Submitted:
19 July 2024
Posted:
19 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material and Curing
2.2. Bacteria Culture Preparation and Isolation
2.3. Hemolysis Test of Isolates
2.4. Bacteria Identification and PCR Amplification
2.5. Phylogenic Analysis of the Bacillus Strains Isolated from Vanilla
2.6. Fermentation and Curing of Vanilla Beans
2.7. Determination of Volatile Compounds
2.7.1. Methanol Extraction
2.7.2. GC-MS Analysis
3. Results and Discussion
3.1. Strain Isolation and 16S rRNA Identification
3.2. Morphological Characteristics of the Bacillus Strains Isolated from Vanilla
3.3. Hemolysis Physiognomies of the Bacillus Strains
3.4. Phylogenic Tree of the Isolated Bacillus Strains from Vanilla
3.5. Volatile Compounds Analysis by GC-MS
3.5.1. Gas Chromatography Profiles
3.5.2. Analysis of Volatile Compounds
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iftikhar, T.; Majeed, H.; Waheed, M.; Zahra, S.S.; Niaz, M.; AL-Huqail, A.A. Vanilla. In Essentials of Medicinal and Aromatic Crops; Springer: 2023; pp. 341–371.
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.; Clements, M.A.; Arroyo, M.T.; Leebens-Mack, J. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proceedings of the Royal Society B: Biological Sciences 2015, 282, 20151553. [Google Scholar] [CrossRef] [PubMed]
- Dionísio, A.P.; Molina, G.; de Carvalho, D.S.; Dos Santos, R.; Bicas, J.; Pastore, G. Natural flavourings from biotechnology for foods and beverages. In Natural food additives, ingredients and flavourings; Elsevier: 2012; pp. 231–259.
- Frenkel, C.; Ranadive, A.S.; Vázquez, J.T.; Havkin-Frenkel, D. Curing of vanilla. Handbook of vanilla science and technology 2018, 191–221. [Google Scholar] [CrossRef]
- Ahmad, H.; Khera, R.A.; Hanif, M.A.; Ayub, M.A.; Jilani, M.I. Vanilla. In Medicinal plants of South Asia; Elsevier: 2020; pp. 657–669.
- Menon, S.; Nayeem, N. Vanilla planifolia: a review of a plant commonly used as flavouring agent. International Journal of Pharmaceutical Sciences Review and Research 2013, 20, 225–228. [Google Scholar]
- Hasing, T.; Tang, H.; Brym, M.; Khazi, F.; Huang, T.; Chambers, A.H. A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat Food 2020, 1, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Vijayalakshmi, S.; Disalva, X.; Srivastava, C.; Arun, A. Vanilla-natural vs artificial: a review. Research Journal of Pharmacy and Technology 2019, 12, 3068–3072. [Google Scholar] [CrossRef]
- Anuradha, K.; Shyamala, B.N.; Naidu, M.M. Vanilla--its science of cultivation, curing, chemistry, and nutraceutical properties. Crit Rev Food Sci Nutr 2013, 53, 1250–1276. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra Rao, S.; Ravishankar, G.A. Vanilla flavour: production by conventional and biotechnological routes. Journal of the Science of Food and Agriculture 2000, 80, 289–304. [Google Scholar] [CrossRef]
- Hernández-Hernández, J. Mexican vanilla production. Handbook of Vanilla science and technology 2018, 1–26. [Google Scholar]
- Coban, H.B. Organic acids as antimicrobial food agents: applications and microbial productions. Bioprocess and Biosystems Engineering 2020, 43, 569–591. [Google Scholar] [CrossRef]
- Liszkowska, W.; Berlowska, J. Yeast fermentation at low temperatures: Adaptation to changing environmental conditions and formation of volatile compounds. Molecules 2021, 26, 1035. [Google Scholar] [CrossRef]
- Palama, T.L.; Khatib, A.; Choi, Y.H.; Payet, B.; Fock, I.; Verpoorte, R.; Kodja, H. Metabolic changes in different developmental stages of Vanilla planifolia pods. J Agric Food Chem 2009, 57, 7651–7658. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Chen, K.Y.; Chou, C.Y.; Liao, H.Y.; Chen, H.C. New Insights on Volatile Components of Vanilla planifolia Cultivated in Taiwan. Molecules 2021, 26, 3608. [Google Scholar] [CrossRef]
- Gallage, N.J.; Møller, B.L. Vanilla: The most popular flavour. Biotechnology of natural products 2018, 3–24. [Google Scholar] [CrossRef]
