Submitted:
01 August 2024
Posted:
01 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Alignment
2.2. Evolutionary Analysis in Mammals
2.3. Correlation with Meiotic Gene Expression
2.4. Prediction of Disordered Regions and Functional Motifs
3. Results
3.1. Evolutionary Analysis in Mammals: SMC Complexes Evolve at Different Rates
3.2. Positive Selection Drives the Evolution of Meiosis-Specific Cohesins
3.3. Analysis of Positively Selected Sites
3.4. Meiotic Cohesin Evolutionary Rates Correlate with Expression during Female Meiosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Uhlmann, F. SMC Complexes: From DNA to Chromosomes. Nature reviews.Molecular cell biology 2016, 17, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Haering, C.H.; Gruber, S. SnapShot: SMC Protein Complexes Part I. Cell 2016, 164, 326–326.e1. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, J.-J.; Ryu, J.-K. Mechanism of Phase Condensation for Chromosome Architecture and Function. Exp Mol Med 2024, 56, 809–819. [Google Scholar] [CrossRef]
- Ryu, J.-K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; De Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-Induced Phase Separation Induced by Cohesin SMC Protein Complexes. Sci. Adv. 2021, 7, eabe5905. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophysical Journal 2018, 114, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Beverley, R.; Snook, M.L.; Brieño-Enríquez, M.A. Meiotic Cohesin and Variants Associated With Human Reproductive Aging and Disease. Front. Cell Dev. Biol. 2021, 9, 710033. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.K.; Kim, J.-S.; Waldman, T. Cohesin Mutations in Human Cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2016, 1866, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pati, D. Role of Chromosomal Cohesion and Separation in Aneuploidy and Tumorigenesis. Cell. Mol. Life Sci. 2024, 81, 100. [Google Scholar] [CrossRef]
- Di Nardo, M.; Pallotta, M.M.; Musio, A. The Multifaceted Roles of Cohesin in Cancer. J Exp Clin Cancer Res 2022, 41, 96. [Google Scholar] [CrossRef]
- Liu, J.; Krantz, I.D. Cornelia de Lange Syndrome, Cohesin, and Beyond. Clinical genetics 2009, 76, 303–314. [Google Scholar] [CrossRef]
- Barbero, J.L. Genetic Basis of Cohesinopathies. The application of clinical genetics 2013, 6, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Kline, A.D.; Moss, J.F.; Selicorni, A.; Bisgaard, A.-M.; Deardorff, M.A.; Gillett, P.M.; Ishman, S.L.; Kerr, L.M.; Levin, A.V.; Mulder, P.A.; et al. Diagnosis and Management of Cornelia de Lange Syndrome: First International Consensus Statement. Nat Rev Genet 2018, 19, 649–666. [Google Scholar] [CrossRef]
- Kline, A.D.; Krantz, I.D.; Sommer, A.; Kliewer, M.; Jackson, L.G.; FitzPatrick, D.R.; Levin, A.V.; Selicorni, A. Cornelia de Lange Syndrome: Clinical Review, Diagnostic and Scoring Systems, and Anticipatory Guidance. American J of Med Genetics Pt A 2007, 143A, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Neira, J.; Pehlivan, D.; Santiago-Sim, T.; Song, X.; Rosenfeld, J.; Posey, J.E.; Patel, V.; Jin, W.; Adam, M.P.; et al. Clinical Exome Sequencing Reveals Locus Heterogeneity and Phenotypic Variability of Cohesinopathies. Genetics in Medicine 2019, 21, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell 2016, 164, 847–857. [Google Scholar] [CrossRef]
- Hoencamp, C.; Rowland, B.D. Genome Control by SMC Complexes. Nat Rev Mol Cell Biol 2023, 24, 633–650. [Google Scholar] [CrossRef]
- Ono, T.; Fang, Y.; Spector, D.L.; Hirano, T. Spatial and Temporal Regulation of Condensins I and II in Mitotic Chromosome Assembly in Human Cells. MBoC 2004, 15, 3296–3308. [Google Scholar] [CrossRef]
- Cuylen, S.; Haering, C.H. Deciphering Condensin Action during Chromosome Segregation. Trends in Cell Biology 2011, 21, 552–559. [Google Scholar] [CrossRef]
- Kinoshita, K.; Hirano, T. Dynamic Organization of Mitotic Chromosomes. Current Opinion in Cell Biology 2017, 46, 46–53. [Google Scholar] [CrossRef]
