Submitted:
27 June 2024
Posted:
28 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
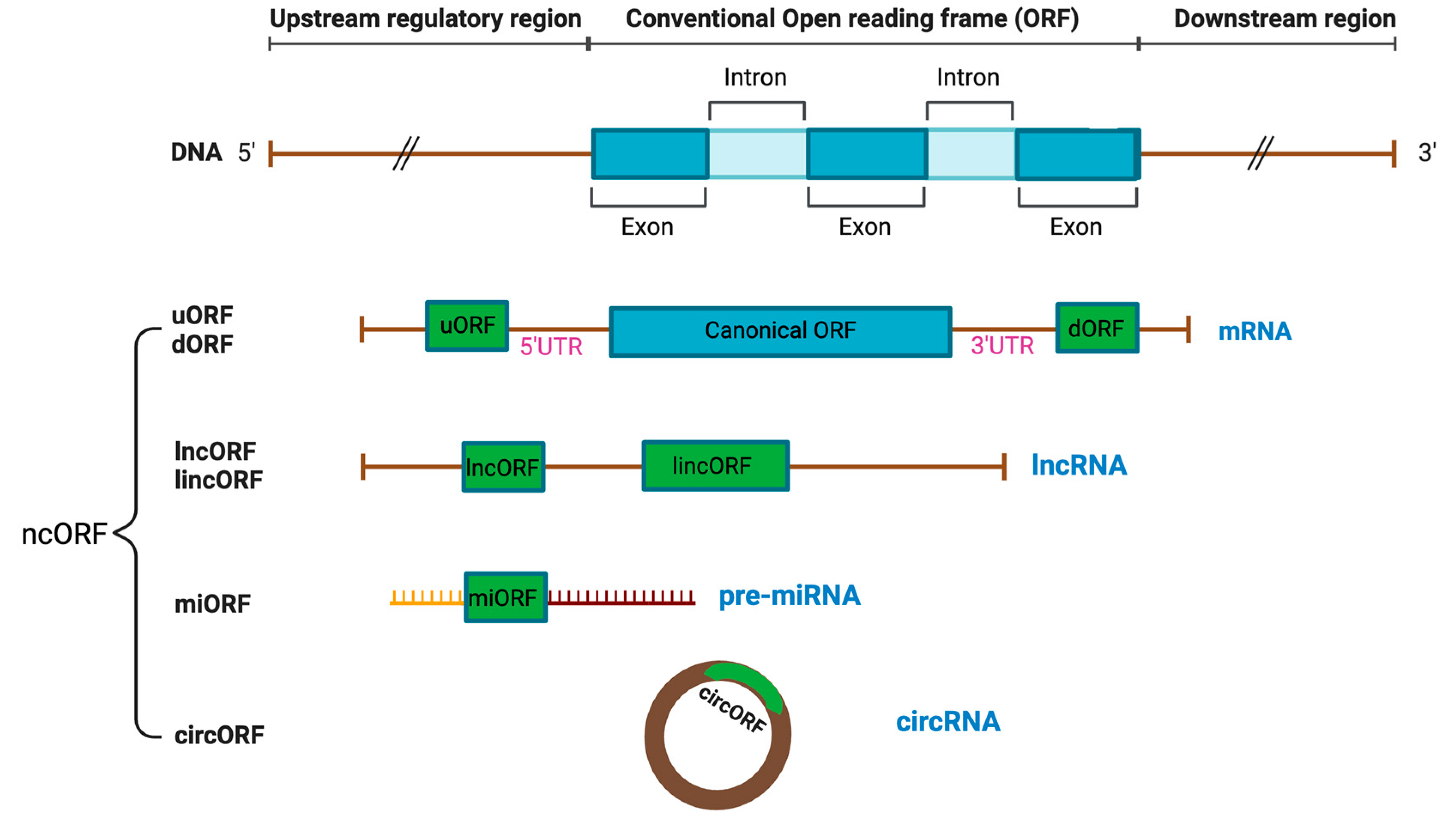
1.1. Definition of ncORF
1.2. Classification of ncORF
1.3. Identification of ncORF
2. ncORFs in Cancer Diagnosis and Prognosis
2.1. Colorectal Cancer
2.2. Breast Cancer
2.3. Glioblastoma
2.4. Hepatocellular Carcinoma (HCC)
2.5. Ovarian Cancer (OC)
2.6. Prostate Cancer
2.7. Other Types of Cancer
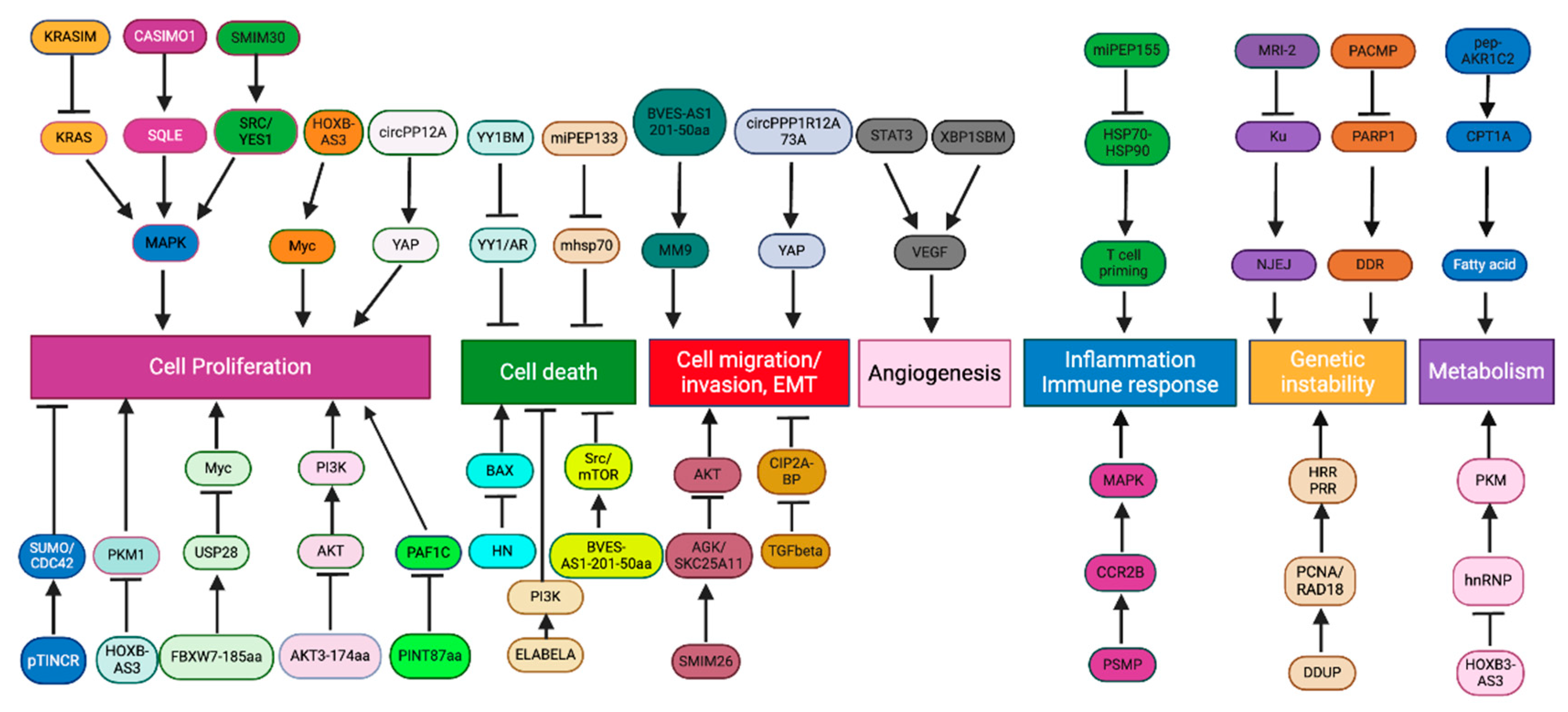
3. ncORFs as Regulators of Cancer Hallmarks
3.1. Cell Proliferation and Death
3.1.1. Cell Proliferation
3.1.2. Cell Death
3.2. Metastasis
3.2.1. Cell Migration and Invasion
3.2.2. Angiogenesis
3.3. Inflammation and Immune Responses
3.4. DNA Damage Response and Genetic Instability
3.5. Metabolism
4. ncORF in Cancer Therapy
5. Conclusion and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y., S. Zeng, and M. Wu, Novel insights into noncanonical open reading frames in cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 2022. 1877. [CrossRef]
- Consortium, I.H.G.S., Finishing the euchromatic sequence of the human genome. Nature, 2004. 431: p. 931-945.
