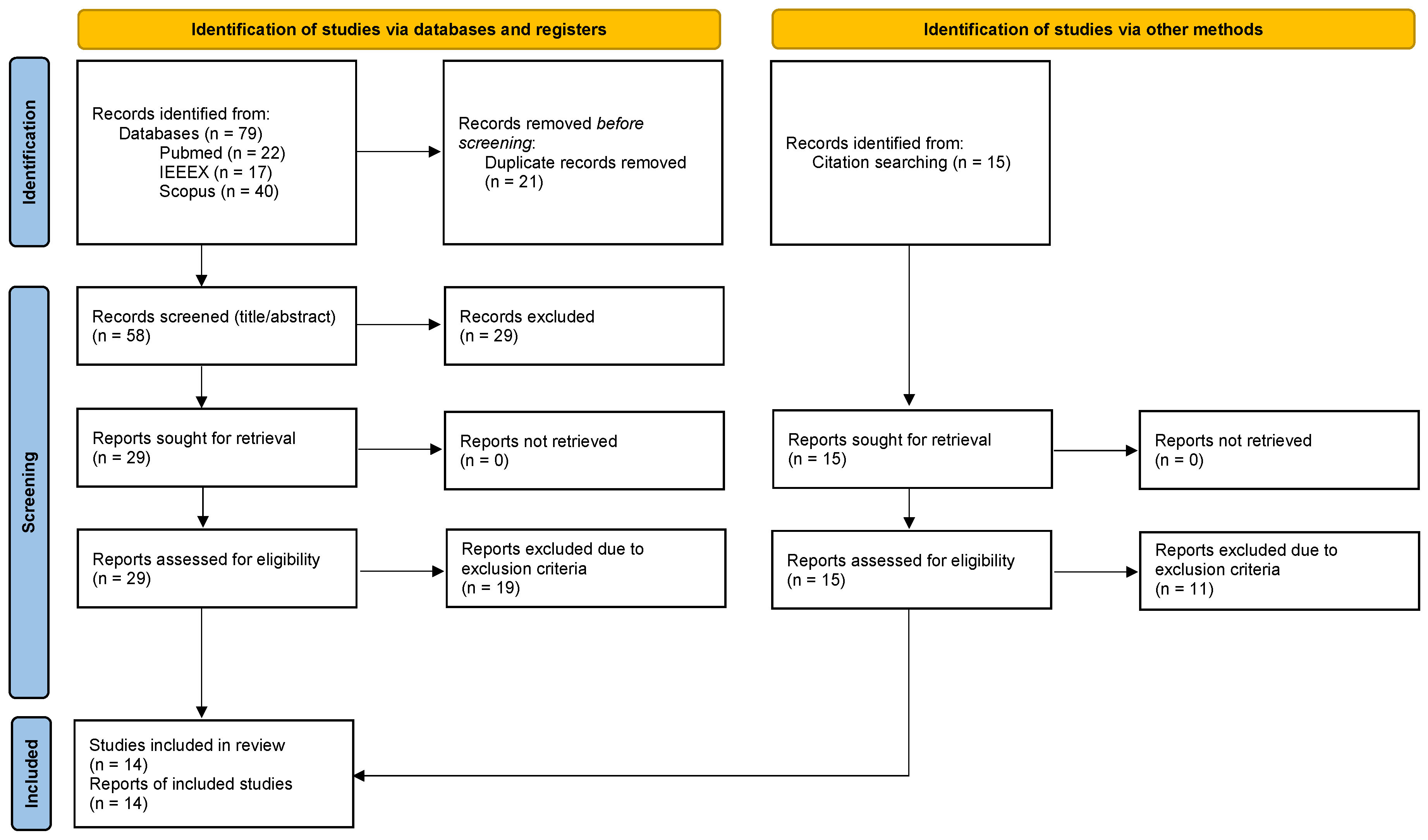
3.1. Study Selection
3.2. Study Characteristics
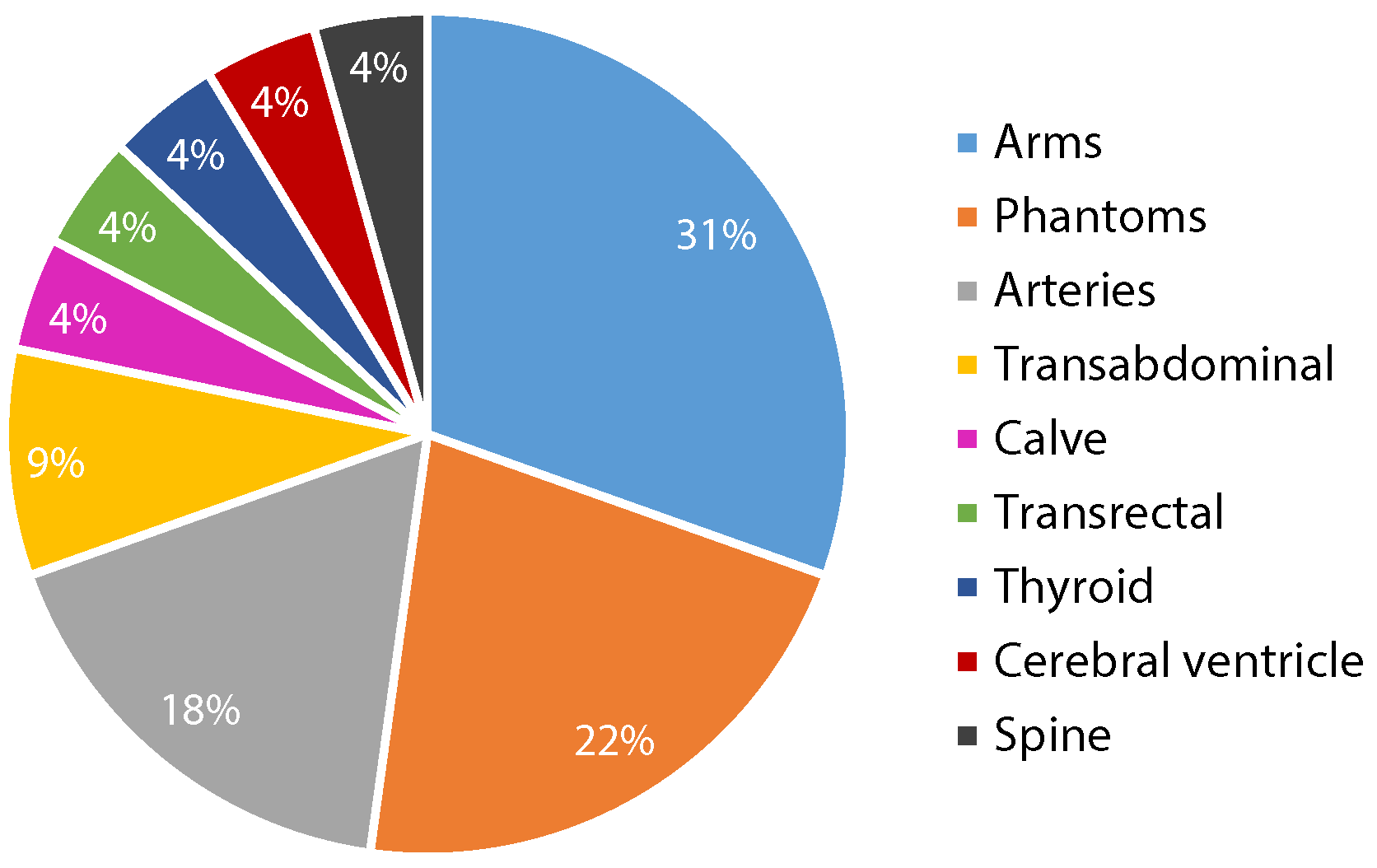
3.2.1. Data Acquisition And Datasets
3.2.2. Reconstruction Methods
Preprocessing
Network architectures
Loss functions
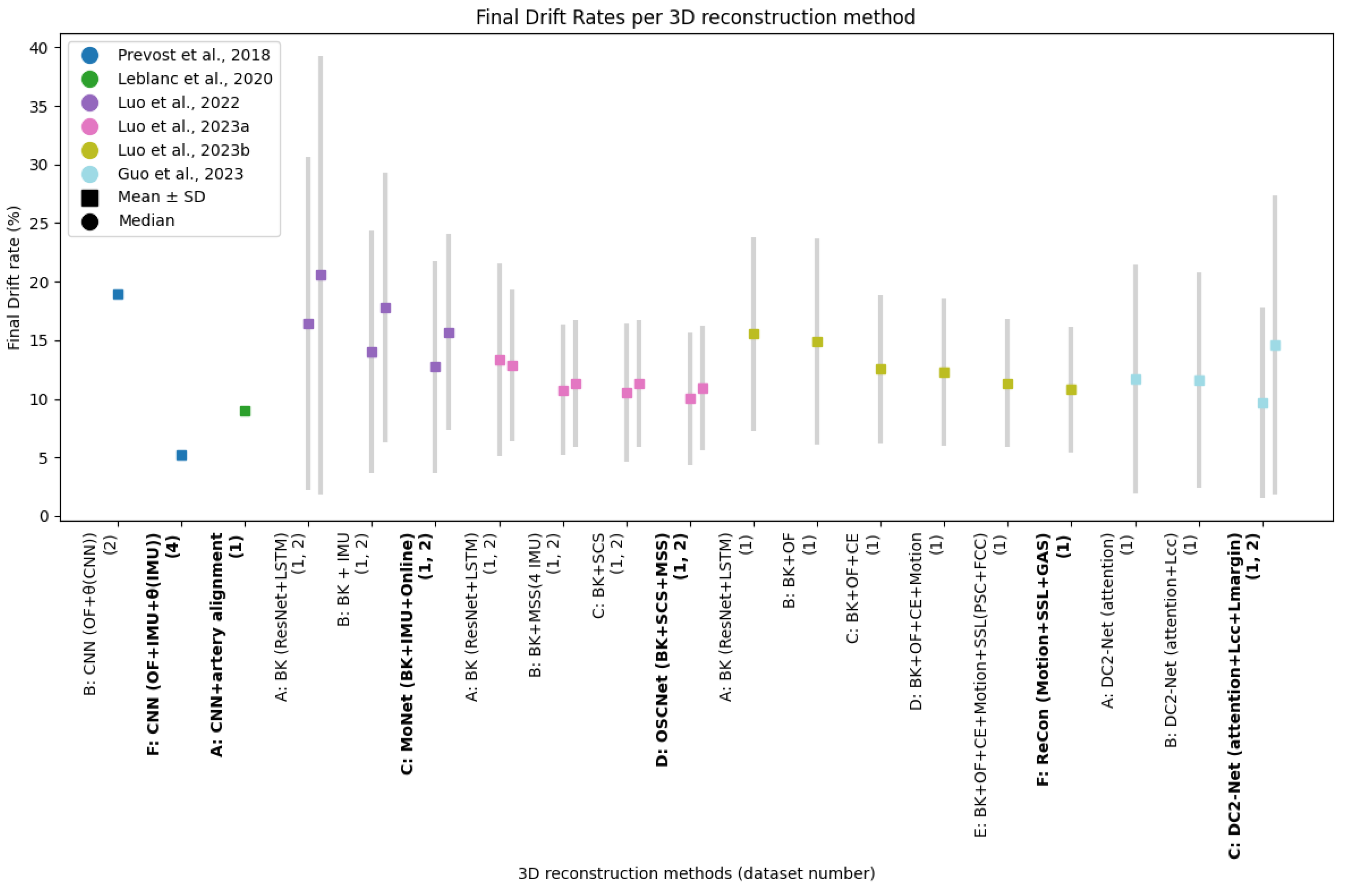
3.3. Quantitative Outcomes
3.3.1. Analysis Of Ablation Experiments
Incorporation of multiple data inputs
Contextual and temporal information
3.3.2. Generalization And Robustness Of Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Search strings
References
- Fenster, A.; Downey, D.B. Three-Dimensional Ultrasound Imaging. Annual Review of Biomedical Engineering 2000, 2, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Mozaffari, M.H.; Lee, W.S. Freehand 3-D Ultrasound Imaging: A Systematic Review. Ultrasound in Medicine & Biology 2017, 43, 2099–2124. [Google Scholar] [CrossRef]
- Lang, A.; Parthasarathy, V.; Jain, A.K. Calibration of EM Sensors for Spatial Tracking of 3 D Ultrasound Probes. In Data Acquisition Applications; IntechOpen, 2012; chapter 0.
- Huang, Q.; Zeng, Z. A Review on Real-Time 3D Ultrasound Imaging Technology. BioMed Research International 2017, 2017, 6027029. [Google Scholar] [CrossRef] [PubMed]
- Clarius Website. http://www.clarius.me 2024.
- Chen, J.F.; Fowlkes, J.B.; Carson, P.L.; Rubin, J.M. Determination of scan-plane motion using speckle decorrelation: Theoretical considerations and initial test. International Journal of Imaging Systems and Technology 1997, 8, 38–44. [Google Scholar] [CrossRef]
- Chang, R.F.; Wu, W.J.; Chen, D.R.; Chen, W.M.; Shu, W.; Lee, J.H.; Jeng, L.B. 3-D US frame positioning using speckle decorrelation and image registration. Ultrasound in Medicine & Biology 2003, 29, 801–812. [Google Scholar] [CrossRef]
- Gee, A.H.; James Housden, R.; Hassenpflug, P.; Treece, G.M.; Prager, R.W. Sensorless freehand 3D ultrasound in real tissue: Speckle decorrelation without fully developed speckle. Medical Image Analysis 2006, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Yung, L.S.; Yu, W. On Feature Motion Decorrelation in Ultrasound Speckle Tracking. IEEE Transactions on Medical Imaging 2013, 32, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Afsham, N.; Rasoulian, A.; Najafi, M.; Abolmaesumi, P.; Rohling, R. Nonlocal means filter-based speckle tracking. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control 2015, 62, 1501–1515. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021, n71. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annual Review of Biomedical Engineering 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Tetrel, L.; Chebrek, H.; Laporte, C. Learning for graph-based sensorless freehand 3D ultrasound. Machine Learning in Medical Imaging (MLMI) 2016, 2016, Vol. 10019 LNCS, pp. 205–212. [Google Scholar] [CrossRef]
