Submitted:
04 July 2024
Posted:
05 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Structural Variations of PSI Complexes in Cyanobacteria
2.1. Oligomers of PSI Complexes in Cyanobacteria
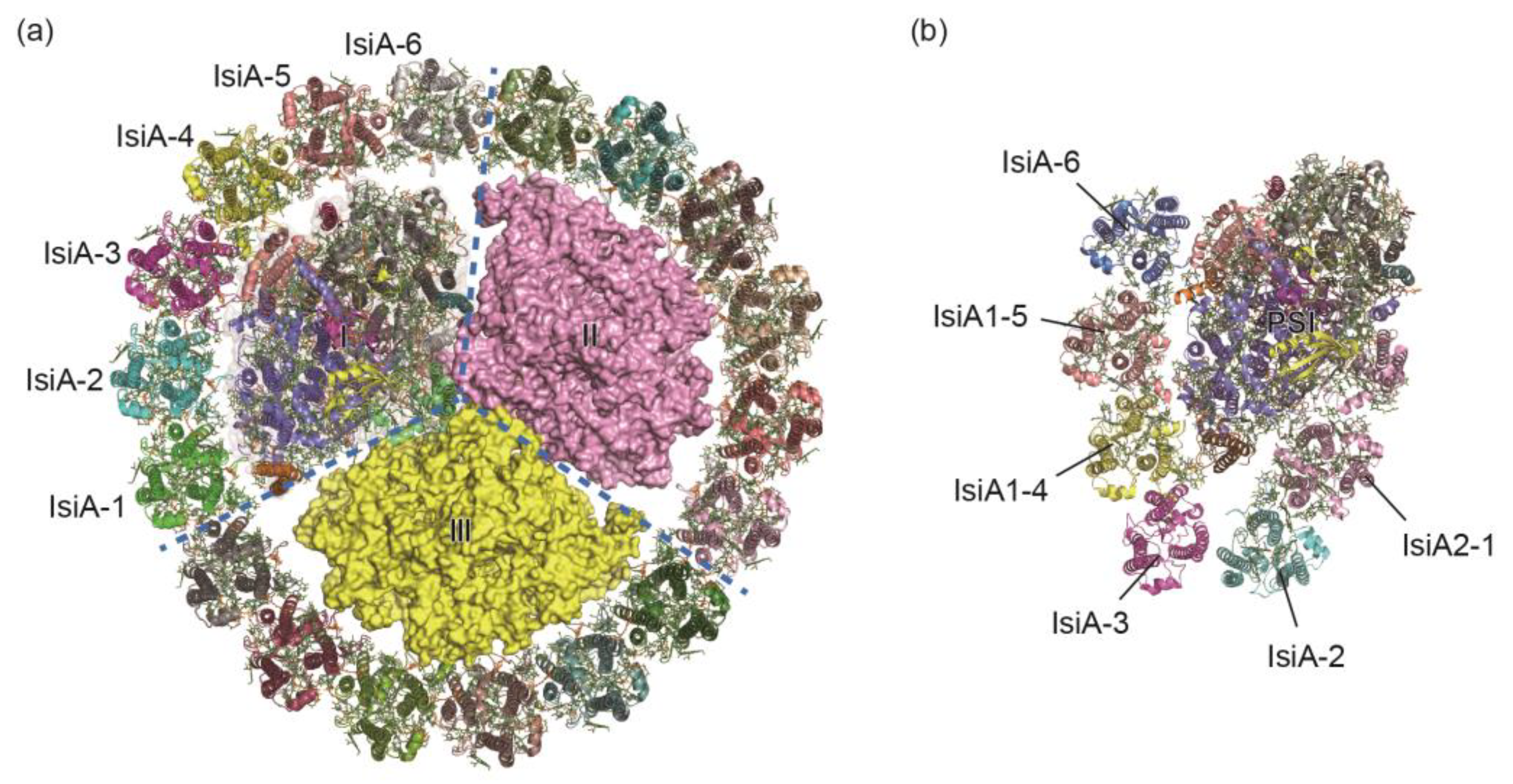
2.2. PSI-IsiA in Iron–Deficient Environment
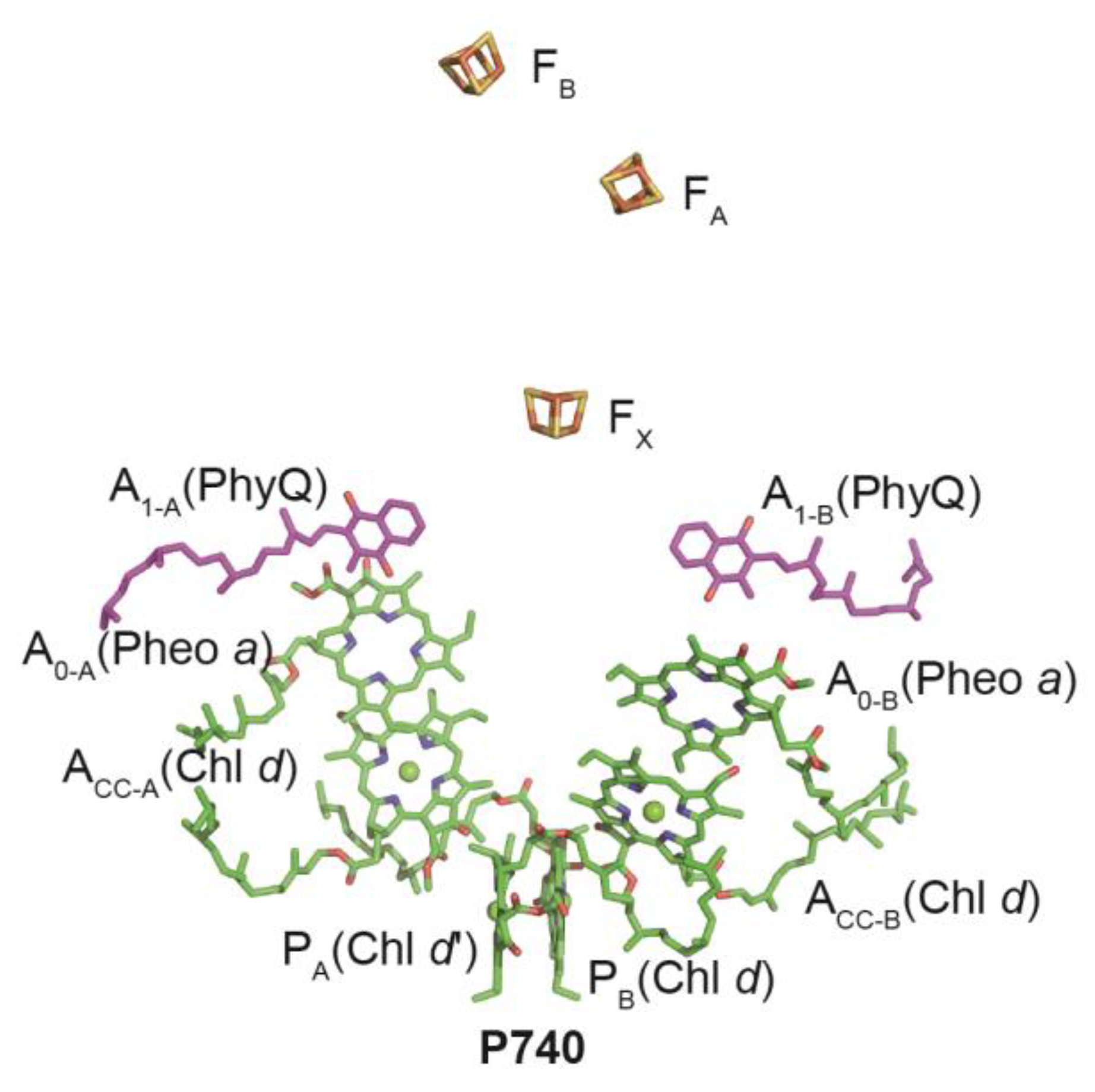
2.3. PSI Complexes from Chls d/f–Containing Cyanobacteria
3. Structural Variations of Algal PSI-LHCI Complexes
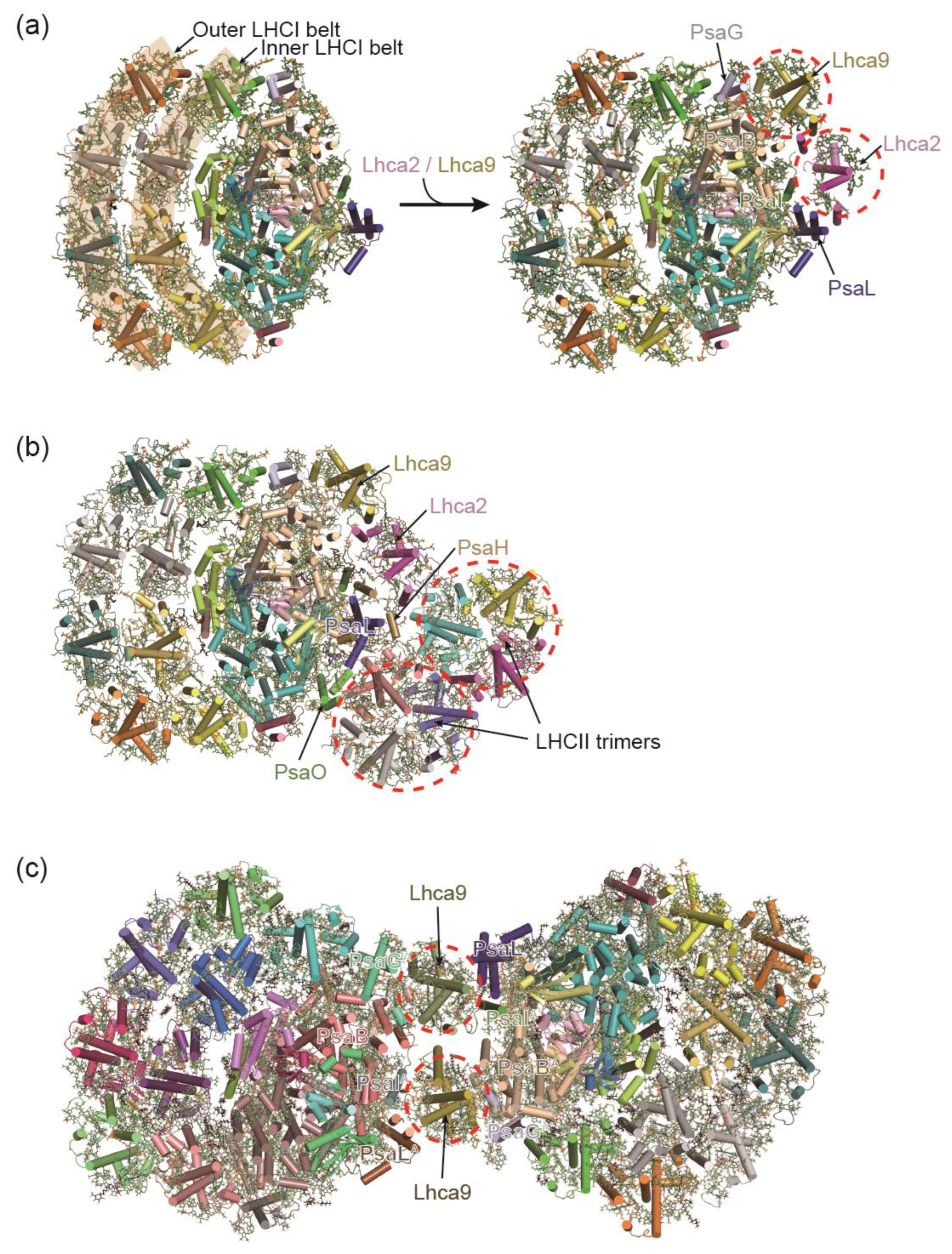
3.1. PSI-LHCI Complexes of Chlamydomonas reinhardtii
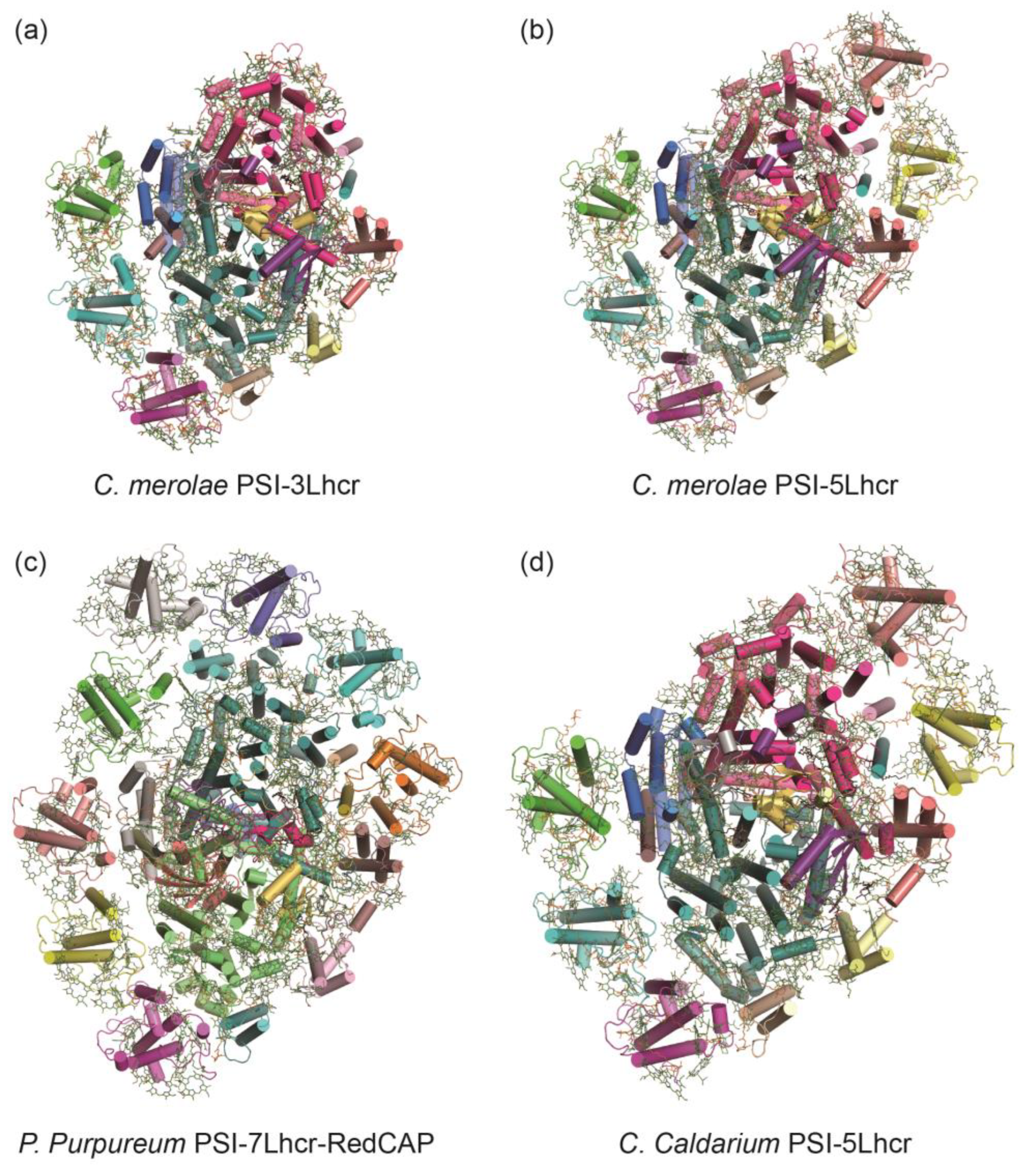
3.2. PSI-LHCI Complexes of Red Algae
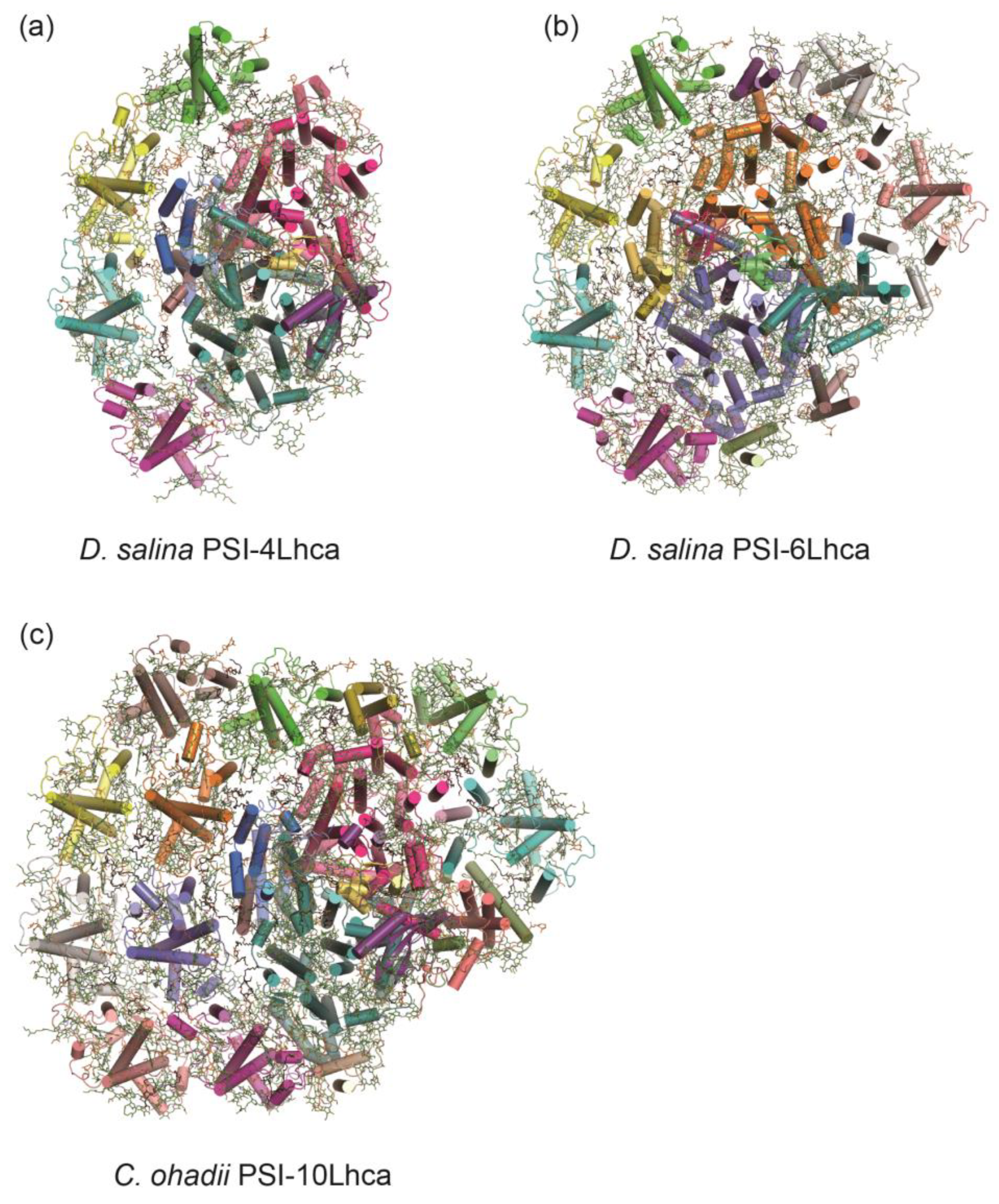
3.3. A Minimal PSI from Salt-Tolerant Green Alga Dunaliella salina
3.4. PSI–LHCI in Desert Algae Chlorella ohadii
3.5. Diatom PSI-FCPI Complex
3.6. Tetrameric PSI from Glaucophyte Algae
3.7. PSI-ACPI Complex in Cryptophytes
3.8. PSI-ACPPCI Complex in Symbiotic Dinoflagellates
4. Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nelson, N.; Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004, 5, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, P.R. Photosystem I: function and physiology. Annu. Rev. Plant Biol. 2001, 52, 593–626. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F. Photosynthesis of ATP-electrons, proton pumps, rotors, and poise. Cell 2002, 110, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, Y. Function and Structure of Cyanobacterial Photosystem I. In Photosynthesis: Structures, Mechanisms, and Applications; Chapter 7; Hou, H., Najafpour, M., Moore, G., Allakhverdiev, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 111–168. ISBN 978-3-319-48873-8. [Google Scholar]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauss, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Hippler, M.; Nelson, N. The plasticity of photosystem I. Plant Cell Physiol. 2021, 62, 1073–1081. [Google Scholar] [CrossRef]
- Bai, T.; Guo, L.; Xu, M.; Tian, L. Structural diversity of photosystem I and its light-harvesting system in eukaryotic algae and plants. Front. Plant Sci. 2021, 12, 781035. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.-R. Structure, function, and variations of the photosystem I-antenna supercomplex from different photosynthetic organisms, in Macromolecular Protein Complexes IV. Subcellular Biochemistry, Harris J. R., Marles-Wright J., Eds. 2022, 99, 351–377. [Google Scholar]