- Ravier, A.; Chalut, P.; Belarbi, S.; Santerre, C.; Vallet, N.; Nhouchi, Z. Impact of the Post-Harvest Period on the Chemical and Sensorial Properties of planifolia and pompona Vanillas. Molecules 2024, 29, 839. [Google Scholar] [CrossRef] [PubMed]
- Roling, W.F.; Kerler, J.; Braster, M.; Apriyantono, A.; Stam, H.; van Verseveld, H.W. Microorganisms with a taste for vanilla: microbial ecology of traditional Indonesian vanilla curing. Appl Environ Microbiol 2001, 67, 1995–2003. [Google Scholar] [CrossRef]
- Khoyratty, S.; Kodja, H.; Verpoorte, R. Vanilla flavor production methods: a review. Industrial crops and products 2018, 125, 433–442. [Google Scholar] [CrossRef]
- Chen, Y.G.; Gu, F.L.; Li, J.H.; Xu, F.; He, S.Z.; Fang, Y.M. Bacillus vanillea sp. nov., Isolated from the Cured Vanilla Bean. Curr Microbiol 2015, 70, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Pak, F.E.; Gropper, S.; Dai, W.D.; Havkin-Frenkel, D.; Belanger, F.C. Characterization of a multifunctional methyltransferase from the orchid Vanilla planifolia. Plant Cell Rep 2004, 22, 959–966. [Google Scholar] [CrossRef]
- Xu, F.; Chen, Y.; Cai, Y.; Gu, F.; An, K. Distinct Roles for Bacterial and Fungal Communities During the Curing of Vanilla. Front Microbiol 2020, 11, 552388. [Google Scholar] [CrossRef]
- Peña-Barrientos, A.; Perea-Flores, M.d.J.; Martínez-Gutiérrez, H.; Patrón-Soberano, O.A.; González-Jiménez, F.E.; Vega-Cuellar, M.Á.; Dávila-Ortiz, G. Physicochemical, microbiological, and structural relationship of vanilla beans (Vanilla planifolia, Andrews) during traditional curing process and use of its waste. Journal of Applied Research on Medicinal and Aromatic Plants 2023, 32, 100445. [Google Scholar] [CrossRef]
- Lavine, B.K.; Corona, D.T.; Perera, U.D.N.T. Analysis of vanilla extract by reversed phase liquid chromatography using water rich mobile phases. Microchemical Journal 2012, 103, 49–61. [Google Scholar] [CrossRef]
- Ali, L.; Perfetti, G.; Diachenko, G. Rapid method for the determination of coumarin, vanillin, and ethyl vanillin in vanilla extract by reversed-phase liquid chromatography with ultraviolet detection. J AOAC Int 2008, 91, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Gurnani, N.; Kapoor, N.; Mehta, D.; Gupta, M.; Mehta, B. Characterization of chemical groups and identification of novel volatile constituents in organic solvent extracts of cured Indian vanilla beans by GC-MS. Middle-East Journal of Scientific Research 2014, 22, 769–776. [Google Scholar]
- Sharma, A.; Verma, S.C.; Saxena, N.; Chadda, N.; Singh, N.P.; Sinha, A.K. Microwave- and ultrasound-assisted extraction of vanillin and its quantification by high-performance liquid chromatography in Vanilla planifolia. J Sep Sci 2006, 29, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Langford, V.S.; Padayachee, D.; McEwan, M.J.; Barringer, S.A. Comprehensive odorant analysis for on-line applications using selected ion flow tube mass spectrometry (SIFT-MS). Flavour and fragrance journal 2019, 34, 393–410. [Google Scholar] [CrossRef]
- Gu, F.; Chen, Y.; Hong, Y.; Fang, Y.; Tan, L. Comparative metabolomics in vanilla pod and vanilla bean revealing the biosynthesis of vanillin during the curing process of vanilla. AMB Express 2017, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Inai, Y.; Miyazawa, N.; Kurobayashi, Y.; Fujita, A. Key odorants in cured Madagascar vanilla beans (Vanilla planiforia) of differing bean quality. Biosci Biotechnol Biochem 2013, 77, 606–611. [Google Scholar] [CrossRef]
- Silva, A.P.; Gunata, Z.; Lepoutre, J.-P.; Odoux, E. New insight on the genesis and fate of odor-active compounds in vanilla beans (Vanilla planifolia G. Jackson) during traditional curing. Food Research International 2011, 44, 2930–2937. [Google Scholar] [CrossRef]
- Ghadiriasli, R.; Lorber, K.; Wagenstaller, M.; Buettner, A. Smoky, Vanilla, or Clove-Like? In Sex, Smoke, and Spirits: The Role of Chemistry; ACS Symposium Series; ACS Publications: 2019; pp. 43–54.