- Kakui, Y.; Uhlmann, F. SMC Complexes Orchestrate the Mitotic Chromatin Interaction Landscape. Curr Genet 2018, 64, 335–339. [Google Scholar] [CrossRef]
- Martin, C.-A.; Murray, J.E.; Carroll, P.; Leitch, A.; Mackenzie, K.J.; Halachev, M.; Fetit, A.E.; Keith, C.; Bicknell, L.S.; Fluteau, A.; et al. Mutations in Genes Encoding Condensin Complex Proteins Cause Microcephaly through Decatenation Failure at Mitosis. Genes Dev. 2016, 30, 2158–2172. [Google Scholar] [CrossRef] [PubMed]
- Pang, D.; Yu, S.; Yang, X. A Mini-Review of the Role of Condensin in Human Nervous System Diseases. Front. Mol. Neurosci. 2022, 15, 889796. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.P.; Zhao, X. The Multi-Functional Smc5/6 Complex in Genome Protection and Disease. Nat Struct Mol Biol 2023, 30, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Aragón, L. The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative. Annu. Rev. Genet. 2018, 52, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Sun, F.; O’Brien, M.; Eppig, J.J.; Handel, M.A.; Jordan, P.W. SMC5/6 Is Required for the Formation of Segregation-Competent Bivalent Chromosomes during Meiosis I in Mouse Oocytes. Development 2017, dev.145607. [Google Scholar] [CrossRef] [PubMed]
- Payne, F.; Colnaghi, R.; Rocha, N.; Seth, A.; Harris, J.; Carpenter, G.; Bottomley, W.E.; Wheeler, E.; Wong, S.; Saudek, V.; et al. Hypomorphism in Human NSMCE2 Linked to Primordial Dwarfism and Insulin Resistance. J. Clin. Invest. 2014, 124, 4028–4038. [Google Scholar] [CrossRef] [PubMed]
- Van Der Crabben, S.N.; Hennus, M.P.; McGregor, G.A.; Ritter, D.I.; Nagamani, S.C.S.; Wells, O.S.; Harakalova, M.; Chinn, I.K.; Alt, A.; Vondrova, L.; et al. Destabilized SMC5/6 Complex Leads to Chromosome Breakage Syndrome with Severe Lung Disease. Journal of Clinical Investigation 2016, 126, 2881–2892. [Google Scholar] [CrossRef]
- Decorsière, A.; Mueller, H.; Van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.K.; Livingston, C.M.; Niu, C.; Fletcher, S.P.; Hantz, O.; et al. Hepatitis B Virus X Protein Identifies the Smc5/6 Complex as a Host Restriction Factor. Nature 2016, 531, 386–389. [Google Scholar] [CrossRef]
- Murphy, C.M.; Xu, Y.; Li, F.; Nio, K.; Reszka-Blanco, N.; Li, X.; Wu, Y.; Yu, Y.; Xiong, Y.; Su, L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Reports 2016, 16, 2846–2854. [Google Scholar] [CrossRef]
- Irwan, I.D.; Cullen, B.R. The SMC5/6 Complex: An Emerging Antiviral Restriction Factor That Can Silence Episomal DNA. PLoS Pathog 2023, 19, e1011180. [Google Scholar] [CrossRef]
- Xu, W.; Ma, C.; Zhang, Q.; Zhao, R.; Hu, D.; Zhang, X.; Chen, J.; Liu, F.; Wu, K.; Liu, Y.; et al. PJA1 Coordinates with the SMC5/6 Complex To Restrict DNA Viruses and Episomal Genes in an Interferon-Independent Manner. J Virol 2018, 92, e00825-18. [Google Scholar] [CrossRef]
- Gibson, R.T.; Androphy, E.J. The SMC5/6 Complex Represses the Replicative Program of High-Risk Human Papillomavirus Type 31. Pathogens 2020, 9, 786. [Google Scholar] [CrossRef]
- Bentley, P.; Tan, M.J.A.; McBride, A.A.; White, E.A.; Howley, P.M. The SMC5/6 Complex Interacts with the Papillomavirus E2 Protein and Influences Maintenance of Viral Episomal DNA. J Virol 2018, 92, e00356-18. [Google Scholar] [CrossRef]
- Yiu, S.P.T.; Guo, R.; Zerbe, C.; Weekes, M.P.; Gewurz, B.E. Epstein-Barr Virus BNRF1 Destabilizes SMC5/6 Cohesin Complexes to Evade Its Restriction of Replication Compartments. Cell Reports 2022, 38, 110411. [Google Scholar] [CrossRef]
- Dupont, L.; Bloor, S.; Williamson, J.C.; Cuesta, S.M.; Shah, R.; Teixeira-Silva, A.; Naamati, A.; Greenwood, E.J.D.; Sarafianos, S.G.; Matheson, N.J.; et al. The SMC5/6 Complex Compacts and Silences Unintegrated HIV-1 DNA and Is Antagonized by Vpr. Cell Host & Microbe 2021, 29, 792–805.e6. [Google Scholar] [CrossRef]