- Suga, M.; Shen, J.-R. Structural variations of photosystem I-antenna supercomplex in response to adaptations to different light environments. Curr. Opin. Struct. Biol. 2020, 63, 10–17. [Google Scholar] [CrossRef]
- Grotjohann, I.; Fromme, P. Structure of cyanobacterial photosystem I. Photosynth. Res. 2005, 85, 51–72. [Google Scholar] [CrossRef]
- Çoruh, O.; Frank, A.; Tanaka, H.; Kawamoto, A.; El-Mohsnawy, E.; Kato, T.; Namba, K.; Gerle, C.; Nowaczyk, M.M.; Kurisu, G. Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster. Commun. Biol. 2021, 4, 304. [Google Scholar] [CrossRef]
- Nelson, N. Investigating the balance between structural conservation and functional flexibility in Photosystem I. Int. J. Mol. Sci. 2024, 25, 5073. [Google Scholar] [CrossRef] [PubMed]
- Rögner, M.; Mühlenhoff, U.; Boekema, E.J.; Witt, H.T. Mono-, di- and trimeric PS I reaction center complexes isolated from the thermophilic cyanobacterium Synechococcus sp.: Size, shape and activity. Biochim. Biophys. Acta. 1990, 1015, 415–424. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; He, Y.; Li, N.; He, J.; Zhang, Y. Diversity among cyanobacterial Photosystem I oligomers. Front. Microbiol. 2022, 12, 781826. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nagao, R.; Jiang, T.Y.; Ueno, Y.; Yokono, M.; Chan, S.K.; Watanabe, M.; Ikeuchi, M.; Shen, J.R.; Akimoto, S.; et al. Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy. Nat. Commun. 2019, 10, 4929. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Li, Y.; Li, X.; Zhong, Q.; Li, N.; Zhang, K.; Zhang, Y.; Chu, H.; Ma, C.; Li, G.; et al. Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria. Nat. Plants 2019, 5, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Perez-Boerema, A.; Zhang, L.; Li, Y.; Yang, M.; Li, S.; Amunts, A. Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly. Nat. Plants. 2020, 6, 314–320. [Google Scholar] [CrossRef]
- Watanabe, M.; Kubota, H.; Wada, H.; Narikawa, R.; Ikeuchi, M. Novel supercomplex organization of photosystem I in Anabaena and Cyanophora paradoxa. Plant Cell Physiol. 2011, 52, 162–168. [Google Scholar] [CrossRef]
- Li, M.; Semchonok, D.A.; Boekema, E.J.; Bruce, B.D. Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp TS-821. Plant Cell. 2014, 26, 1230–1245. [Google Scholar] [CrossRef]
- Li, M.; Calteau, A.; Semchonok, D.A.; Witt, T.A.; Nguyen, J.T.; Sassoon, N.; Boekema, E.J.; Whitelegge, J.; Gugger, M.; Bruce, B.D. Physiological and evolutionary implications of tetrameric photosystem I in cyanobacteria. Nat. Plants 2019, 5, 1309–1319. [Google Scholar] [CrossRef]