- Balakrishnan, S.; Patel, R.; Illanes, A.; Friebe, M. Novel Similarity Metric for Image-Based Out-Of-Plane Motion Estimation in 3D Ultrasound. Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), 2019, pp. 5739–5742. [CrossRef]
- Guo, H.; Xu, S.; Wood, B.; Yan, P. Sensorless Freehand 3D Ultrasound Reconstruction via Deep Contextual Learning. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, 2020, pp. 463–472. [Google Scholar] [CrossRef]
- Miura, K.; Ito, K.; Aoki, T.; Ohmiya, J.; Kondo, S. Localizing 2D Ultrasound Probe from Ultrasound Image Sequences Using Deep Learning for Volume Reconstruction. Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis. ASMUS PIPPI 2020, 2020, 97–105. [Google Scholar] [CrossRef]
- Guo, H.; Xu, S.; Wood, B.J.; Yan, P. Transducer adaptive ultrasound volume reconstruction. IEEE 18th International Symposium on Biomedical Imaging (ISBI), 2021, pp. 511–515. [CrossRef]
- Luo, M.; Yang, X.; Wang, H.; Du, L.; Ni, D. Deep Motion Network for Freehand 3D Ultrasound Reconstruction. International Conference on Medical Image Computing and Computer-Assisted Intervention, 2022, pp. 290–299. [CrossRef]
- Luo, M.; Yang, X.; Wang, H.; Dou, H.; Hu, X.; Huang, Y.; Ravikumar, N.; Xu, S.; Zhang, Y.; Xiong, Y.; et al. RecON: Online learning for sensorless freehand 3D ultrasound reconstruction. Medical Image Analysis 2023, 87. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yang, X.; Yan, Z.; Li, J.; Zhang, Y.; Chen, J.; Hu, X.; Qian, J.; Cheng, J.; Ni, D. Multi-IMU with online self-consistency for freehand 3D ultrasound reconstruction. International Conference on Medical Image Computing and Computer-Assisted Intervention, 2023, pp. 342–351. [CrossRef]
- Li, Q.; Shen, Z.; Li, Q.; Barratt, D.C.; Dowrick, T.; Clarkson, M.J.; Vercauteren, T.; Hu, Y. Long-Term Dependency for 3D Reconstruction of Freehand Ultrasound Without External Tracker. IEEE Transactions on Biomedical Engineering 2024, 71, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Prevost, R.; Salehi, M.; Jagoda, S.; Kumar, N.; Sprung, J.; Ladikos, A.; Bauer, R.; Zettinig, O.; Wein, W. 3D freehand ultrasound without external tracking using deep learning. Medical Image Analysis 2018, 48, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, T.; Lalys, F.; Tollenaere, Q.; Kaladji, A.; Lucas, A.; Simon, A. Stretched reconstruction based on 2D freehand ultrasound for peripheral artery imaging. International Journal of Computer Assisted Radiology and Surgery 2022, 17, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Wein, W.; Lupetti, M.; Zettinig, O.; Jagoda, S.; Salehi, M.; Markova, V.; Zonoobi, D.; Prevost, R. Three-Dimensional Thyroid Assessment from Untracked 2D Ultrasound Clips. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, 2020, 514–523. [Google Scholar] [CrossRef]