- Chen, C.-E.; Lin, Y.-S.; Lo, H.-C.; Hsu, T.-H. Structural Characterization with Laser Scanning Microscopy and an Analysis of Volatile Components Using GC-MS in Vanilla Pods Coated with Edible Microorganisms. Fermentation 2023, 9, 724. [Google Scholar] [CrossRef]
- Gu, F.; Chen, Y.; Fang, Y.; Wu, G.; Tan, L. Contribution of Bacillus isolates to the flavor profiles of vanilla beans assessed through aroma analysis and chemometrics. Molecules 2015, 20, 18422–18436. [Google Scholar] [CrossRef]
- Toth, S.; Lee, K.J.; Havkin-Frenkel, D.; Belanger, F.C.; Hartman, T.G. Volatile compounds in vanilla. Handbook of vanilla science and technology 2018, 285–347. [Google Scholar]
- Chen; Gu, F. L.; Li, J.H.; Xu, F.; He, S.Z.; Fang, Y.M. Bacillus vanillea sp. nov., Isolated from the Cured Vanilla Bean. Curr Microbiol 2015, 70, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Manyatsi, T.S.; Lin, Y.H.; Jou, Y.T. The isolation and identification of Bacillus velezensis ZN-S10 from vanilla (V. planifolia), and the microbial distribution after the curing process. Sci Rep 2024, 14, 16339. [Google Scholar] [CrossRef] [PubMed]