- Semchonok, D.A.; Mondal, J.; Cooper, C.J.; Schlum, K.; Li, M.; Amin, M.; Sorzano, C.O.S.; Ramı´rez-Aportela, E.; Kastritis, P.L.; Boekema1, E.J.; Guskov, A.; Bruce, B.D. Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium. Plant Commun. 2022, 3, 100248. [Google Scholar] [CrossRef]
- Netzer-El, S.Y.; Caspy, I.; Nelson, N. Crystal structure of Photosystem I monomer from Synechocystis PCC 6803. Front. Plant Sci. 2019, 9, 1865. [Google Scholar] [CrossRef]
- Malavath, T.; Caspy, I.; Netzer-El, S.Y.; Klaiman, D.; Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta. 2018, 1859, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, V.P.; Chitnis, P.R. PsaL subunit is required for the formation of Photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1993, 336, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Kłodawska, K.; Kovács, L.; Vladkova, R.; Rzaska, A.; Gombos, Z.; Laczkó-Dobos, H.; Malec, P. Trimeric organization of photosystem I is required to maintain the balanced photosynthetic electron flow in cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 2020, 143, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Cao, D.; Si, L.; Su, X.; Tian, L.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structural basis for energy and electron transfer of the photosystem I–IsiA–flavodoxin supercomplex. Nat. Plants. 2020, 6, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Toporik, H.; Li, J.; Williams, D.; Chiu, P.-L.; Mazor, Y. The structure of the stress-induced photosystem I–IsiA antenna supercomplex. Nat. Struct. Mol. Biol. 2019, 26, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Nagao, R.; Yokono, M.; Ueno, Y.; Suzuki, T.; Kato, K.; Kato, K.; Tsuboshita, N.; Jiang, T.; Dohmae, N.; Shen, J.-R.; Ehira, S.; Akimoto, S. Molecular organizations and function of iron-stress-induced-A protein family in Anabaena sp. PCC 7120. Biochim. Biophys. Acta. 2021, 862, 148327. [Google Scholar] [CrossRef]
- Nagao, R.; Kato, K.; Hamaguchi, T.; Ueno, Y.; Tsuboshita, N.; Shimizu, S.; Furutani, M.; Ehira, S.; Nakajima, Y.; Kawakami, K.; et al. Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120. Nat. Commun. 2023, 14, 920. [Google Scholar] [CrossRef]
- Akita, F.; Nagao, R.; Kato, K.; Nakajima, Y.; Yokono, M.; Ueno, Y.; Suzuki, T.; Dohmae, N.; Shen, J.-R.; Akimoto, S.; et al. Structure of a cyanobacterial photosystem I surrounded by octadecameric isiA antenna proteins. Commun. Biol. 2020, 3, 232. [Google Scholar] [CrossRef]