- Guo, H.; Chao, H.; Xu, S.; Wood, B.J.; Wang, J.; Yan, P. Ultrasound Volume Reconstruction From Freehand Scans Without Tracking. IEEE Transactions on Biomedical Engineering 2023, 70, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Sciolla, B.; Sdika, M.; Wang, X.; Quetin, P.; Delachartre, P. Automatic Segmentation of the Cerebral Ventricle in Neonates Using Deep Learning with 3D Reconstructed Freehand Ultrasound Imaging. 2018 IEEE International Ultrasonics Symposium (IUS), 2018. [CrossRef]
| Study | Aim | Dataset (n=sequences) | Sweep characteristics | US Probe | Data inputs | Reconstruction methods | Loss function | Ground truth |
|---|---|---|---|---|---|---|---|---|
| Tetrel et al. [14] | Trajectory reconstruction withspeckle decorrelation and graph-based optimization | Phantom:1) Speckle phantom (n=1) 2) Phantom (n=9) | 1) Motorized translation (50 mm), precision of 5 µm 2) Elevational displacement (20-56 mm) | ATL HDI5000 US scanner, linear 4–7 MHz probe, depth 3 cm | US sequence and phantom based speckle decorrelation curve | GMeA: graph-based trajectory estimation based on speckle decorrelation, optimized by weighted graph edges by learned error of tracked sequence using Gaussian process regressor | N/A | Micron Tracker optical sensor, 97% accuracy volume measurements |
| Martin et al. [27] | CVS segmentation after landmark based 3D reconstruction of 2D US images | In vivo:1) Cerebral ventricle (n=15, 14 subjects) | Angular sweep, 136-306 frames per sequence | Siemens Acuson 9L4 probe | 2 freehand 2D US sequences in coronal and sagittal orientation | Reconstruction: landmark based registration of coronal and sagittal views, model parameters optimization, mapped to 3D grid, voxel-based volume interpolation. Segmentation: CNN U-Net | Reconstruction: Gradient descent Segmentation: Soft-Dice loss | Segmentation: manual CVS segmentation in sagittal view |
| Prevost et al. [23] | Trajectory reconstruction incorporating IMU | Phantoms:1) BluePhantom US biopsy (n=20, 7168 frames) In-vivo: 2) Forearms (n=88, 41869 frames, 12 subjects) 3) Calves (n=12, 6647 frames) 4) Forearms + IMU (n=600, 307200 frames, 15 subjects) 5) Carotids + IMU (n=100, 21.945 frames, 10 subjects) | 1) Basic, average length 131 mm 2) Basic, average length 190 mm 3) Basic, average length 175 mm 4) Basic, shift, wave, tilt, average length 202 mm 5) Basic, tilt, average length 75 mm | Cicada research US machine, linear probe, 128 elements, 5 MHz, 35 FPS | Pairs adjacent 2D US frames, optical flow, IMU orientation data | CNN (ablation experiments with input channels optical flow, IMU and based on CNN or IMU) | MSE loss | Optical tracking system Stryker NAV3 Camera, translation accuracy 0.2 |
