Submitted:
28 June 2024
Posted:
01 July 2024
You are already at the latest version
Abstract

Keywords:
1. Introduction
Materials and Methods
Materials
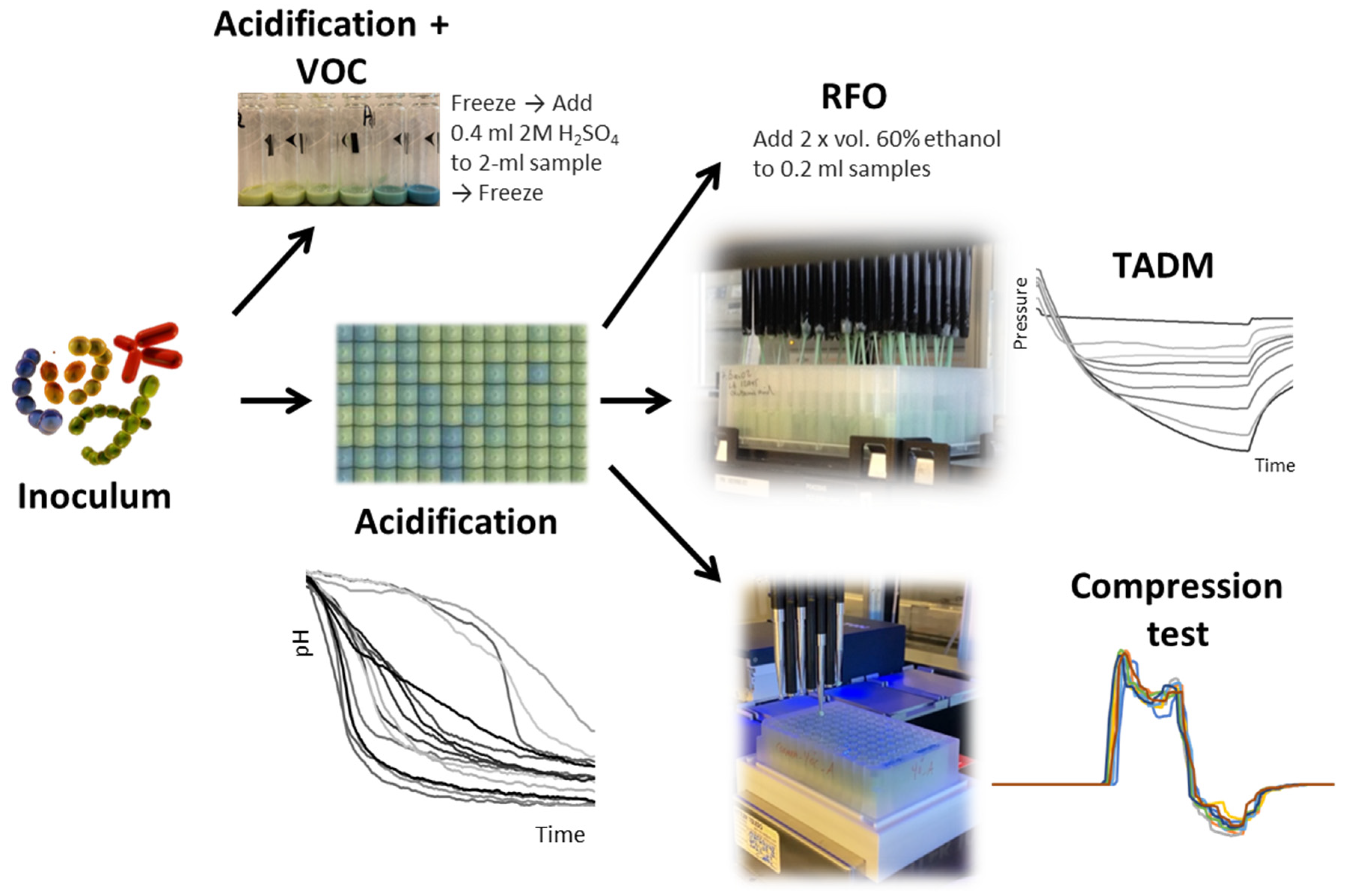
Base Preparation and Fermentation
Analysis of Volatile Organic Compounds
Raffinose Family Type, Alpha-Galactooligosaccharide Analysis
Carotenoid Production
Results and Discussion