- Chen, H.-Y.S.; Bandyopadhyay, A.; Pakrasi, H.B. Function, regulation and distribution of IsiA, a membrane-bound chlorophyll a-antenna protein in cyanobacteria. Photosynthetica 2018, 56, 322–333. [Google Scholar] [CrossRef]
- Adir, N.; Bar-Zvi, S.; Harris, D. The amazing phycobilisome. Biochim. Biophys. Acta. 2020, 1861, 148047. [Google Scholar] [CrossRef]
- Harris, D.; Toporik, H.; Schlau-Cohen, G.S.; Mazor, Y. Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex. Nat. Commun. 2023, 14, 4650. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef]
- La Roche, J.; Van der Staay, G.W.M.; Partensky, F.; Ducret, A.; Aebersold, R.; Li, R.; Golden, S. S.; Hiller, R. G.; Wrench, P. M.; Larkum, A.W.; Green, B.R. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. USA. 1996, 93, 15244–15248. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Semchonok, D.A.; Webber-Birungi, M.T.; Ehira, S.; Kondo, K.; Narikawa, R.; Ohmori, M.; Boekema, E.J.; Ikeuchi, M. Attachment of phycobilisomes in an antenna–photosystem I supercomplex of cyanobacteria. Proc. Natl. Acad. Sci. USA. 2014, 111, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Zhang, X.; Cheng, J.; Xiao, Y.; Ma, J.; Sun, S.; Zhang, X.; Wang, H.W.; Sui, S.F. In situ structure of the red algal phycobilisome–PSII–PSI–LHC megacomplex. Nature 2023, 616, 199–206. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, J.; Liu, D.; Qin, S.; Sun, S.; Zhao, J.; Sui, S.-F. Structure of phycobilisome from the red alga Griffitshia pacifica. Nature 2017, 551, 57–63. [Google Scholar] [CrossRef]
- Boekema, E.J.; Hifney, A.; Yakushevska, A.E.; Piotrowski, M.; Keegstra, W.; Berry, S.; Michel, K.P.; Pistorius, E.K.; Kruip, J. A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 2001, 412, 745–748. [Google Scholar] [CrossRef]
- Bibby, T.S.; Nield, J.; Barber, J. Iron defciency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 2001, 412, 743–745. [Google Scholar] [CrossRef]
- Melkozernov, A.N.; Bibby, T.S.; Lin, S.; Barber, J.; Blankenship, R.E. Time-resolved absorption and emission show that the CP43′ antenna ring of iron-stressed Synechocystis sp. PCC6803 is efficiently coupled to the Photosystem I reaction center core. Biochemistry 2003, 42, 3893–3903. [Google Scholar] [CrossRef]