- Savardi, M.; Ferrari, A.; Signoroni, A. Automatic hemolysis identification on aligned dual-lighting images of cultured blood agar plates. Comput Methods Programs Biomed 2018, 156, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Anh, H.T.L.; Linh, B.N.H.; Dung, N.H.T.; Minh, D.D.; Le Quyen, T.T.; Trung, T.T. In vitro safety evaluation of Bacillus subtilis species complex isolated from Vietnam and their additional beneficial properties. Vietnam Journal of Biotechnology 2022, 20, 727–740. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol 2006, 72, 3832–3845. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rincón-Molina, C.I.; Martínez-Romero, E.; Aguirre-Noyola, J.L.; Manzano-Gómez, L.A.; Zenteno-Rojas, A.; Rogel, M.A.; Rincón-Molina, F.A.; Ruíz-Valdiviezo, V.M.; Rincón-Rosales, R. Bacterial Community with Plant Growth-Promoting Potential associated to pioneer plants from an active mexican volcanic complex. Microorganisms 2022, 10, 1568. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Wang, H.; Yang, F.; Chen, L.; Hao, F.; Lv, X.; Du, H.; Xu, Y. Effects of initial temperature on microbial community succession rate and volatile flavors during Baijiu fermentation process. Food Res Int 2021, 141, 109887. [Google Scholar] [CrossRef]