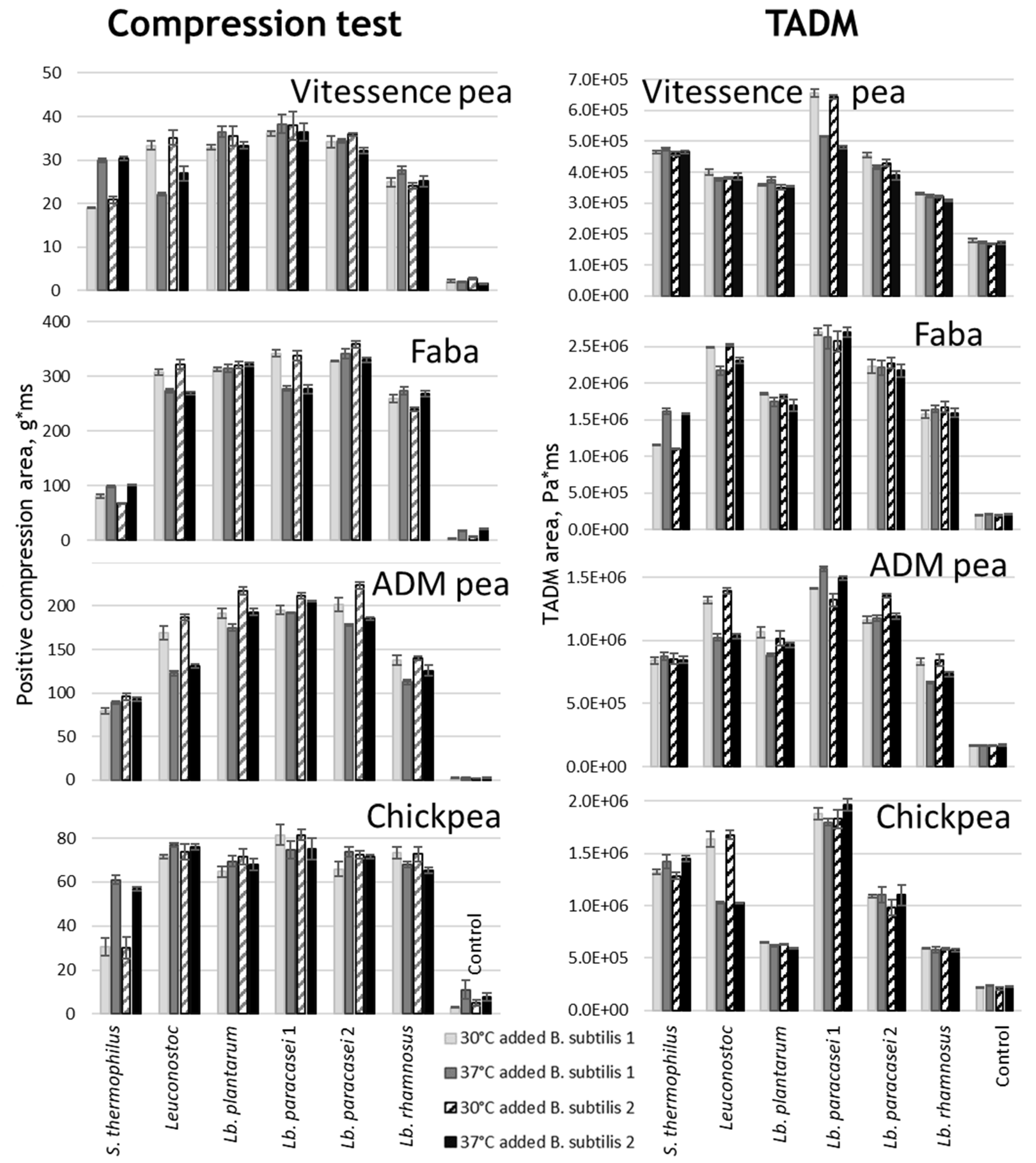
Texture Formation
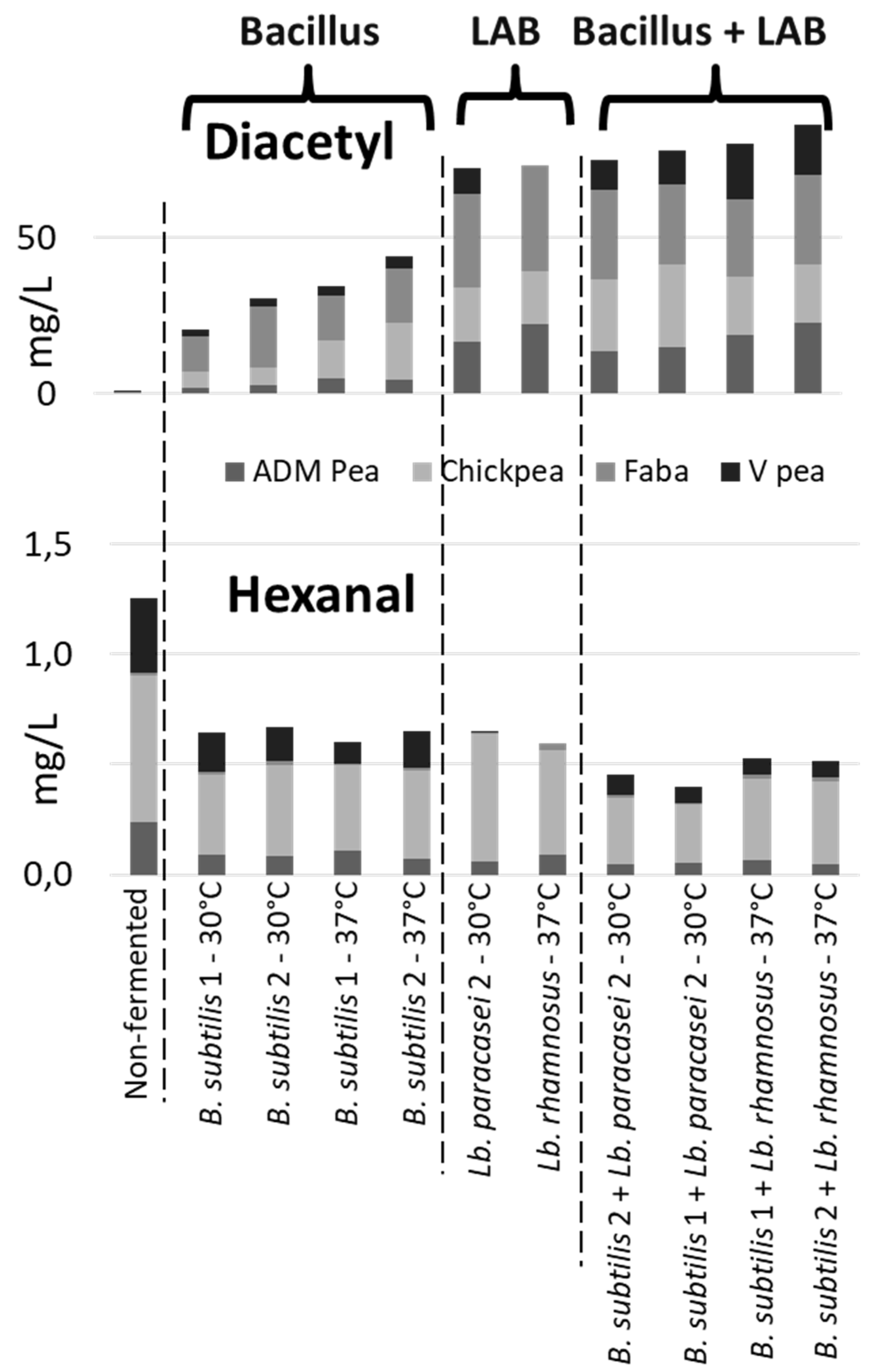
Reduction of Off-Flavors
Formation of Desirable Dairy Note Compounds
Ethanol and Ester Formation
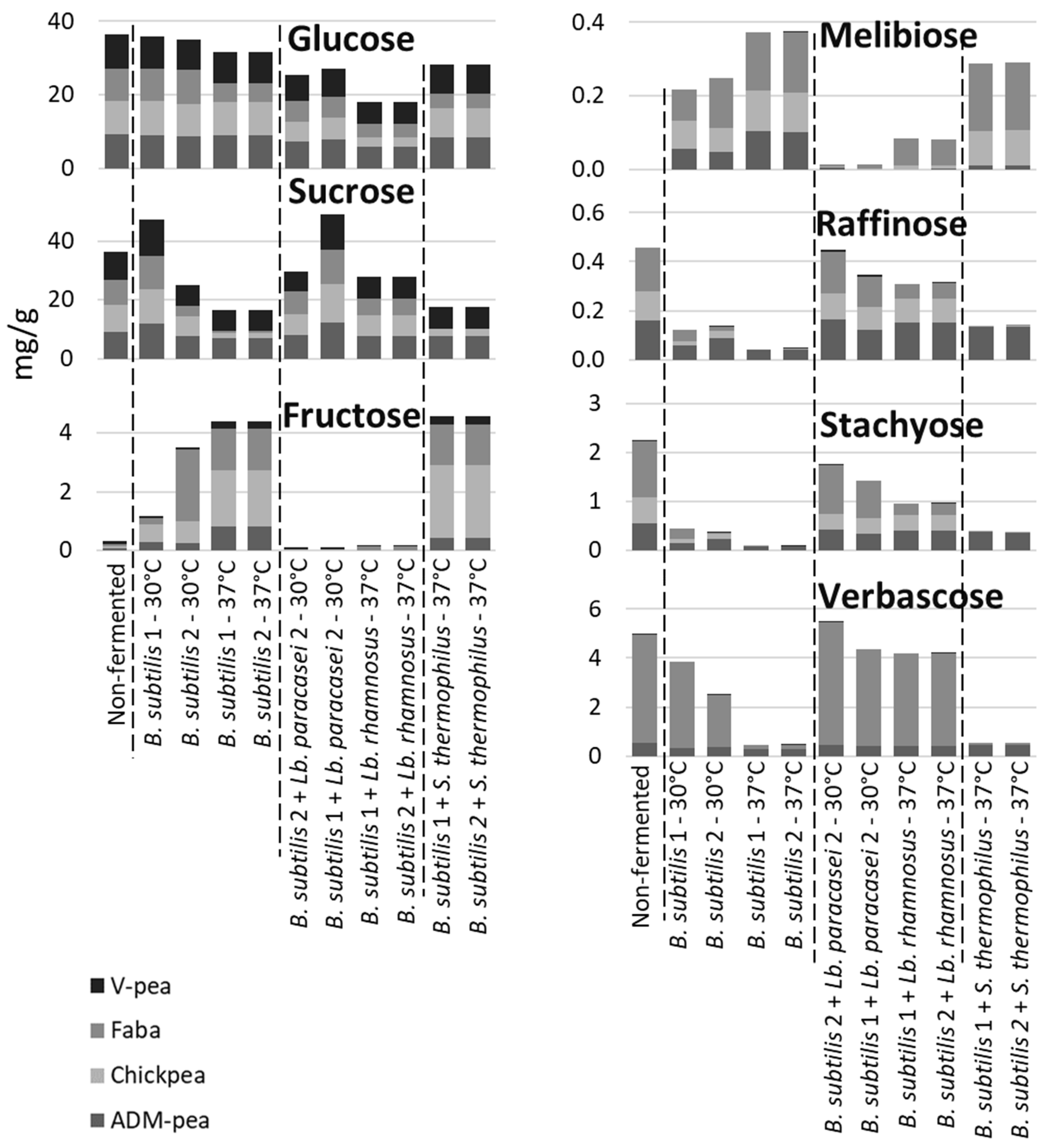
Degradation of Raffinose-Family Oligosaccharides
Pigment Production
Conclusion
Supplementary Materials
Acknowledgments
Author Details
Competing Interests
References
- D. J. McClements, Meat Less: The next food revolution, vol. 30, no. 1. 2023.
- A. A. Nolden and C. G. Forde, “The Nutritional Quality of Plant-Based Foods,” Sustainability (Switzerland), vol. 15, no. 4. 2023. doi: 10.3390/su15043324.
- V. K. Poulsen et al., “Versatile Lactococcus lactis strains improve texture in both fermented milk and soybean matrices,” FEMS Microbiol Lett, vol. 369, no. 1, 2022, doi: 10.1093/femsle/fnac117.
- M. Tangyu et al., “Flavour by design: food-grade lactic acid bacteria improve the volatile aroma spectrum of oat milk, sunflower seed milk, pea milk, and faba milk towards improved flavour and sensory perception,” Microb Cell Fact, vol. 22, no. 1, 2023, doi: 10.1186/s12934-023-02147-6.