- Andrizhiyevskaya, E.G.; Frolov, D.; Van Grondelle, R.; Dekker, J.P. Energy transfer and trapping in the Photosystem I complex of Synechococcus PCC 7942 and in its supercomplex with IsiA. Biochim. Biophys. Acta. 2004, 1656, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Ryan-Keogh, T.J.; Macey, A.I.; Cockshutt, A.M.; Moore, C.M.; Bibby, T.S. The cyanobacterial chlorophyll-binding-protein isiA acts to increase the in vivo effective absorption cross-section of PSI under iron limitation. J Phycol. 2011, 48, 145–154. [Google Scholar] [CrossRef]
- Sandström, S.; Park, Y.-I.; Öquist, G.; Gustafsson, P. CP43′, the IsiA gene product, functions as an excitation energy dissipator in the cyanobacterium Synechococcus Sp. PCC 7942. Photochem. Photobiol. 2007, 74, 431–437. [Google Scholar] [CrossRef]
- Ihalainen, J.A.; D’Haene, S.; Yeremenko, N.; van Roon, H.; Arteni, A.A.; Boekema, E.J.; van Grondelle, R.; Matthijs, H.C.P.; Dekker, J.P. Aggregates of the chlorophyll-binding protein IsiA (CP43′ ) dissipate energy in cyanobacteria. Biochemistry 2005, 44, 10846–10853. [Google Scholar] [CrossRef] [PubMed]
- Yeremenko, N.; Kouřil, R.; Ihalainen, J.A.; D'haene, S.; Van Oosterwijk, N.; Andrizhiyevskaya, E.G.; Keegstra, W.; Dekker, H.L.; Hagemann, M.; Boekema, E.J.; et al. Supramolecular organization and dual function of the isia chlorophyll-binding protein in cyanobacteria. Biochemistry 2004, 43, 10308–10313. [Google Scholar] [CrossRef] [PubMed]
- Goñi, G.; Herguedas, B.; Hervás, M.; Peregrina, J.R.; De La Rosa, M.A.; Gómez-Moreno, C.; Navarro, J.A.; Hermoso, J.A.; Martínez-Júlvez, M.; Medina, M. Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP+ reductase. Biochim. Biophys. Acta. 2009, 1787, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Deng, J. Photosynthetic pigments: A study of photosynthetic pigments; John McCrae Secondary School, 2018. [Google Scholar]
- Chen, M.; Schliep, M.; Willows, R.D.; Cai, Z.L.; Neilan, B.A.; Scheer, H. A red-shifted chlorophyll. Science 2010, 329, 1318–1319. [Google Scholar] [CrossRef] [PubMed]
- Melkozernov, A.N.; Blankenship, R.E. Photosynthetic Functions of Chlorophylls. In Chlorophylls and Bacteriochlorophylls. Advances in Photosynthesis and Respiration; Grimm, B., Porra, R.J., Rüdiger, W., Scheer, H., Eds.; Springer: Dordrecht, 2006; pp. 397–412. ISBN 978-1-4020-4516-5. [Google Scholar]
- Motten, A.F. Diversity of Photosynthetic Pigments. in: Tested studies for laboratory teaching, Volume 16 (C. A. Goldman, Editor). Proceedings of the 16thWorkshop/Conference of the Association for Biology Laboratory Education (ABLE), 1995; pp. 81–98.