- Wright, B.W., et al., The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol, 2022. 32: p. 243-258. [CrossRef]
- Hofman, D.A., et al., Translation of non-canonical open reading frames as a cancer cell survival mechanism in childhood medulloblastoma. Mol Cell., 2024. 84: p. 261-276.
- Ferreira, J.P., et al., Engineering ribosomal leaky scanning and upstream open reading frames for precise control of protein translation. Bioengineered., 2014. 5: p. 186-192.
- Samandi, S., et al., Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. Elife 2017. 6: p. e27860. [CrossRef]
- Jayaram, D.R., et al., Unraveling the hidden role of a uORF-encoded peptide as a kinase inhibitor of PKCs. Proc Natl Acad Sci U S A., 2021. 118: p. e2018899118.
- Wu, Q., et al., Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J., 2020. 39: p. e104763.
- Brunet, M.A., et al., The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity. EMBO Rep., 2021. 22: p. e50640. [CrossRef]
- Valdivia-Francia, F. and A. Sendoel, No country for old methods: New tools for studying microproteins. iScience, 2024. 27: p. 108972.
- Hann, S.R., et al., A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell, 1988. 52: p. 185-195. [CrossRef]
- Mattick, J.S., et al., Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol., 2023. 24: p. 430-447.
- Statello, L., et al., Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol., 2021. 22: p. 96-118.
- Patraquim, P., et al., Translation and natural selection of micropeptides from long non-canonical RNAs. Nat Commun, 2022. 13: p. 6515. [CrossRef]
- Ge, Q., et al., Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J Clin Invest., 2021. 131: p. e152911.
- Wang, Y., et al., LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med., 2020. 217: p. jem.20190950.
- Sun, L., et al., The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol Cell., 2021. 81: p. 4493-4508.
- Guo, B, et al., Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J., 2020. 39: p. e102190.
- Ho, L., et al., ELABELA Is an Endogenous Growth Factor that Sustains hESC Self-Renewal via the PI3K/AKT Pathway. Cell Stem Cell., 2015. 17: p. 435-447.
- Li, M., et al., Micropeptide MIAC Inhibits HNSCC Progression by Interacting with Aquaporin 2. J. Am. Chem. Soc., 2020. 142: p. 6708-6716.
- Lanz, R.B., et al., A Steroid Receptor Coactivator, SRA, Functions as an RNA and Is Present in an SRC-1 Complex. Cell, 1999. 97: p. 17-27.
- Yan, Y., et al., The steroid receptor RNA activator protein (SRAP) controls cancer cell migration/motility. FEBS Lett., 2015. 589: p. 4010-4018.
- Cheetham, S.W., G.J. Faulkner, and M.E. Dinger, Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat Rev Genet., 2020. 21: p. 191-201.
- Ji, Z., et al., Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife, 2015. 4: p. e08890.
- Ransohoff, J.D., Y. Wei, and P.A. Khavari, The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol., 2018. 19: p. 143-157.
- Apcher, S., et al., Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci U S A., 2013. 110: p. 17951-17956.
- Ratti, M., et al., MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target Oncol., 2020. 15: p. 261-278.
- Motameny, S., et al., Next Generation Sequencing of miRNAs - Strategies, Resources and Methods. Genes (Basel)., 2010. 1: p. 70-84.
- Chen, R., et al., Engineering circular RNA for enhanced protein production. Nat Biotechnol., 2023. 41: p. 262-272.
- Misir, S., N. Wu, and B.B. Yang, Specific expression and functions of circular RNAs. Cell Death Differ., 2022. 29: p. 481-491.
- Yang, Y., et al., Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res., 2017. 27: p. 626-641.
- Jiang, T., et al., A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer., 2021. 20: p. 66.
- Merry, T.L., et al., Mitochondrial-derived peptides in energy metabolism. Am J Physiol Endocrinol Metab., 2020. 319: p. E659-E666.
- Yang, H., et al., Widespread stable noncanonical peptides identified by integrated analyses of ribosome profiling and ORF features. Nat Commun, 2024. 15: p. 1932.
- Martinez, T.F., et al., Accurate annotation of human protein-coding small open reading frames. Nat Chem Biol., 2020. 16: p. 458-468.