- Han, C.; Zhang, D.; Gui, C.; Huang, L.; Chang, S.; Dong, L.; Bai, L.; Wu, S.; Lan, K. KSHV RTA Antagonizes SMC5/6 Complex-Induced Viral Chromatin Compaction by Hijacking the Ubiquitin-Proteasome System. PLoS Pathog 2022, 18, e1010744. [Google Scholar] [CrossRef] [PubMed]
- King, T.D.; Leonard, C.J.; Cooper, J.C.; Nguyen, S.; Joyce, E.F.; Phadnis, N. Recurrent Losses and Rapid Evolution of the Condensin II Complex in Insects. Molecular Biology and Evolution 2019, 36, 2195–2204. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, Duplication-Aware Phylogenetic Trees in Vertebrates. Genome research 2009, 19, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, R. RevTrans: Multiple Alignment of Coding DNA from Aligned Amino Acid Sequences. Nucleic Acids Research 2003, 31, 3537–3539. [Google Scholar] [CrossRef]
- Guindon, S.; Delsuc, F.; Dufayard, J.F.; Gascuel, O. Estimating Maximum Likelihood Phylogenies with PhyML. Methods in molecular biology (Clifton, N.J.) 2009, 537, 113–137. [Google Scholar] [CrossRef]
- Anisimova, M.; Nielsen, R.; Yang, Z. Effect of Recombination on the Accuracy of the Likelihood Method for Detecting Positive Selection at Amino Acid Sites. Genetics 2003, 164, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Sironi, M.; Cagliani, R.; Forni, D.; Clerici, M. Evolutionary Insights into Host-Pathogen Interactions from Mammalian Sequence Data. Nat Rev Genet 2015, 16, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Molecular biology and evolution 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis Testing Using Phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Bielawski, J.P.; Yang, Z. Accuracy and Power of Bayes Prediction of Amino Acid Sites Under Positive Selection. Molecular Biology and Evolution 2002, 19, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS genetics 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Molecular Biology and Evolution 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Synonymous and Nonsynonymous Rate Variation in Nuclear Genes of Mammals. J Mol Evol 1998, 46, 409–418. [Google Scholar] [CrossRef]
- Soh, Y.Q.; Junker, J.P.; Gill, M.E.; Mueller, J.L.; Oudenaarden, A. van; Page, D.C. A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary. PLoS genetics 2015, 11, e1005531. [Google Scholar] [CrossRef]
- Margolin, G.; Khil, P.P.; Kim, J.; Bellani, M.A.; Camerini-Otero, R.D. Integrated Transcriptome Analysis of Mouse Spermatogenesis. BMC genomics 2014, 15, 39–39. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Griffith, D.; Holehouse, A.S. Metapredict V2: An Update to Metapredict, a Fast, Accurate, and Easy-to-Use Predictor of Consensus Disorder and Structure 2022.
- Emenecker, R.J.; Griffith, D.; Holehouse, A.S. Metapredict: A Fast, Accurate, and Easy-to-Use Predictor of Consensus Disorder and Structure. Biophysical Journal 2021, 120, 4312–4319. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Research 2012, 41, D344–D347. [Google Scholar] [CrossRef]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A Simple Hidden Markov Model for Nuclear Localization Signal Prediction. BMC Bioinformatics 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.Y.; Khaodeuanepheng, N.P.; Amarasekara, D.L.; Correia, J.J.; Lewis, K.A.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. Intrinsically Disordered Regions That Drive Phase Separation Form a Robustly Distinct Protein Class. Journal of Biological Chemistry 2023, 299, 102801. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Lewis, K.A.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. ParSe 2.0: A Web Tool to Identify Drivers of Protein Phase Separation at the Proteome Level. Protein Science 2023, 32, e4756. [Google Scholar] [CrossRef]
- Tesei, G.; Trolle, A.I.; Jonsson, N.; Betz, J.; Pesce, F.; Johansson, K.E.; Lindorff-Larsen, K. Conformational Ensembles of the Human Intrinsically Disordered Proteome: Bridging Chain Compaction with Function and Sequence Conservation 2023.