| Balakrishnan et al. [15] | Trajectory reconstruction with a texture-based similarity metric | In vivo:1) Forearm (n=13, 7503 frames, 3 subjects) | Varying acquisition speeds, forearm sizes and shapes, axial resolution 400x457 | GE Logiq E Ultrasound System, 9L-RS linear probe | US, optical flow, texture-based similarity values | Gaussian (SVM) based regression model, similarity metric TexSimAR | N/A | EM Tracking System Ascension trakSTAR |
| Miura et al. [17] | Trajectory reconstruction incorporating motion features and consistency loss | In vivo:1) Forearm (n=190, 30801 frames, 5 subjects) Phantom: 2) Breast phantom (n=60, 8940 frames) 3) Hypogastric phantom (n=40, 6242 frames) | 1, 2, 3) Sweeps of 6 seconds | 1, 2) SONIMAGE HS1, L18-4 linear probe 3) C5-2 convex probe, 30 FPS | Pairs adjacent 2D US frames, optical flow | CNN (ResNet45) + FlownetS (optical flow), with ablation experiments adding FlowNetS (motion features) and consistency loss | MSE loss, consistency loss | Optical tracking V120: Trio OptiTrack, with 5 markers attached on US probe |
| Wein et al. [25] | Trajectory reconstruction and thyroid volume segmentation | In-vivo:1) Thyroid (n=180, 9 subjects). | Variations in acquisition speed, captured anatomy and tilt angles | Cicada research US machine, linear, 128 elements, 5 MHz | Pairs adjacent 2D US frames and optical flow, 1 transverse (TRX) and 1 sagittal (SAG) direction | Segmentation: 2D U-Net + union of segmentations in SAG+TRX. Trajectory: CNN, optimized by joint sweep reconstruction through co-registration of orthogonal sweeps | MSE loss | 3D: Based on 2D U-Net, dice 0.73 Trajectory: optical tracking system Stryker NAV3 Camera, translation accuracy 0.2 |
| Guo et al. [16] | Trajectory reconstruction with contextual learning | In vivo:1) Transrectal US (n=640, 640 subjects)a | Axial images, steadily through prostate from base to apex | Philips iU22 scanner in varied lengths, end firing C95 transrectal US probe | Transrectal US, N-neighboring frames | Deep contextual learning network (DCL-Net) (3D ResNext) with self-attention module focusing on speckle-rich areasb | MSE loss + case-wise correlation loss | EM tracking (mean over N neighboring frames) |
| Guo et al. [18] | Trajectory reconstruction with two US transducers by domain adaptation techniques | In vivo:1) Transrectal US for training (n=640, 640 subjects) 2) Transabdominal US (n=12, 12 subjects) | 1) Axial images, steadily sweeping through the prostate from base to apex 2) N/A | 1) End-firing C95 transrectal US probe 2) C51 US probe | Videosubsequence of transrectal and transabdominal US | CNN with novel paired-sampling strategy to transfer task specific feature learning from source (transrectal) to target (transabdominal) domain | MSE loss, discrepancy loss (L2 norm between feature outputs of generators of both domains) | EM tracking (mean over N neighboring frames) |