- C. Masiá, R. Fernández-Varela, V. K. Poulsen, P. E. Jensen, and K. I. Sørensen, “Composition of bacterial blends for fermentation-induced pea protein emulsion gels using multi-property screening of lactic acid bacteria,” Food Biosci, vol. 56, 2023, doi: 10.1016/j.fbio.2023.103333.
- M. Tangyu, J. Muller, C. J. Bolten, and C. Wittmann, “Fermentation of plant-based milk alternatives for improved flavour and nutritional value,” Applied Microbiology and Biotechnology, vol. 103, no. 23–24. 2019. doi: 10.1007/s00253-019-10175-9.
- F. Beck, N. R. Pedersen, and D. S. Nielsen, “Fermented Rapeseed and Soybean Alone and in Combination with Macro Algae Inhibit Human and Pig Pathogenic Bacteria In Vitro,” Microorganisms, vol. 12, no. 5, 2024, doi: 10.3390/microorganisms12050891.
- A. R. Harper, R. C. J. Dobson, V. K. Morris, and G. J. Moggré, “Fermentation of plant-based dairy alternatives by lactic acid bacteria,” Microbial Biotechnology, vol. 15, no. 5. 2022. doi: 10.1111/1751-7915.14008.
- W. Engels, J. Siu, S. van Schalkwijk, W. Wesselink, S. Jacobs, and H. Bachmann, “Metabolic Conversions by Lactic Acid Bacteria during Plant Protein Fermentations,” Foods, vol. 11, no. 7, 2022, doi: 10.3390/foods11071005.
- K. M. Heravi, H. Watzlawick, and J. Altenbuchner, “The melredca operon encodes a utilization system for the raffinose family of oligosaccharides in bacillus subtilis,” J Bacteriol, vol. 201, no. 15, 2019, doi: 10.1128/JB.00109-19.
- D. Elango et al., “Raffinose Family Oligosaccharides: Friend or Foe for Human and Plant Health?,” Frontiers in Plant Science, vol. 13. 2022. doi: 10.3389/fpls.2022.829118.
- S. van Vliet et al., “A metabolomics comparison of plant-based meat and grass-fed meat indicates large nutritional differences despite comparable Nutrition Facts panels,” Sci Rep, vol. 11, no. 1, 2021, doi: 10.1038/s41598-021-93100-3.
- M. I. Ahmad, S. Farooq, Y. Alhamoud, C. Li, and H. Zhang, “Soy Leghemoglobin: A review of its structure, production, safety aspects, and food applications,” Trends in Food Science and Technology, vol. 141. 2023. doi: 10.1016/j.tifs.2023.104199.
- H. E. Khoo, A. Azlan, S. T. Tang, and S. M. Lim, “Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits,” Food and Nutrition Research, vol. 61. 2017. doi: 10.1080/16546628.2017.1361779.
- B. Coleman et al., “Potential of microalgae as flavoring agents for plant-based seafood alternatives,” Future Foods, vol. 5, 2022, doi: 10.1016/j.fufo.2022.100139.
- M. Kazir and Y. D. Livney, “Plant-based seafood analogs,” Molecules, vol. 26, no. 6. 2021. doi: 10.3390/molecules26061559.
- M. García-Vaquero, N. Brunton, and T. Lafarga, “Microalgae as a source of pigments for food applications,” in Cultured Microalgae for the Food Industry: Current and Potential Applications, 2021. doi: 10.1016/B978-0-12-821080-2.00014-9.
- V. Balasubramaniam, L. June Chelyn, S. Vimala, M. N. Mohd Fairulnizal, I. A. Brownlee, and I. Amin, “Carotenoid composition and antioxidant potential of Eucheuma denticulatum, Sargassum polycystum and Caulerpa lentillifera,” Heliyon, vol. 6, no. 8, 2020, doi: 10.1016/j.heliyon.2020.e04654.
- M. Kauser-Ul-alam, Y. Toba, S. Hioki, T. Hayakawa, H. Kumura, and J. I. Wakamatsu, “Lactococcus lactis subsp. cremoris Produces Zinc Protoporphyrin IX Both Aerobically and Anaerobically and Improves the Bright Red Color of Fermented Meat Products,” Foods, vol. 9, no. 11, 2020, doi: 10.3390/foods9111583.