- Gisriel, C.J. Recent structural discoveries of photosystems I and II acclimated to absorb far-red light. Biochim. Biophys. Acta. 2024, 1865, 149032. [Google Scholar] [CrossRef] [PubMed]
- Büchel, C. Light harvesting complexes in chlorophyll c-containing algae. Biochim. Biophys. Acta. 2020, 1861, 148027. [Google Scholar] [CrossRef]
- Croce, R.; van Amerongen, H. Light harvesting in oxygenic photosynthesis: Structural biology meets spectroscopy. Science 2020, 369, eaay2058. [Google Scholar] [CrossRef]
- Gan, F.; Zhang, S.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Bryant, D.A. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 2014, 345, 1312–1317. [Google Scholar] [CrossRef]
- Airs, R.L.; Temperton, B.; Sambles, C.; Farnham, G.; Skill, S.C.; Llewellyn, C.A. Chlorophyll f and chlorophyll d are produced in the cyanobacterium Chlorogloeopsis fritschii when cultured under natural light and near–infrared radiation. FEBS Lett. 2014, 588, 3770–3777. [Google Scholar] [CrossRef] [PubMed]
- Behrendt, L.; Brejnrod, A.; Schliep, M.; Sørensen, S.J.; Larkum, A.W.; Kühl, M. Chlorophyll f-driven photosynthesis in a cavernous cyanobacterium. ISME J. 2015, 9, 2108–2111. [Google Scholar] [CrossRef]
- Miyashita, H.; Ikemoto, H.; Kurano, N.; Adachi, K.; Chihara, M.; Miyachi, S. Chlorophyll d as a major pigment. Nature 1996, 383, 402. [Google Scholar] [CrossRef]
- Miyashita, H.; Ohkubo, S.; Komatsu, H.; Sorimachi, Y.; Fukayama, D.; Fujinuma, D.; Akutsu, S.; Kobayashi, M. Discovery of chlorophyll d in Acaryochloris marina and chlorophyll f in a unicellular cyanobacterium, Strain KC1, isolated from Lake Biwa. J Phys Chem Biophys 2014, 4, 149. [Google Scholar] [CrossRef]
- Loughlin, P.; Lin, Y.; Chen, M. Chlorophyll d and Acaryochloris marina: current status. Photosynth. Res. 2013, 116, 277–293. [Google Scholar] [CrossRef]
- Hamaguchi, T.; Kawakami, K.; Shinzawa-Itoh, K.; Inoue-Kashino, N.; Itoh, S.; Ifuku, K.; Yamashita, E.; Maeda, K.; Yonekura, K.; Kashino, Y. Structure of the far-red light utilizing photosystem I of Acaryochloris marina. Nat. Commun. 2021, 12, 2333. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, Q.; Chen, J.; Shen, L.; Yi, X.; Huang, Z.; Wang, W.; Chen, M.; Kuang, T.; Shen, J.; et al. A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina. J. Integr. Plant Biol. 2021, 63, 1740–1752. [Google Scholar] [CrossRef]
- Chen, M.; Li, Y.; Birch, D.; Willows, R.D. A cyanobacterium that contains chlorophyll f–a red-absorbing photopigment. FEBS Lett. 2012, 586, 3249–3254. [Google Scholar] [CrossRef]
- Kühl, M.; Trampe, E.; Mosshammer, M.; Johnson, M.; Larkum, A.W.; Frigaard, N.U.; Koren, K. Substantial near-infrared radiation-driven photosynthesis of chlorophyll f-containing cyanobacteria in a natural habitat. eLife 2020, 9, e50871. [Google Scholar] [CrossRef]
- Shen, G.; Canniffe, D.P.; Ho, M.Y.; Kurashov, V.; van der Est, A.; Golbeck, J.H.; Bryant, D.A. Characterization of chlorophyll f synthase heterologously produced in Synechococcus sp. PCC 7002. Photosynth. Res. 2019, 140, 77–92. [Google Scholar] [CrossRef]
- Nürnberg, D.J.; Morton, J.; Santabarbara, S.; Telfer, A.; Joliot, P.; Antonaru, L.A.; Ruban, A.V.; Cardona, T.; Krausz, E.; Boussac, A.; Fantuzzi, A.; Rutherford, A.W. Photochemistry beyond the red limit in chlorophyll f-containing photosystems. Science 2018, 360, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Gisriel, C.; Shen, G.; Kurashov, V.; Ho, M.-Y.; Zhang, S.; Williams, D.; Golbeck, J.H.; Fromme, P.; Bryant, D.A. The structure of Photosystem I acclimated to far-red light illuminates an ecologically important acclimation process in photosynthesis. Sci. Adv. 2020, 6, eaay6415. [Google Scholar] [CrossRef]
- Kato, K.; Shinoda, T.; Nagao, R.; Akimoto, S.; Suzuki, T.; Dohmae, N.; Chen, M.; Allakhverdiev, S.I.; Shen, J.-R.; Akita, F.; et al. Structural basis for the adaptation and function of chlorophyll f in photosystem I. Nat. Commun. 2020, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-Y.; Shen, G.; Canniffe, D.P.; Zhao, C.; Bryant, D.A. Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 2016, 353, aaf9178. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Gan, F.; Shen, G.; Bryant, D.A. RfpA, RfpB, and RfpC are the master control elements of far-red light photoacclimation (FaRLiP). Front. Microbiol. 2015, 6, 1303. [Google Scholar] [CrossRef]
- Chen, M.; Hernandez-Prieto, M.A.; Loughlin, P.C.; Li, Y.; Willows, R.D. Genome and proteome of the chlorophyll f-producing cyanobacterium Halomicronema hongdechloris: adaptative proteomic shifts under different light conditions. BMC Genomics. 2019, 20, 207. [Google Scholar] [CrossRef]
- Dupuis, S.; Merchant, S.S. Chlamydomonas reinhardtii: a model for photosynthesis and so much more. Nat Methods 2023, 20, 1441–1442. [Google Scholar] [CrossRef]