- Tesei, G.; Lindorff-Larsen, K. Improved Predictions of Phase Behaviour of Intrinsically Disordered Proteins by Tuning the Interaction Range. Open Res Europe 2023, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Ebel, E.R.; Telis, N.; Venkataram, S.; Petrov, D.A.; Enard, D. High Rate of Adaptation of Mammalian Proteins That Interact with Plasmodium and Related Parasites. PLoS genetics 2017, 13, e1007023. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A Program Package for Phylogenetic Analysis by Maximum Likelihood. Computer applications in the biosciences : CABIOS 1997, 13, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Abdul, F.; Filleton, F.; Gerossier, L.; Paturel, A.; Hall, J.; Strubin, M.; Etienne, L. Smc5/6 Antagonism by HBx Is an Evolutionarily Conserved Function of Hepatitis B Virus Infection in Mammals. J Virol 2018, 92, e00769-18. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat Rev Mol Cell Biol 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase Separation Drives Heterochromatin Domain Formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Mirny, L.A.; Imakaev, M.; Abdennur, N. Two Major Mechanisms of Chromosome Organization. Current Opinion in Cell Biology 2019, 58, 142–152. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid Droplet Formation by HP1α Suggests a Role for Phase Separation in Heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Kragelund, B.B. The Molecular Basis for Cellular Function of Intrinsically Disordered Protein Regions. Nat Rev Mol Cell Biol 2024, 25, 187–211. [Google Scholar] [CrossRef] [PubMed]
- Afanasyeva, A.; Bockwoldt, M.; Cooney, C.R.; Heiland, I.; Gossmann, T.I. Human Long Intrinsically Disordered Protein Regions Are Frequent Targets of Positive Selection. Genome research 2018, 28, 975–982. [Google Scholar] [CrossRef]
- Brown, C.J.; Johnson, A.K.; Dunker, A.K.; Daughdrill, G.W. Evolution and Disorder. Current opinion in structural biology 2011, 21, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Molteni, C.; Forni, D.; Cagliani, R.; Mozzi, A.; Clerici, M.; Sironi, M. Evolution of the Orthopoxvirus Core Genome. Virus Research 2023, 323, 198975. [Google Scholar] [CrossRef]
- Mozzi, A.; Forni, D.; Cagliani, R.; Clerici, M.; Pozzoli, U.; Sironi, M. Intrinsically Disordered Regions Are Abundant in Simplexvirus Proteomes and Display Signatures of Positive Selection. Virus evolution 2020, 6, veaa028. [Google Scholar] [CrossRef]
- Zarin, T.; Strome, B.; Nguyen Ba, A.N.; Alberti, S.; Forman-Kay, J.D.; Moses, A.M. Proteome-Wide Signatures of Function in Highly Diverged Intrinsically Disordered Regions. eLife 2019, 8, e46883. [Google Scholar] [CrossRef]
- Cagliani, R.; Forni, D.; Mozzi, A.; Fuchs, R.; Tussia-Cohen, D.; Arrigoni, F.; Pozzoli, U.; De Gioia, L.; Hagai, T.; Sironi, M. Evolution of virus-like features and intrinsically disordered regions in retrotransposon-derived mammalian genes. Molecular Biology and Evolution 2024, in press. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Y.; Gao, J. Phase Separation in Controlling Meiotic Chromosome Dynamics. In Current Topics in Developmental Biology; Elsevier, 2023; Vol. 151, pp. 69–90 ISBN 978-0-12-820156-5.