- Leong, A.Z.-X., et al., Short open reading frames (sORFs) and microproteins: an update on their identification and validation measures. J Biomed Sci., 2022. 29: p. 19.
- Hanada, K., et al., A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res., 2007. 17: p. 632-640.
- Brito-Estrada, O., K. R. Hassel, and C.A. Makarewich, An Integrated Approach for Microprotein Identification and Sequence Analysis. J Vis Exp., 2022.
- Zhu, M. and M. Gribskov, MiPepid: MicroPeptide identification tool using machine learning. BMC Bioinformatics, 2019. 20: p. 559.
- Skarshewski, A. , et al., uPEPperoni: An online tool for upstream open reading frame location and analysis of transcript conservation. BMC Bioinformatics, 2014. 15: p. 36.
- Lin, M.F. I. Jungreis, and M. Kellis, PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics, 2011. 27: p. i275-282.
- Ingolia, N.T., et al., Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science, 2009. 324: p. 218-223.
- Giacomini, G., et al., A Neural Network Approach for the Analysis of Reproducible Ribo–Seq Profile. Algorithms, 2022. 15: p. 274.
- Kearse, M.G. and J.E. Wilusz, Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev., 2017. 31: p. 1717-1731.
- Slavoff, S.A., et al., Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat Chem Biol., 2013. 9: p. 59-64.
- Guttman, M., et al., Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell, 2013. 154: p. 240-251.
- Brar, G.A. and J.S. Weissman, Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol., 2015. 16: p. 651-664.
- Aspden, J.L., et al., Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife, 2014. 3: p. e03528.
- Mohsen, J.J., A.A. Martel, and S.A. Slavoff, Microproteins-Discovery, structure, and function. Proteomics, 2023. 23(23-24): p. e2100211.
- López, E., et al., Functional phosphoproteomic mass spectrometry-based approaches. Clin Transl Med., 2012. 1: p. 20.
- Chen, J., et al., Pervasive functional translation of noncanonical human open reading frames. Science, 2020. 367: p. 1140-1146.
- Prensner, J.R., et al., Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat Biotechnol, 2021. 39(6): p. 697-704.
- Schlesinger, D. and S.J. Elsässer, Revisiting sORFs: overcoming challenges to identify and characterize functional microproteins. FEBS J., 2022. 289: p. 53-74.
- Hofman, D.A., et al., Translation of non-canonical open reading frames as a cancer cell survival mechanism in childhood medulloblastoma. Mol Cell, 2024. 84(2): p. 261-276.e18.
- Zhu, S., et al., Peptides/Proteins Encoded by Non-coding RNA: A Novel Resource Bank for Drug Targets and Biomarkers. Front Pharmacol., 2018. 9: p. 1295.
- Erady, C., et al., Pan-cancer analysis of transcripts encoding novel open-reading frames (nORFs) and their potential biological functions. NPJ Genom Med., 2021. 6: p. 4.
- Zhou, H., et al., Micropeptides: potential treatment strategies for cancer. Cancer Cell International. Cancer Cell Int., 2024. 24: p. 134.
- Setrerrahmane, S., et al., Cancer-related micropeptides encoded by ncRNAs: Promising drug targets and prognostic biomarkers. Cancer Lett., 2022. 547: p. 215723.
- Carlomagno, N., et al., Diagnostic, Predictive, Prognostic, and Therapeutic Molecular Biomarkers in Third Millennium: A Breakthrough in Gastric Cancer. Biomed Res Int., 2017. 2017: p. 7869802.
- Huang, J.-Z., et al., A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell., 2017. 68: p. 171-184.
- Meng, N., et al., Small Protein Hidden in lncRNA LOC90024 Promotes “Cancerous” RNA Splicing and Tumorigenesis. Adv Sci (Weinh). 2020. 7: p. 1903233.
- Zhu, S., et al., An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nat Commun, 2020. 11: p. 1685.
- Zhang, M., et al., A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene, 2018. 37: p. 1805-1814.
- Zhang, M., et al., A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun, 2018. 9: p. 4475.
- Yang, Y., et al., Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst., 2018. 110: p. 304-315.
- Pang, Y., et al., Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J Hepatol., 2020. 73: p. 1155-1169.
- Xiao, M.-H., et al., Downregulation of a mitochondrial micropeptide, MPM, promotes hepatoma metastasis by enhancing mitochondrial complex I activity. Mol Ther., 2022. 30: p. 714-725.