- Qin, X.; Suga, M.; Kuang, T.; Shen, J.-R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 2015, 348, 989–995. [Google Scholar] [CrossRef]
- Suga, M.; Ozawa, S.-I.; Yoshida-Motomura, K.; Akita, F.; Miyazaki, N.; Takahashi, Y. Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat. Plants 2019, 5, 626–636. [Google Scholar] [CrossRef]
- Mullineaux, C.W. State transitions: an example of acclimation to low-light stress. J. Exp. Botany 2004, 56, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F. Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta. 1992, 1098, 275–335. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, W.J.; Santabarbara, S.; Mosebach, L.; Wollman, F.-A.; Rappaport, F. State transitions redistribute rather than dissipate energy between the two photosystems in Chlamydomonas. Nat. Plants 2016, 2, 16031. [Google Scholar] [CrossRef] [PubMed]
- Bellafiore, S.; Barneche, F.; Peltier, G.; Rochaix, J.-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 2005, 433, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Wollman, F.A. State-transitions-reveal-the-dynamics-and-flexibility-of-the-photosynthetic-apparatus. EMBO J. 2001, 20, 3623–3630. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, S.; Wang, C.; Lin, W.; Huang, C.; Fan, C.; Han, D.; Lu, D.; Xu, X.; Sui, S.; Zhang, L. Regulatory dynamics of the higher-plant PSI–LHCI supercomplex during state transitions. Mol. Plant. 2023, 16, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Li, M.; Pan, X. Dynamic regulation of the light-harvesting system through state transitions in land plants and green algae. Plants 2023, 2023. 12, 1173. [Google Scholar] [CrossRef]
- Huang, Z.; Shen, L.; Wang, W.; Mao, Z.; Yi, X.; Kuang, T.; Shen, J.-R.; Zhang, X.; Han, G. Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2. Nat. Commun. 2021, 12, 1100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tang, K.; Yan, Q.; Li, X.; Shen, L.; Wang, W.; He, Y.-K.; Kuang, T.; Han, G.; Shen, J.R.; Zhang, X. Structural insights into a unique PSI–LHCI–LHCII–Lhcb9 supercomplex from moss Physcomitrium patens. Nat. Plants 2023, 9, 832–846. [Google Scholar] [CrossRef]
- Lima-Melo, Y.; Kiliç, M.; Aro, E.-M.; Gollan, P.J. Photosystem I inhibition, protection and signalling: knowns and unknowns. Front. Plant Sci. 2021, 12, 791124. [Google Scholar] [CrossRef]
- Zavafer, A.; Mancilla, C. Concepts of photochemical damage of Photosystem II and the role of excessive excitation. J Photoch. Photobio. C. 2021, 47, 100421. [Google Scholar] [CrossRef]
- Su, J.; Jiao, Q.; Jia, T.; Hu, X. The photosystem-II repair cycle: updates and open questions. Planta 2023, 259, 20. [Google Scholar] [CrossRef]
- Naschberger, A.; Mosebach, L.; Tobiasson, V.; Kuhlgert, S.; Scholz, M.; Perez-Boerema, A.; Ho, T.T.H.; Vidal-Meireles, A.; Takahashi, Y.; Hippler, M.; et al. Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex. Nat. Plants 2022, 8, 1191–1201. [Google Scholar] [CrossRef]
- Su, X.; Ma, J.; Pan, X.; Zhao, X.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 2019, 5, 273–281. [Google Scholar] [CrossRef]
- Pi, X.; Tian, L.; Dai, H.-E.; Qin, X.; Cheng, L.; Kuang, T.; Sui, S.-F.; Shen, J.-R. Unique organization of photosystem I–light-harvesting supercomplex revealed by Cryo-EM from a red alga. Proc. Natl. Acad. Sci. USA. 2018, 115, 4423–4428. [Google Scholar] [CrossRef]
- Chang, L.; Tian, L.; Ma, F.; Mao, Z.; Liu, X.; Han, G.; Wang, W.; Yang, Y.; Kuang, T.; Pan, J.; et al. Regulation of photosystem I-light-harvesting complex I from a red alga Cyanidioschyzon Merolae in response to light intensities. Photosynth. Res. 2020, 146, 287–297. [Google Scholar] [CrossRef]
- Qin, X.; Pi, X.; Wang, W.; Han, G.; Zhu, L.; Liu, M.; Cheng, L.; Shen, J.-R.; Kuang, T.; Sui, S.-F. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 2019, 5, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Perez-Boerema, A.; Klaiman, D.; Caspy, I.; Netzer-El, S.Y.; Amunts, A.; Nelson, N. Structure of a minimal photosystem I from the green alga Dunaliella salina. Nat. Plants 2020, 6, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef] [PubMed]
- Treves, H.; Raanan, H.; Kedem, I.; Murik, O.; Keren, N.; Zer, H.; Berkowicz, S.M.; Giordano, M.; Norici, A.; Shotland, Y.; et al. The mechanisms whereby the green alga Chlorella ohadii, isolated from desert soil crust, exhibits unparalleled photodamage resistance. New Phytol. 2016, 210, 1229–1243. [Google Scholar] [CrossRef]
- Kedem, I.; Milrad, Y.; Kaplan, A.; Yacoby, I. Juggling lightning: how Chlorella ohadii handles extreme energy inputs without damage. Photosynth. Res. 2021, 147, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Caspy, I.; Neumann, E.; Fadeeva, M.; Liveanu, V.; Savitsky, A.; Frank, A.; Kalisman, Y.L.; Shkolnisky, Y.; Murik, O.; Treves, H.; et al. Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii. Nat. Plants 2021, 7, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Malviya, S.; Scalco, E.; Audic, S.; Vincent, F.; Veluchamy, A.; Poulain, J.; Wincker, P.; Iudicone, D.; De Vargas, C.; Bittner, L.; Zingone, A.; Bowler, C. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 2016, 113, 1516–1525. [Google Scholar] [CrossRef] [PubMed]