| Leblanc et al. [24] | 3D stretched reconstruction of femoral artery, DL-based | In vivo:1) Femoral artery (n=111, 40788 frames, 18 subjects) | Thigh to knee following femoral artery, lengths 102-272 mm | Aixplorer echograph, Supersonic Imagine | Pair of 2D US frames, after echograph processing with speckle reduction | In-plane by registration of mask R-CNN based artery segmentation and interpolation, out-of-plane by CNN and linear interpolation to generate final volume | MAE | Optical tracking, NDI Polaris Spectra. Segmentation 2D: manual |
| Luo et al. [19] | Trajectory reconstruction with IMU and online learning, contextual information | In vivo:1) Arm (n=250, 41 subjects) 2) Carotid (n=160, 40 subjects) | 1) Linear, curved, fast and slow, loop, average length 94.83 mm 2) linear, average length 53.71 mm | Linear probe, 10 MHz, image depth 3.5 cm | N-neighboring frames US sweep, IMU | MoNet: BK (ResNet + LSTM) + IMU + online learning (adaptive optimization self-supervised by weak IMU labels) with ablation experiments for IMU and online learning | BK: MAE + Pearson correlation loss, online learning: MAE(°) + Pearson correlation loss (acceleration) | EM tracking, resolution of 1.4 mm positioning and 0.5° orientation |
| Luo et al. [21] | Trajectory reconstruction using 4 IMUs, online learning and contextual information | In vivo:1) Arm (n=288, 36 subjects) 2) Carotid (n=216, 36 subjects) | 1) Linear, curved, loop, sector scan, average length of 323.96 mm 2) Linear, loop, sector scan, average length of 203.25 mm | Linear probe, 10 MHz, image depth 4 cm | N-length US sweep, 4 IMUs | OSCNet: BK (ResNet + LSTM + IMU) + online learning on modal-level self-supervised (MSS) by weak IMU labels and sequence-level self-consistency strategy (SCS) | BK: MAE + Pearson correlation loss, Online learning: MAE(°) + Pearson correlation loss (acceleration) for single- and multi-IMU, self-consistency loss | EM tracking, resolution of 1.4 mm positioning and 0.5° orientation |
| Luo et al. [20] | Trajectory reconstruction with online learning and contextual information | In vivo:1) Spine (n=68, 23 subjects) dataset on a robotic arm with EM positioning | Linear, average length of 186 mm | N/A | US images, canny edge maps and optical flow of 2 adjacent frames | RecON: BK (ResNet + LSTM) + online Self Supervised Learning (SSL): Frame-level Contextual Consistency (FCC), Path-level Similarity Constraint (PSC) and Global Adversarial Shape prior (GAS) | BK: MAE + Pearson correlation loss + motion-weighted training loss. Online learning: MAE + Pearson correlation loss, adversarial loss | Volume based on EM and mechanical tracking |