- A. Solopova et al., “Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate,” Metab Eng, vol. 54, 2019, doi: 10.1016/j.ymben.2019.04.002.
- R. P. Pandey, P. Parajuli, M. A. G. Koffas, and J. K. Sohng, “Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology,” Biotechnology Advances, vol. 34, no. 5. 2016. doi: 10.1016/j.biotechadv.2016.02.012.
- J. Zha, X. Wu, and M. A. Koffas, “Making brilliant colors by microorganisms,” Current Opinion in Biotechnology, vol. 61. 2020. doi: 10.1016/j.copbio.2019.12.020.
- Q. Zhang et al., “De novo biosynthesis of carminic acid in Saccharomyces cerevisiae,” Metab Eng, vol. 76, 2023, doi: 10.1016/j.ymben.2023.01.005.
- R. J. N. Frandsen et al., “Heterologous production of the widely used natural food colorant carminic acid in Aspergillus nidulans,” Sci Rep, vol. 8, no. 1, 2018, doi: 10.1038/s41598-018-30816-9.
- G. Sandmann, H. Pollmann, S. Gassel, and J. Breitenbach, “Xanthophyllomyces dendrorhous, a Versatile Platform for the Production of Carotenoids and Other Acetyl-CoA-Derived Compounds,” in Advances in Experimental Medicine and Biology, vol. 1261, 2021. doi: 10.1007/978-981-15-7360-6_11.
- P. Buzzini, M. Innocenti, B. Turchetti, D. Libkind, M. Van Broock, and N. Mulinacci, “Carotenoid profiles of yeasts belonging to the genera Rhodotorula, Rhodosporidium, Sporobolomyces, and Sporidiobolus,” Can J Microbiol, vol. 53, no. 8, 2007, doi: 10.1139/W07-068.
- M. Legein, S. Wittouck, and S. Lebeer, “Latilactobacillus fragifolii sp. nov., isolated from leaves of a strawberry plant (Fragaria x ananassa),” Int J Syst Evol Microbiol, vol. 72, no. 1, 2022, doi: 10.1099/ijsem.0.005193.
- V. Kuzina and E. Cerdá-Olmedo, “Ubiquinone and carotene production in the Mucorales Blakeslea and Phycomyces,” Appl Microbiol Biotechnol, vol. 76, no. 5, 2007, doi: 10.1007/s00253-007-1069-7.
- E. Cerdá-Olmedo, “ Phycomyces and the biology of light and color ,” FEMS Microbiol Rev, vol. 25, no. 5, 2001, doi: 10.1111/j.1574-6976.2001.tb00588.x.
- M. J. Ruiz-Hidalgo, M. A. López-Matas, A. Velayos, P. D. Fraser, P. M. Bramley, and A. P. Eslava, “Carotenoid Mutants of Mucor circinelloides,” Botanica Acta, vol. 108, no. 4, 1995, doi: 10.1111/j.1438-8677.1995.tb00511.x.
- R. Rocchi, J. Zwinkels, M. Kooijman, A. Garre, and E. J. Smid, “Development of novel natto using legumes produced in Europe,” Heliyon, vol. 10, no. 5, 2024, doi: 10.1016/j.heliyon.2024.e26849.
- K. Bjerre, M. D. Cantor, T. Janzen, and P. Derkx, “Fermented milk inoculated with both lactic acid bacteria (LAB) and Bacillus,” US11684073B2, 2017.
- V. K. Poulsen, P. Derkx, and G. Oregaard, “High-throughput screening for texturing Lactococcus strains,” FEMS Microbiol Lett, vol. 366, no. 2, 2019, doi: 10.1093/femsle/fnz001.
- S. Lara-Abia, G. Lobo-Rodrigo, J. Welti-Chanes, and M. Pilar Cano, “Carotenoid and carotenoid ester profile and their deposition in plastids in fruits of new papaya (Carica papaya l.) varieties from the canary islands,” Foods, vol. 10, no. 2, 2021, doi: 10.3390/foods10020434.
- S. Saffarionpour, “Off-Flavors in Pulses and Grain Legumes and Processing Approaches for Controlling Flavor-Plant Protein Interaction: Application Prospects in Plant-Based Alternative Foods,” Food and Bioprocess Technology. 2023. doi: 10.1007/s11947-023-03148-4.