- Toy, J.Y.H.; Lu, Y.; Huang, D.; Matsumura, K.; Liu, S.Q. Enzymatic treatment, unfermented and fermented fruit-based products: current state of knowledge. Crit Rev Food Sci Nutr 2022, 62, 1890–1911. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, M.; Liu, F.; Zeng, S.; Hu, J. Identification of 2, 3-dihydro-3, 5-dihydroxy-6-methyl-4H-pyran-4-one as a strong antioxidant in glucose–histidine Maillard reaction products. Food research international 2013, 51, 397–403. [Google Scholar] [CrossRef]
- Murata, M. Browning and pigmentation in food through the Maillard reaction. Glycoconjugate Journal 2021, 38, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, S.; Zhang, X.; Meng, N.; Chai, X.; Wang, Y. Fermentation characteristics and the dynamic trend of chemical components during fermentation of Massa Medicata Fermentata. Arabian Journal of Chemistry 2022, 15, 103472. [Google Scholar] [CrossRef]
- van Schijndel, J.; Molendijk, D.; van Beurden, K.; Canalle, L.A.; Noël, T.; Meuldijk, J. Preparation of bio-based styrene alternatives and their free radical polymerization. European Polymer Journal 2020, 125, 109534. [Google Scholar] [CrossRef]