- Pontremoli, C.; Forni, D.; Cagliani, R.; Pozzoli, U.; Clerici, M.; Sironi, M. Evolutionary Rates of Mammalian Telomere-Stability Genes Correlate with Karyotype Features and Female Germline Expression. Nucleic acids research 2018, 46, 7153–7168. [Google Scholar] [CrossRef]
- Pontremoli, C.; Forni, D.; Pozzoli, U.; Clerici, M.; Cagliani, R.; Sironi, M. Kinetochore Proteins and Microtubule-destabilizing Factors Are Fast Evolving in Eutherian Mammals. Molecular Ecology 2021, 30, 1505–1515. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science (New York, N.Y.) 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- Wu, C.-I.; Yujun Xu, E. Sexual Antagonism and X Inactivation – the SAXI Hypothesis. Trends in Genetics 2003, 19, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Tenthorey, J.L.; Emerman, M.; Malik, H.S. Evolutionary Landscapes of Host-Virus Arms Races. Annu. Rev. Immunol. 2022, 40, 271–294. [Google Scholar] [CrossRef]
- Abdul, F.; Diman, A.; Baechler, B.; Ramakrishnan, D.; Kornyeyev, D.; Beran, R.K.; Fletcher, S.P.; Strubin, M. Smc5/6 Silences Episomal Transcription by a Three-Step Function. Nat Struct Mol Biol 2022, 29, 922–931. [Google Scholar] [CrossRef]