| Guo et al. [26] | Trajectory reconstruction utilizing contextual information and volume segmentation prostate | In vivo:1) Transrectal US (n=618, 618 subjects) + segmentations 2) Transabdominal (n=100) | 1) Axial images, steady sweep through prostate from base to apex 2) Varying lengths and resolutions | 1) Philips C9-5 transrectal US probe 2) Philips mc7-2 US probe | Video subsequence of transrectal and transperinal US, n=7 frames | Deep contextual-contrastive network (DC2-Net) (3D ResNext) with self-attention module to focus on speckle-rich areas and a contrastive feature learning strategy | MSE loss, case wise correlation loss and margin ranking loss for contrastive feature learning | EM tracking (mean over N neighboring frames) |
| Li et al. [22] | Trajectory reconstruction incorporating long-term temporal information | In vivo:1) Forearm (n=228 19 subjects)a | Left and right arms, straight, c-shape, and s-shape, distal-to-proximal, 36-430 frames/sequence, 20 fps, lengths 100-200 mm | Ultrasonix machine, curvilinear probe (4DC7-3/40), 6 MHz, depth 9 cm | US sequence with number M of past (i) and future (j) frames, subsets of trainset to evaluate anatomical and protocol dependency | 1) RNN + LSTM 2) Feedforward CNN (EfficientNet) leveraging multi-task learning framework designed to exploit long-term dependenciesb | Multi-transformation loss based on MSE (consistency at multiple frame intervals) | Optical tracking, NDI Polaris Vicra |
| Method | Final drift (mm) | FDR (%) | Errors between successive frames | Volume measures |
|---|---|---|---|---|
| Tetrel et al. [14] | Mean | |||
| A: Graph based - MeA | 1.1) 1.803, 1.2) 0.731, 1.3) 3.039, 1.4) 1.995, 1.5) 3.735, 1.6) 9.345, 1.7) 9.154, 1.8) 9.999 | N/A | N/A | N/A |
| Martin et al. [27] | MAD between SAG/TRX (mm) | HD (mm), Dice | ||
| A: landmark based – CVS segmentation (U-Net) | N/A | N/A | 1.55 ± 1.59 | 13.6 ± 4.7, 0.82 ± 0.04 |
| Prevost et al. [23] | Median (min - max) | Mean | MAE of tx, ty, tz, x, y, z (mm/°) | |
| A: Standard CNN | 1) 26.17 (14.31 - 65.10) 2) 25.16 (3.72 - 63.26) 3) 54.72 (27.11 - 116.64) | N/A | 1) 2.25, 5.67, 14.37, 2.13, 1.86, 0.98 2) 6.30, 5.97, 6.15, 2.82, 2.78, 2.40 3) 4.91, 8.95, 25.89, 2.01, 2.54, 2.90 | N/A |
| B: CNN (OFa + by CNN) | 1) 18.30 (1.70 - 36.90) 2) 14.44 (3.35 - 41.93 3) 19.69 (8.53 - 30.11) 4) 27.34 (3.22 - 139.02) | 2) 19 | 1) 1.32, 2.13, 7.79, 2.32, 1.21, 0.90 2) 3.54, 3.05, 4.19, 2.63, 2.52, 1.93 3) 3.11, 5.86, 5.63, 2.75, 3.17, 5.24 4) 8.89, 6.61, 5.73, 5.21, 7.38, 4.01 | N/A |
| C: CNN (IMU + by CNN) | 4) 29.22 (3.12 - 186.83) | N/A | 4) 6.56, 7.23, 16.70, 0.94, 2.65, 2.80 | N/A |