- Rehman, N.U.; Ansari, M.N.; Ahmad, W.; Amir, M. The Detailed Pharmacodynamics of the Gut Relaxant Effect and GC-MS Analysis of the Grewia tenax Fruit Extract: In Vivo and Ex Vivo Approach. Molecules 2022, 27, 8880. [Google Scholar] [CrossRef]
- McCormick, D. Characterisation of vanilla extracts based on sensory properties and chemical composition: a thesis presented in partial fulfilment of the requirements for the degree of Doctor of Philosophy in Food Technology at Massey University, New Zealand. Massey University, 2018.
- Hernández-Fernández, M.Á.; Rojas-Avila, A.; Vazquez-Landaverde, P.A.; Cornejo-Mazón, M.; Dávila-Ortiz, G. Volatile compounds and fatty acids in oleoresins from Vanilla Planifolia Andrews obtained by extraction with supercritical carbon dioxide. CyTA-Journal of Food 2019, 17, 419–430. [Google Scholar] [CrossRef]
- Al-Amin, M.; Rahiman, S.S.F.; Hossain, C.F.; Khairuddean, M.; Salhimi, S.M. Natural products from Rhynchostylis retusa (Orchidaceae), their chemophenetic significance and bioactivity. Biochemical Systematics and Ecology 2023, 111, 104737. [Google Scholar] [CrossRef]

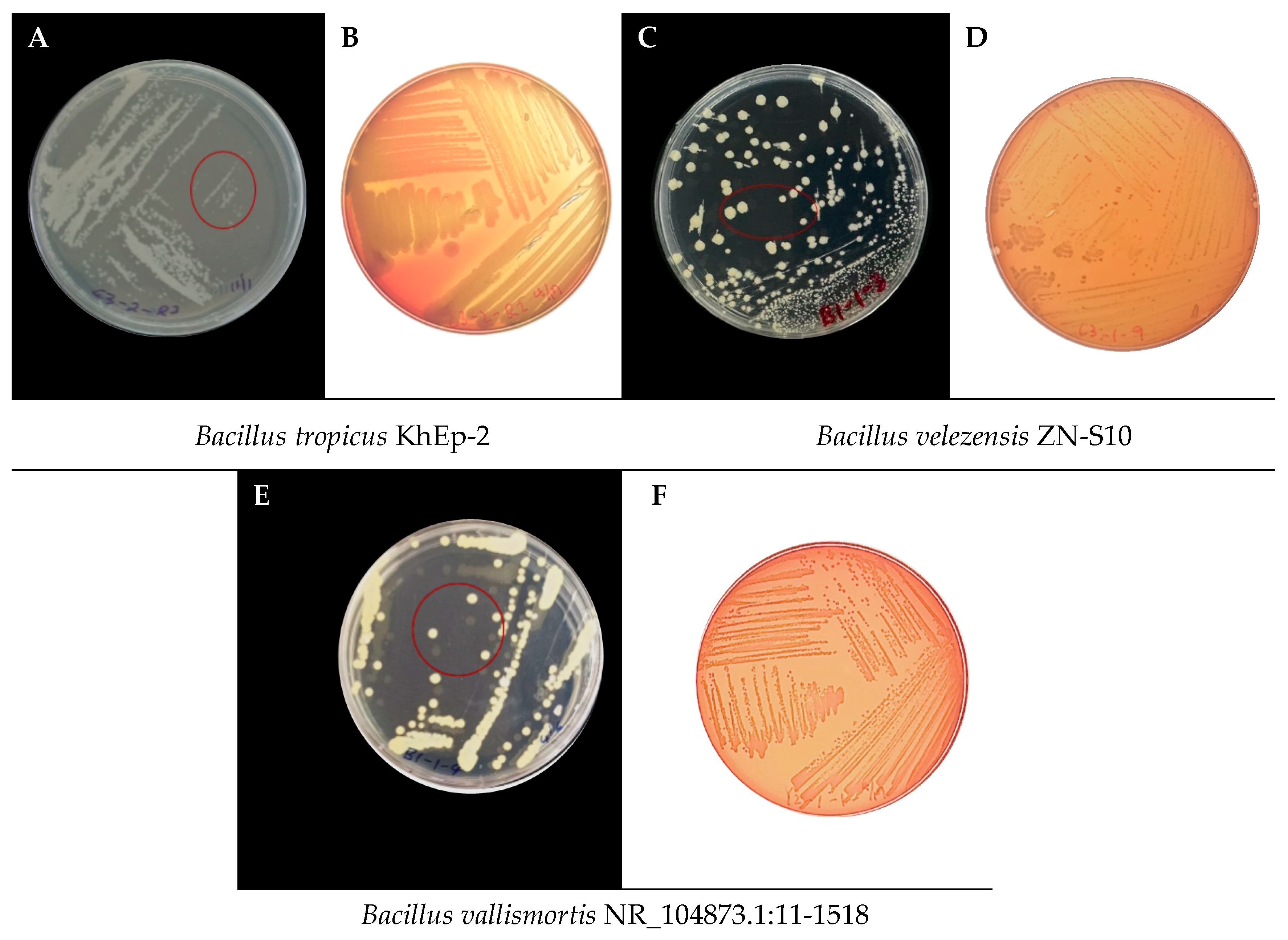
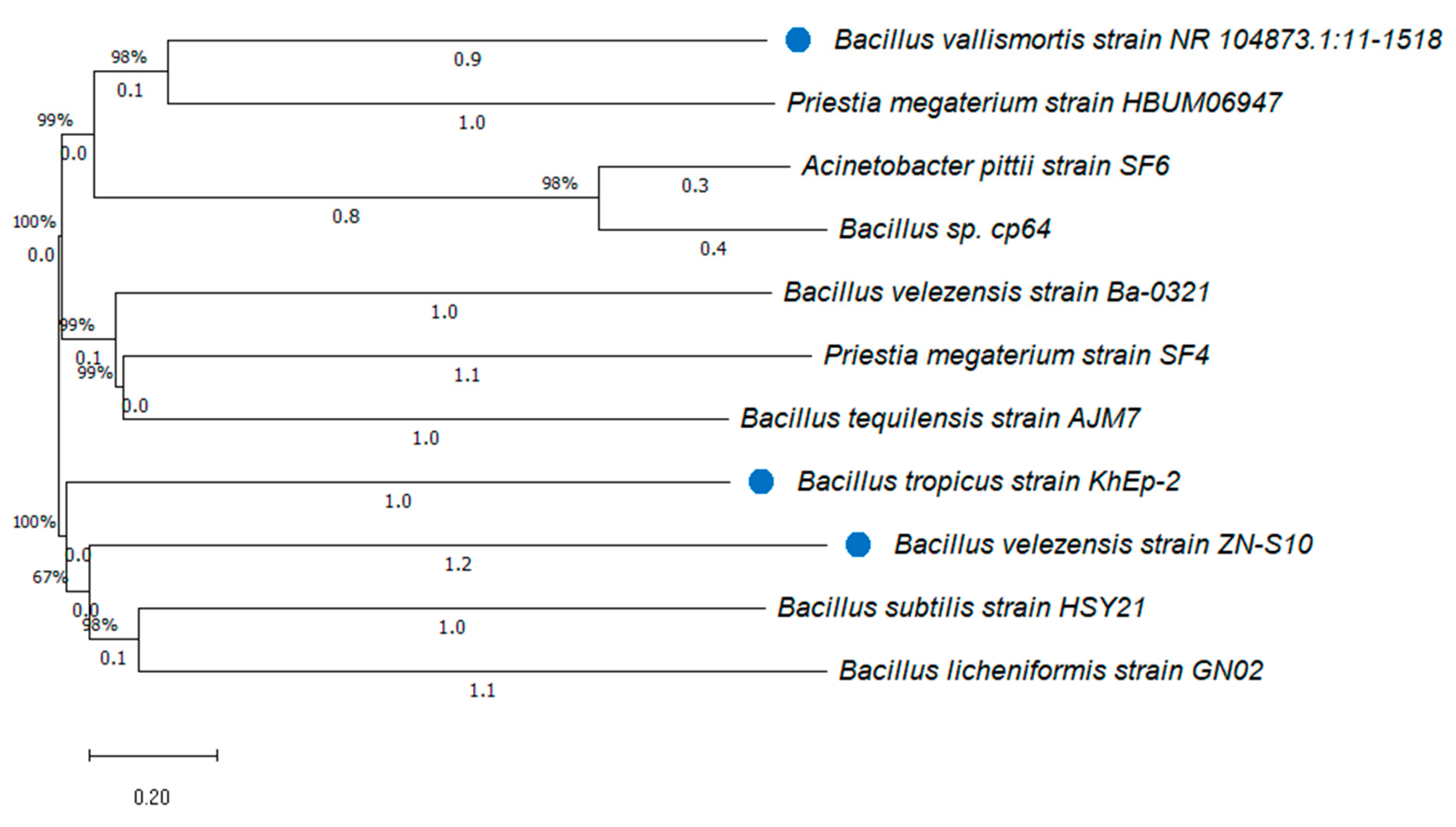
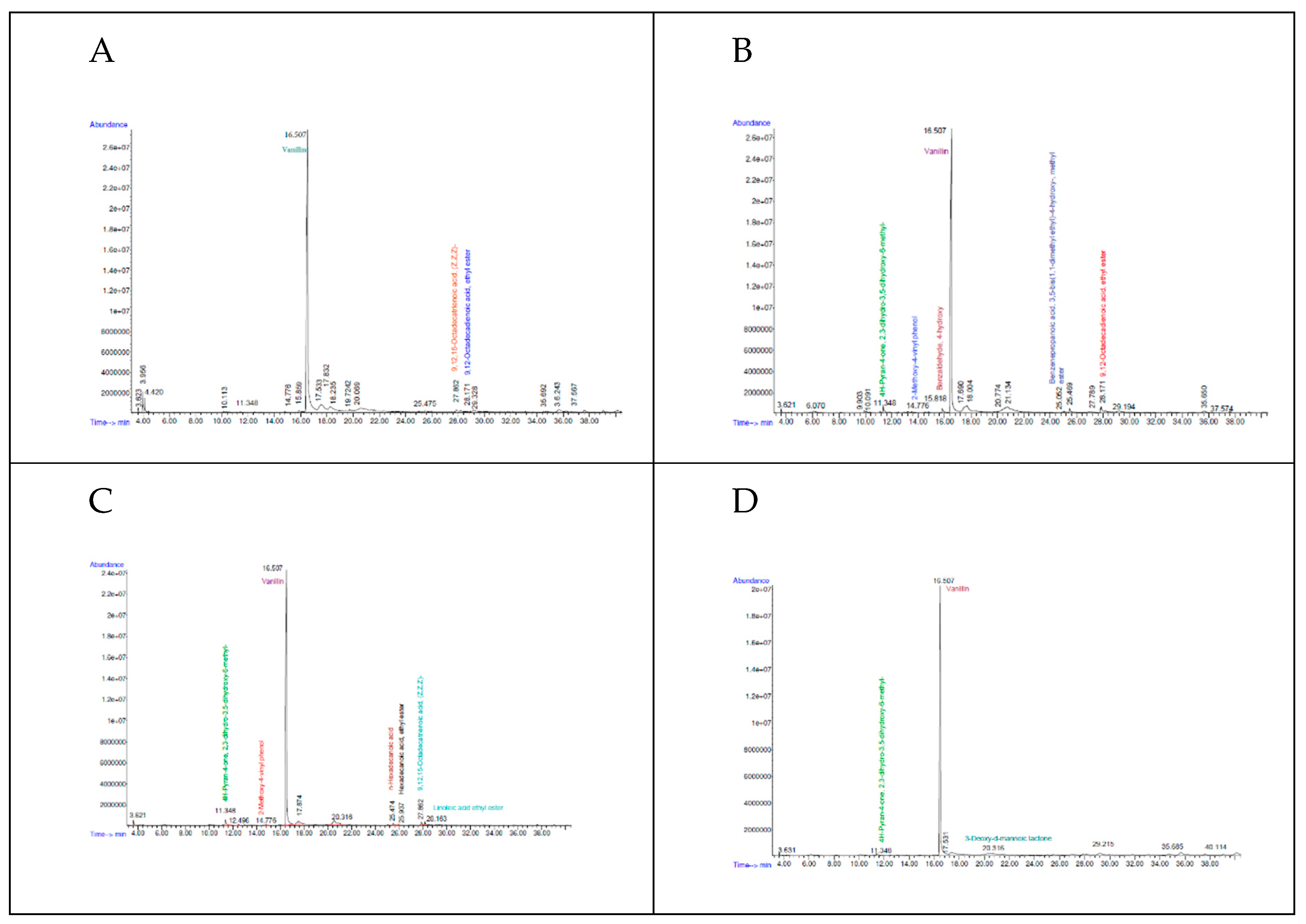