- Lee, C.-P.; Huang, Y.-H.; Lin, S.-F.; Chang, Y.; Chang, Y.-H.; Takada, K.; Chen, M.-R. Epstein-Barr Virus BGLF4 Kinase Induces Disassembly of the Nuclear Lamina To Facilitate Virion Production. J Virol 2008, 82, 11913–11926. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, A.; Kimura, K.; Yanagisawa, J.; Yokoyama, S.; Hanaoka, F. Negative Regulation of Condensin I by CK2-Mediated Phosphorylation. EMBO J 2006, 25, 5339–5348. [Google Scholar] [CrossRef]
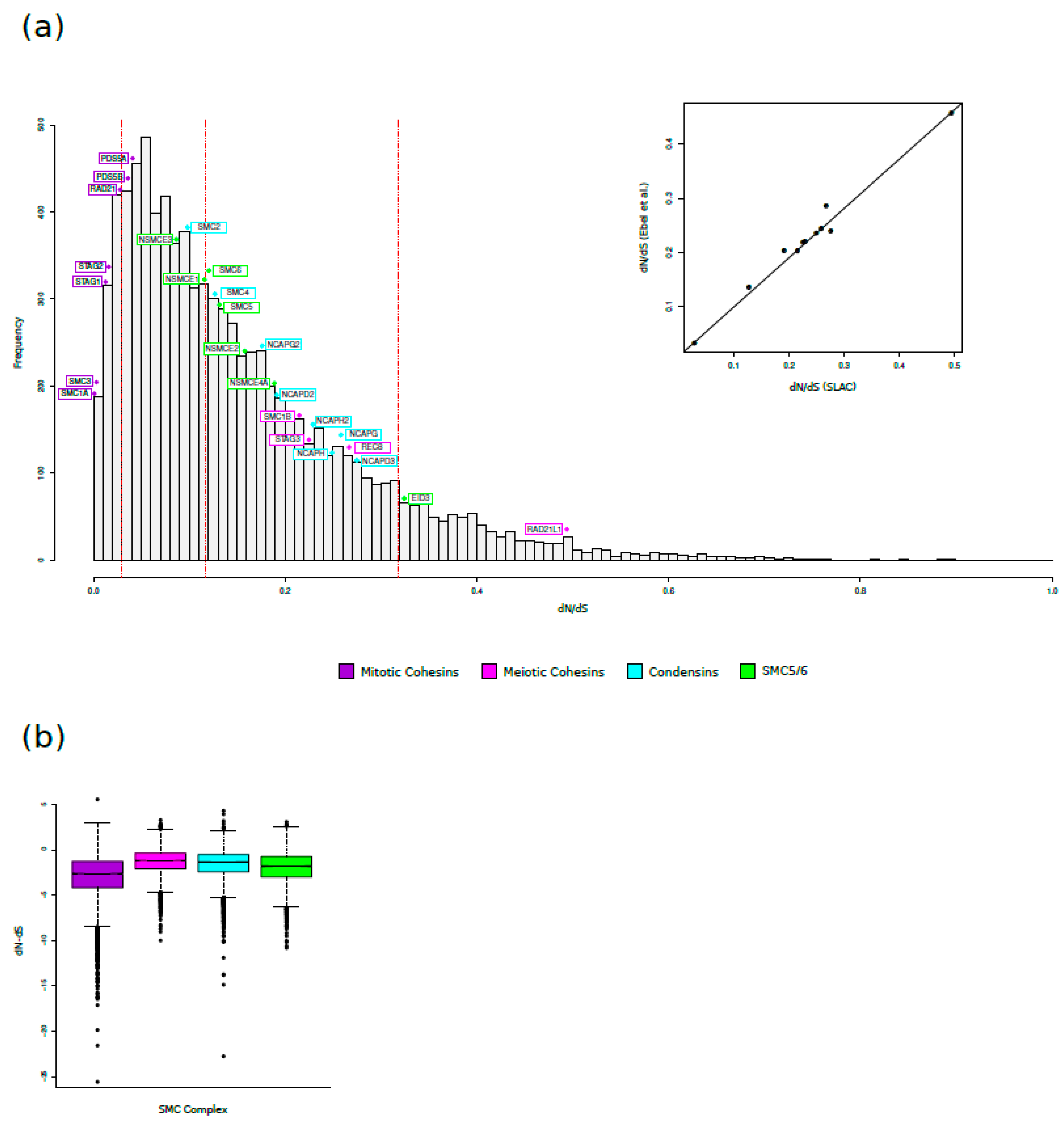
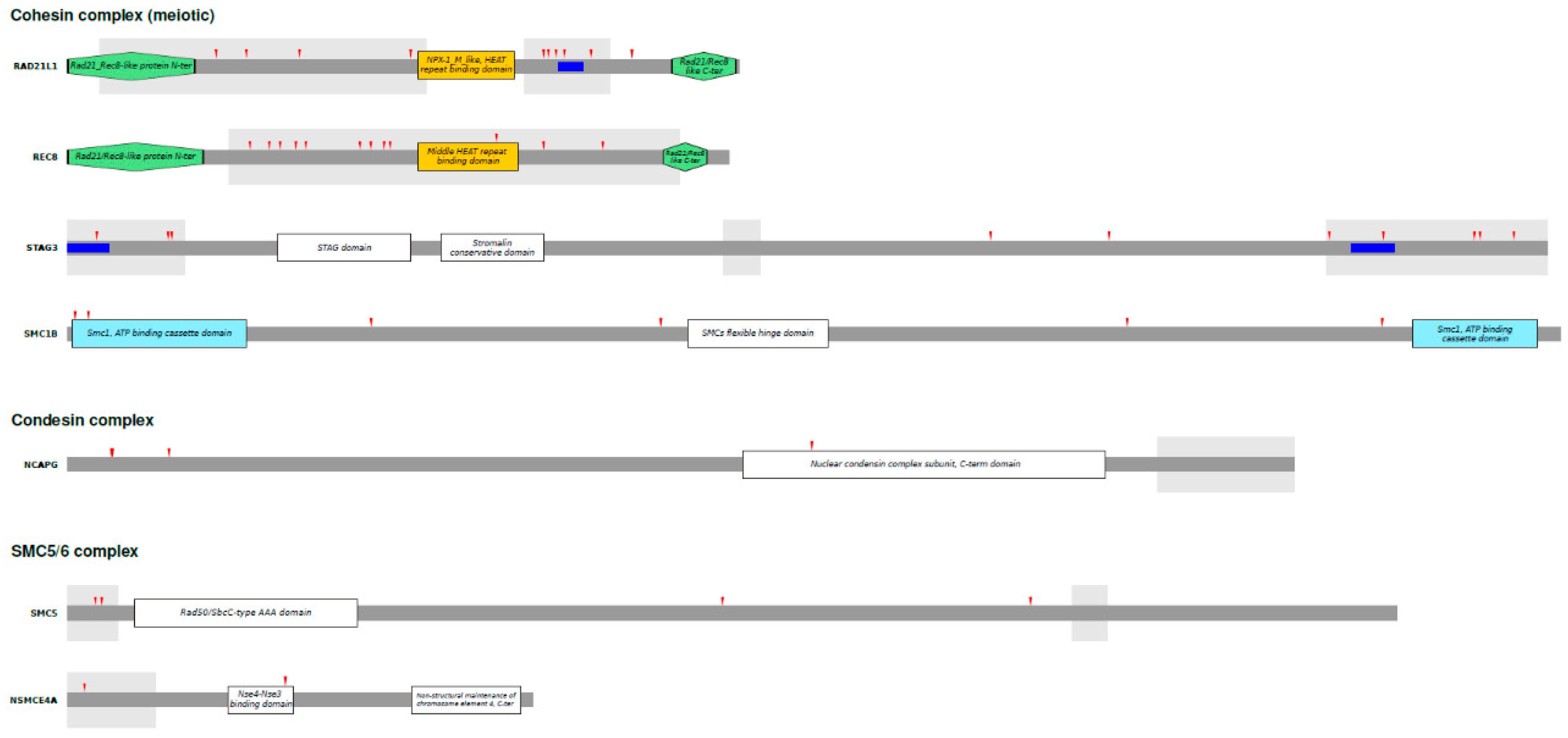
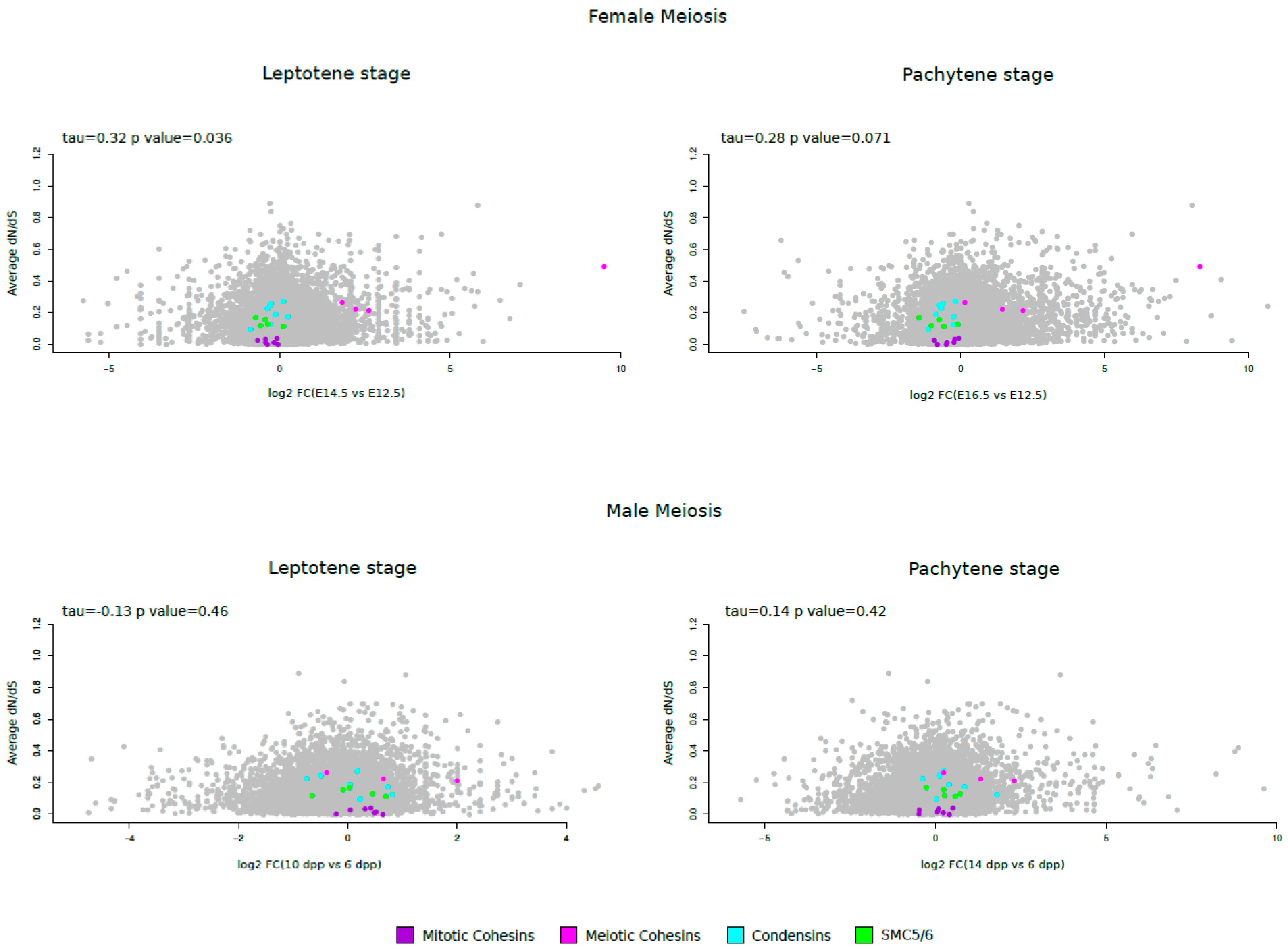
| Gene | Alias gene symbol | Subunits | n. of species | dN/dS | |
|---|---|---|---|---|---|
| Cohesin complex | |||||
| RAD21 | SCC1 | Kleisin | 63 | 0.028 | |
| RAD21L1* | RAD21L | Kleisin | 63 | 0.494 | |
| REC8* | - | Kleisin | 63 | 0.267 | |
| SMC1A | - | SMC | 61 | 0.003 | |
| SMC1B* | - | SMC | 60 | 0.215 | |
| SMC3 | - | SMC | 63 | 0.001 | |
| PDS5A | SCC112 | HEAT-A | 57 | 0.041 | |
| PDS5B | APRIN, AS3 | HEAT-A | 60 | 0.036 | |
| STAG1 | SA1 | HEAT-B | 63 | 0.013 | |