| D: CNN (OF + IMU + by CNN) | 4) 15.07 (2.54 - 55.20) | N/A | 4) 5.16, 2.67, 4.43, 0.96, 3.54, 2.85 | N/A |
| E: CNN (OF + by IMU) | 4) 11.43 (1.33 - 42.94) | N/A | 4) 2.98 2.57 4.79 0.19 0.21 0.13 | N/A |
| F: CNN (OF + IMU + by IMU) | 4) 10.42 (0.76 - 35.22) | 5.2 | 4) 2.75 2.41 4.36 0.19 0.21 0.13 | N/A |
| Balakrishnan et al. [15] | Median (min - max) | MAE of tz, x, y (mm/°), out of plane | ||
| A: Gaussian SVM regressor | 6.59 (5.550 - 23.02) | N/A | 9.11, 1.95, 1.66 | N/A |
| Miura et al. [17] | MAE of tx, ty, tz, x, y, z (mm/°) | |||
| A: CNN (ResNet, OF) | N/A | N/A | 0.72, 0.18, 0.76, 0.60, 1.26, 0.52 | N/A |
| B: CNN (ResNet, OF + FlowNetS) | N/A | N/A | 0.74, 0.18, 0.78, 0.61, 1.28, 0.52 | N/A |
| C: CNN (ResNet, OF + Lconsistency) | N/A | N/A | 0.66, 0.15, 0.82, 0.56, 1.23, 0.47 | N/A |
| D: CNN (ResNet, OF + FlowNetS + Lconsistency) | N/A | N/A | 0.64, 0.15, 0.80, 0.53, 1.21, 0.47 | N/A |
| Wein et al. [25] | Relative trajectory error, mean ± SD: cumulative in-plane translation/length | Volume error (ml) | ||
| A: CNN + joint co-registration 54-DOF | N/A | N/A | 0.16 ± 0.09 | 1.15 ± 0.12 |
| Guo et al. [26] | Median (min - max), mean | |||
| A: DCL-Net (attention, n=5, LMSE + LCC) | 17.40 (1.09 - 55.50), 17.39 | N/A | N/A | N/A |
| B: DCL-Net (attention, n=2 to n=8, LMSE + LCC) | Visualized in boxplot per n input frames, n=5 significant improvement to n=2 (p<0.05) | N/A | N/A | N/A |
| Guo et al. [18] | Median (min - max), mean | |||
| A: Target transabdominal | 21.21 (5.88 - 32.94), 20.01 | N/A | N/A | N/A |
| B: TAUVR | 22.02 (6.87 - 32.13), 20.34 | N/A | N/A | N/A |
| Leblanc et al. [24] | Median (min - max), mean | Median | MAE (mm), out-of-plane translation | |
| A: CNN + artery alignment | 13.42 (0.18 - 68.31), 17.22 | 8.98 | 0.28 | N/A |
| Luo et al. [19] | Mean ± SD | MAE (x, y, z) (°), mean ± SD | ||
| A: BK (ResNet + LSTM) | N/A | 1) 16.42 ± 14.24, 2) 20.55 ± 18.73 | 1) 2.29 ± 2.50, 2) 2.61 ± 1.72 | N/A |
| B: BK + IMU | N/A | 1) 14.05 ± 10.36, 2) 17.78 ± 11.50 | 1) 1.75 ± 1.57, 2) 2.18 ± 1.43 | N/A |
| C: MoNet (CNN + IMU + Online) | N/A | 1) 12.75 ± 9.05, 2) 15.67 ± 8.37 | 1) 1.55 ± 1.46, 2) 1.50 ± 0.98 | N/A |
| Luo et al. [21] | Mean ± SD | MAE (x, y, z) (°), mean ± SD | ||
| A: BK (ResNet + LSTM) | N/A | 1) 13.32 ± 8.2, 2) 12.85 ± 6.5 | 1) 4.32 ± 1.7, 2) 3.83 ± 2.0 | N/A |
| B: BK + MSS (4 IMU) | N/A | 1) 10.78 ± 5.6, 2) 11.31 ± 5.4 | 1) 3.18 ± 2.76, 2) 3.16 ± 1.8 | N/A |
| C: BK + SCS (Lconsistency) | N/A | 1) 10.56 ± 5.9, 2) 11.30 ± 5.4 | 1) 3.65 ± 1.9, 2) 3.36 ± 1.8 | N/A |
| D: OSCNet (BK + SCS + MSS) | N/A | 1) 10.01 ± 5.7, 2) 10.90 ± 5.3 | 1) 2.76 ± 1.3, 2) 2.60 ± 1.6 | N/A |
| Luo et al. [20] | Mean ± SD | MAE (x, y, z) (°), mean ± SD | ||