| Sample code | Strain name | Identity (%) | Accession length (bp) | Accession ID | Genome Sequence |
|---|---|---|---|---|---|
|
Bacillus tropicus KhEp-2 | 100.00 | 1510 | OP422217.1 | partial |
|
Bacillus velezensis ZN-S10 | 100.00 | 3929792 | CP102933.1 | complete |
|
Bacillus vallismortis NR_104873.1:11-1518 | 100.00 | 1508 | OP104906.1 | partial |
| Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compounds | RT (min) | Qualitative Score (%) | Mol weight (amu) | CAS no. | Control |
B. tropicus KhEp-2 |
B. velezensis ZN-S10 |
B. vallismortis NR_104873.1:11-1518 |
|
11.348 | 90 | 144.042 | 028564-83-2 | - | ✓ | ✓ | ✓ |
|
14.776 | 91 | 150.068 | 007786-61-0 | - | ✓ | ✓ | - |
|
15.816 | 93 |
122.037 | 006386-38-5 | - | ✓ | - | - |
|
16.507 | 96 | 152.047 | 000121-33-5 | ✓ | ✓ | ✓ | ✓ |
|
20.316 | 89 | 162.053 | 1000127-87-1 | - | - | - | ✓ |
|
25.474 | 98 | 256.240 | 000057-10-3 | - | - | ✓ | - |
|
25.937 | 96 | 278.272 | 000628-97-7 | - | - | ✓ | - |
|
25.052 | 87 | 292.204 | 006386-38-5 | - | ✓ | - | - |
|
27.862 | 99 | 278.225 | 000463-40-1 | ✓ | - | ✓ | - |
|
28.171 | 97 | 280.24 | 007619-08-1 | ✓ | ✓ | - | - |
|
28.163 | 99 | 308.272 | 000544-35-4 | - | - | ✓ | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