| STAG2 | SA2 | HEAT-B | 63 | 0.016 | |
| STAG3* | SA3 | HEAT-B | 63 | 0.225 | |
| Condensin complex | |||||
| NCAPD2 | CAP-D2 | HEAT-A (I) | 62 | 0.191 | |
| NCAPD3 | CAP-D3 | HEAT-A (II) | 63 | 0.275 | |
| NCAPG | CAP-G | HEAT-B (I) | 63 | 0.258 | |
| NCAPG2 | CAP-G2 | HEAT-B (II) | 59 | 0.176 | |
| NCAPH | CAP-H | Kleisin (I) | 63 | 0.249 | |
| NCAPH2 | CAP-H2 | Kleisin (II) | 62 | 0.229 | |
| SMC2 | CAP-E | SMC | 62 | 0.098 | |
| SMC4 | CAP-C | SMC | 61 | 0.127 | |
| SMC5/6 complex | |||||
| NSMCE1 | NSE1 | Tandem-WHD E3 ligase | 60 | 0.120 | |
| NSMCE2 | NSE2 | SUMO ligase | 63 | 0.158 | |
| NSMCE3 | NSE3/MAGEG1 | Tandem-WHD | 54 | 0.087 | |
| NSMCE4A | NSE4A | Kleisin | 63 | 0.189 | |
| EID3 | NSMCE4B | Kleisin | 46 | 0.342 | |
| SMC5 | - | SMC | 63 | 0.131 | |
| SMC6 | - | SMC | 63 | 0.116 |
| Gene/ LRT model |
n. of species | F3x4 | F61 | Positively selected sitesc | |||
|---|---|---|---|---|---|---|---|
| -2ΔlnLa | p valueb | -2ΔlnLa | p valueb | ||||
| Cohesin Complex | |||||||
| RAD21L* | 63 | ||||||
| M1 vs M2 | 102.59 | 5.28x10-23 | 92.78 | 7.13x10-21 | 122,148,192,284,394,398, 404,411,477,433 |
||