- Büchel, C. Light-Harvesting Complexes of Diatoms: Fucoxanthin-Chlorophyll Proteins. In Photosynthesis in Algae: Biochemical and Physiological Mechanisms. Advances in Photosynthesis and Respiration; Larkum, A., Grossman, A., Raven, J., Eds.; Springer: Cham, Switzerland, 2020; pp. 441–457. ISBN 978-3-030-33397-3. [Google Scholar]
- Xu, C.; Pi, X.; Huang, Y.; Han, G.; Chen, X.; Qin, X.; Huang, G.; Zhao, S.; Yang, Y.; Kuang, T.; et al. Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex. Nat. Commun. 2020, 11, 5081. [Google Scholar] [CrossRef]
- Löffelhardt, W.; Bohnert, H.J.; Bryant, D.A.; Hagemann, R. The cyanelles of Cyanophora paradoxa. Crit. Rev. Plant Sci. 1997, 16, 393–413. [Google Scholar] [CrossRef]
- Kato, K.; Nagao, R.; Ueno, Y.; Yokono, M.; Suzuki, T.; Jiang, T.-Y.; Dohmae, N.; Akita, F.; Akimoto, S.; Miyazaki, N.; et al. Structure of a tetrameric photosystem I from a glaucophyte alga Cyanophora paradoxa. Nat. Commun. 2022, 13, 1679. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.W.; Schreiber, J.; Yue, J.; Guo, H.; Ding, Q.; Huang, J. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 2014, 5, 5764. [Google Scholar] [CrossRef]
- Zimorski, V.; Ku, C.; Martin,W. F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef]
- Kim, J.I.; Moore, C.E.; Archibald, J.M.; Bhattacharya, D.; Yi, G.; Yoon, H.S.; Shin, W. Evolutionary dynamics of cryptophyte plastid genomes. Genome Biol. Evol. 2017, 9, 1859–1872. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Sanchez-Puerta, M.V.; Delwiche, C.F. Evolution of light harvesting complex proteins from Chl c-containing algae. BMC Evol. Biol. 2011, 11, 101. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, P.; Li, K.; Zhang, Q.; He, F.; Li, C.; Su, H.; Chen, X.; Liu, L.; Zhang, Y. Structural basis and evolution of the photosystem I–light-harvesting supercomplex of cryptophyte algae. Plant Cell 2023, 35, 2449–2463. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.R.; McIlvin, M.R.; Moran, D.M.; Held, N.A.; Saunders, J.K.; Hawco, N.J.; Brosnahan, M.; DiTullio, G.R.; Lamborg, C.; McCrow, J.P.; et al. Dinoflagellates alter their carbon and nutrient metabolic strategies across environmental gradients in the central Pacific Ocean. Nat Microbiol. 2021, 6, 173–86. [Google Scholar] [CrossRef]
- Stephens, T.G.; Ragan, M.A.; Bhattacharya, D.; Chan, C.X. Core genes in diverse dinoflagellate lineages include a wealth of conserved dark genes with unknown functions. Sci. Rep. 2018, 8, 17175. [Google Scholar] [CrossRef] [PubMed]
- Janouškovec, J.; Gavelis, G.S.; Burki, F.; Dinh, D.; Bachvaroff, T.R.; Gornik,S. G.; Bright,K. J.; Imanian, B.; Strom, S.L.; Delwiche, C.F.; et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc Natl Acad Sci USA. 2017, 114, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef]
- Jacobovitz, M.R.; Hambleton, E.A.; Guse, A. Unlocking the complex cell biology of coral–dinoflagellate symbiosis: a model systems approach. Annu. Rev. Genet. 2023, 57, 411–434. [Google Scholar] [CrossRef] [PubMed]
- Hehenberger, E.; Burki, F.; Kolisko, M.; Keeling, P.J. Functional relationship between a dinoflagellate host and its diatom endosymbiont. Mol. Biol. Evol. 2016, 33, 2376–2390. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Wu, S.; He, J.; Wang, X.; Grossman, A.R. Shining light on dinoflagellate photosystem I. Nat Commun, 2024, 15, 3337. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Wang, F.; Zhao, S.; Xu, C.; Mao, Z.; Duan, J.; Feng, Y.; Yang, Y.; Shen, L.; et al. Structures and organizations of PSI–AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates. Proc. Natl. Acad. Sci. USA 2024, 121, e2315476121. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, N.; Li, K.; Li, C.; Guo, J.; He, F.; Liu, G.; Chen, X.; Gao, J.; Liu, L.; et al. Architecture of symbiotic dinoflagellate photosystem I–light-harvesting supercomplex in Symbiodinium. Nat. Commun. 2024, 15, 2392. [Google Scholar] [CrossRef]
- Gisriel, C.; Sarrou, I.; Ferlez, B.; Golbeck, J.H.; Redding, K.E.; Fromme, R. Structure of a symmetric photosynthetic reaction center–photosystem. Science 2017, 357, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Wu, H.; Xu, C.; Liu, X.-C.; Huang, Z.; Chang, S.; Wang, W.; Han, G.; Kuang, T.; Shen, J.-R.; et al. ,Architecture of the photosynthetic complex from a green sulfur bacterium. Science, 2020, 370, eabb6350. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Huang, G.; Wang, C.; Wang, J.; Sui, S.-F.; Qin, X. Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c. Nat. Commun. 2022, 13, 7745. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