- E. Fischer, R. Cachon, and N. Cayot, “Impact of Ageing on Pea Protein Volatile Compounds and Correlation with Odor,” Molecules, vol. 27, no. 3, 2022, doi: 10.3390/molecules27030852.
- Y. Wang et al., “Flavor challenges in extruded plant-based meat alternatives: A review,” Compr Rev Food Sci Food Saf, vol. 21, no. 3, 2022, doi: 10.1111/1541-4337.12964.
- C. El Youssef, P. Bonnarme, S. Fraud, A. C. Péron, S. Helinck, and S. Landaud, “Sensory improvement of a pea protein-based product using microbial co-cultures of lactic acid bacteria and yeasts,” Foods, vol. 9, no. 3, 2020, doi: 10.3390/foods9030349.
- M. Xu, Z. Jin, Y. Lan, J. Rao, and B. Chen, “HS-SPME-GC-MS/olfactometry combined with chemometrics to assess the impact of germination on flavor attributes of chickpea, lentil, and yellow pea flours,” Food Chem, vol. 280, 2019, doi: 10.1016/j.foodchem.2018.12.048.
- V. Macciola, G. Candela, and A. De Leonardis, “Rapid gas-chromatographic method for the determination of diacetyl in milk, fermented milk and butter,” Food Control, vol. 19, no. 9, 2008, doi: 10.1016/j.foodcont.2007.08.014.
- M. Zhao et al., “Variation of Aroma Components of Pasteurized Yogurt with Different Process Combination before and after Aging by DHS/GC-O-MS,” Molecules, vol. 28, no. 4, 2023, doi: 10.3390/molecules28041975.
- T. B. Pedersen, D. Ristagno, P. L. H. McSweeney, F. K. Vogensen, and Y. Ardö, “Potential impact on cheese flavour of heterofermentative bacteria from starter cultures,” Int Dairy J, vol. 33, no. 2, 2013, doi: 10.1016/j.idairyj.2013.03.003.
- S. Rajendran, P. Silcock, and P. Bremer, “Flavour Volatiles of Fermented Vegetable and Fruit Substrates: A Review,” Molecules, vol. 28, no. 7. 2023. doi: 10.3390/molecules28073236.
- M. Boulay, M. Al Haddad, and F. Rul, “Streptococcus thermophilus growth in soya milk: Sucrose consumption, nitrogen metabolism, soya protein hydrolysis and role of the cell-wall protease PrtS,” Int J Food Microbiol, vol. 335, 2020, doi: 10.1016/j.ijfoodmicro.2020.108903.
- O. N. Donkor, A. Henriksson, T. Vasiljevic, and N. P. Shah, “α-Galactosidase and proteolytic activities of selected probiotic and dairy cultures in fermented soymilk,” Food Chem, vol. 104, no. 1, 2007, doi: 10.1016/j.foodchem.2006.10.065.
- Y. C. Wang, R. C. Yu, H. Y. Yang, and C. C. Chou, “Sugar and acid contents in soymilk fermented with lactic acid bacteria alone or simultaneously with bifidobacteria,” Food Microbiol, vol. 20, no. 3, 2003, doi: 10.1016/S0740-0020(02)00125-9.
- M. S. Garro, G. F. De Valdez, G. Oliver, and G. S. De Giori, “Growth characteristics and fermentation products of Streptococcus salivarius subsp. thermophilus, Lactobacillus casei and L. fermentum in soymilk,” European Food Research and Technology, vol. 206, no. 1, 1998, doi: 10.1007/s002170050217.
- M Kim M, DH Jung, DH Seo, WH Chung, MJ Seo, “Genome analysis of Lactobacillus plantarum subsp. plantarum KCCP11226 reveals a well-conserved C30 carotenoid biosynthetic pathway,”. 3 Biotech, vol 10, no 4, 2020,. doi: 10.1007/s13205-020-2149-y.
- C. Tanger, J. Engel, and U. Kulozik, “Influence of extraction conditions on the conformational alteration of pea protein extracted from pea flour,” Food Hydrocoll, vol. 107, 2020, doi: 10.1016/j.foodhyd.2020.105949.
- M. Emkani, B. Oliete, and R. Saurel, “Pea protein extraction assisted by lactic fermentation: Impact on protein profile and thermal properties,” Foods, vol. 10, no. 3, 2021, doi: 10.3390/foods10030549.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