| M7 vs M8 | 113.97 | 1.79x10-25 | 108.91 | 2.25x10-24 | |||
| REC8* | 63 | ||||||
| M1 vs M2 | 51.13 | 7.89x10-12 | 10.11 | 0.0064 | 152,168,191,199,253,264, 269, 358,400,449,178,244 |
||
| M7 vs M8 | 88.22 | 6.97x10-20 | 50.28 | 1.21x10-11 | |||
| SMC1B* | 60 | ||||||
| M1 vs M2 | 37.77 | 6.29x10-09 | 16.92 | 0.00021 | 6,18,251,491,877,1088 | ||
| M7 vs M8 | 105.04 | 1.55x10-23 | 55.29 | 9.85x10-13 | |||
| STAG3* | 62 | ||||||
| M1 vs M2 | 27.39 | 1.13x10-06 | 18.02 | 0.00012 | 24,83,86,764,862,1044, 1089,1154,1159,1197 |
||
| M7 vs M8 | 79.88 | 4.51x10-18 | 58.44 | 2.04x10-13 | |||
| Condensin Complex | |||||||
| NCAPG | 63 | ||||||
| M1 vs M2 | 46.98 | 6.29x10-11 | 48.72 | 2.63x10-11 | 36,37,84,616 | ||
| M7 vs M8 | 90.97 | 1.76x10-20 | 102.35 | 5.96x10-23 | |||
| SMC5/6 Complex | |||||||
| SMC5 | 63 | ||||||
| M1 vs M2 | 17.97 | 0.000125 | 7.91 | 0.019 | 797,38,542,33 | ||
| M7 vs M8 | 61.40 | 4.65x10-14 | 45.29 | 1.46x10-10 | |||
| NSMCE4A | 63 | ||||||
| M1 vs M2 | 33.96 | 4.22x10-08 | 22.82 | 1.11x10-05 | 14, 185 | ||
| M7 vs M8 | 45.11 | 1.60x10-10 | 35.79 | 1.69x10-08 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