| A: BK (ResNet + LSTM) | N/A | 15.54 ± 8.29 | 1.36 ± 0.71 | N/A |
| B: BK + OF | N/A | 14.88 ± 8.83 | 1.35 ± 0.46 | N/A |
| C: BK + OF + CE | N/A | 12.53 ± 6.32 | 1.33 ± 0.58 | N/A |
| D: BK + OF + CE + Motion | N/A | 12.30 ± 6.31 | 1.30 ± 0.45 | N/A |
| E: BK + OF + CE + Motion + SSL(PSC+FCC) | N/A | 11.36 ± 5.51 | 1.30 ± 0.44 | N/A |
| F: ReCon (Motion + SSL + GAS) | N/A | 10.82 ± 5.36 | 1.25 ± 0.46 | N/A |
| Guo et al. [26] | Mean ± SD | Mean ± SD | Frame error (mm), mean ± SD Euclidean distance, 4 corner points | Dice, volume error (ml) mean ± SD, (prostate) |
| A: DC2-Net (attention) | 1) 12.20 ± 10.07 | 1) 11.66 ± 9.77 | 1) 0.93 ± 0.28 | 1) 0.83 ± 0.08 |
| B: DC2-Net (attention + LCC) | 1) 11.92 ± 8.89 | 1) 11.59 ± 9.22 | 1) 0.93 ± 0.28 | 1) 0.86 ± 0.05 |
| C: DC2-Net (attention + LCC + Lmargin) | 1) 10.20 ± 8.47, 2) 9.85 ± 5.74 | 1) 9.64 ± 8.14, 2) 14.58 ± 12.76 | 1) 0.90 ± 0.26, 2) 1.12 ± 0.26 | 1) 0.89 ± 0.06, 2) 3.21 ± 1.93 |
| Li et al. [22] | Mean ± SD | Frame error (mm), mean ± SD Euclidean distance, 4 corner points | Dice, mean ± SD (Trajectory) | |
| A: RNN (M = 2) | 34.54 ± 18.10 | N/A | 0.57 ± 0.44 | 0.41 ± 0.33 |
| B: RNN (M = 100) | 6.97 ± 6.79 | N/A | 0.20 ± 0.07 | 0.73 ± 0.23 |
| C: ff-CNN (M = 2) | 29.59 ± 19.53 | N/A | 0.53 ± 0.46 | 0.50 ± 0.29 |
| D: ff-CNN (M = 100) | 7.24 ± 8.33 | N/A | 0.19 ± 0.08 | 0.77 ± 0.17 |
| E: ff-CNN (M = 100, straight sweeps) | 22.30 ± 41.10 | N/A | 0.48 ± 0.25 | 0.64 ± 0.26 |
| F: ff-CNN (M = 100, c- and s-shapes) | 6.74 ± 7.19 | N/A | 0.24 ± 0.13 | 0.80 ± 0.13 |
| G: ff-CNN (M = 100, 25% of subjects) | 13.66 ± 15.94 | N/A | 0.41 ± 0.24 | 0.75 ± 0.17 |
| Method | Addition of | Dataset | Transformation parameter | Improvement MAE (%) | Improvement drift measures (%) |
|---|---|---|---|---|---|
| Prevost et al. (23) | 1 IMU | 1 | 69.07 | FD: 61.89 | |
| B → F | 63.54 | ||||
| 23.91 | |||||
| 96.35 | |||||
| 97.15 | |||||
| 96.76 | |||||
| Luo et al. [19] | 1 IMU | 1 | 23.58 | FDR: 14.43 | |
| A → B | 2 | 42.53 | FDR: 13.48 | ||
| Luo et al. [21] | 4 IMUs | 1 | 26.39 | FDR: 19.07 | |
| A → B | 2 | 17.49 | FDR: 11.98 | ||
| Luo et al. [20] | Optical Flow + Canny Edge | 1 | 2.21 | FDR: 19.37 |
| Method | Addition of | Dataset | Transformation parameter | Improvement (%) | Improvement drift measures (%) |
|---|---|---|---|---|---|
| Luo et al. [21] | Self Consistency Strategy | 1 | MAE: 13.21 | FDR: 7.14 | |
| A → C | 2 | MAE: 17.72 | FDR: 3.63 | ||
| Luo et al. [20] | Frame, path and sequence level online learning | 1 | MAE: 3.85 | FDR: 4.75 | |
| D → E | |||||
| Li et al. [22] | M=2 to M=100 past and future frames | 1 | N/A | Frame error: 64.15 | FD: 75.53 |
| C → D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