- Yu, R., et al., LncRNA CTBP1-DT-encoded microprotein DDUP sustains DNA damage response signalling to trigger dual DNA repair mechanisms. Nucleic Acids Research, 2022. 50: p. 8060-8079.
- Ren, L., et al., The DDUP protein encoded by the DNA damage-induced CTBP1-DT lncRNA confers cisplatin resistance in ovarian cancer. Cell Death Dis., 2023. 14: p. 568.
- Xiao, J., et al., Low circulating levels of the mitochondrial-peptide hormone SHLP2: novel biomarker for prostate cancer risk. Oncotarget., 2017. 8: p. 94900-94909.
- Kang, M., et al., Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol Cancer., 2020. 19: p. 143.
- Li, M., et al., Micropeptide MIAC inhibits the tumor progression by interacting with AQP2 and inhibiting EREG/EGFR signaling in renal cell carcinoma. Mol Cancer., 2022. 21: p. 181.
- Xu, S., et al., LncRNA HOXB-AS3 promotes growth, invasion and migration of epithelial ovarian cancer by altering glycolysis. Life Sci., 2021. 264: p. 118636.
- Jiang, W., et al., lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J Cell Physiol., 2020. 235: p. 7194-7203.
- Papaioannou, D., et al., The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun, 2019. 10: p. 5351.
- Xing, Y., et al., Long non-coding RNA (lncRNA) HOXB-AS3 promotes cell proliferation and inhibits apoptosis by regulating ADAM9 expression through targeting miR-498-5p in endometrial carcinoma. J Int Med Res., 2021. 49: p. 03000605211013548.
- Zheng, X., et al., A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer., 2019. 18: p. 47.
- Cao, X., et al., Comparative Proteomic Profiling of Unannotated Microproteins and Alternative Proteins in Human Cell Lines. J Proteome Res, 2020. 19(8): p. 3418-3426.
- Hanahan, D. and R.A. Weinberg, The hallmarks of cancer. Cell, 2000. 100: p. 57-70.
- Hanahan, D. and R.A. Weinberg, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646-74.
- Merino-Valverde, I., E. Greco, and M. Abad, The microproteome of cancer: From invisibility to relevance. Exp Cell Res, 2020. 392(1): p. 111997.
- Polycarpou-Schwarz, M., et al., The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene, 2018. 37: p. 4750-4768.
- Leng, F., et al., A micro-peptide encoded by HOXB-AS3 promotes the proliferation and viability of oral squamous cell carcinoma cell lines by directly binding with IGF2BP2 to stabilize c-Myc. Oncol Lett., 2021. 22: p. 697.
- Lara, S.B.D., et al., C20orf204, a hepatocellular carcinoma-specific protein interacts with nucleolin and promotes cell proliferation. Oncogenesis, 2021. 10: p. 31.
- Zhang, S., et al., LncRNA-Encoded Micropeptide ACLY-BP Drives Lipid Deposition and Cell Proliferation in Clear Cell Renal Cell Carcinoma via Maintenance of ACLY Acetylation. Mol Cancer Res., 2023. 21: p. 1064-1078.
- Xia, X., et al., A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer., 2019. 18: p. 131.
- Morgado-Palacin, L., et al., The TINCR uniquitin-like microprotein is a tumor suppressor in squamous cell carcinoma. Nat Commun, 2023. 14: p. 1328.
- Xu, W., et al., Ribosome profiling analysis identified a KRAS-interacting microprotein that represses oncogenic signaling in hepatocellular carcinoma cells. Sci China Life Sci., 2020. 63: p. 529-542.
- Vitale, I., et al., Apoptotic cell death in disease - current understanding of the NCCD 2023. Cell Death Differ., 2023. 30: p. 1097-1154.
- Guo, B., et al., Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 2003. 423: p. 456-461.
- Zhang, W., et al., Peptide encoded by lncRNA BVES-AS1 promotes cell viability, migration, and invasion in colorectal cancer cells via the SRC/mTOR signaling pathway. PLoS One, 2023. 18: p. e0287133.
- Ho, L., et al., ELABELA Is an Endogenous Growth Factor that Sustains hESC Self-Renewal via the PI3K/AKT Pathway. Cell Stem Cell, 2015. 17(4): p. 435-47.
- Wu, S., et al., A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res., 2020. 80: p. 2790-2803.
- Fares, J., et al., Molecular principles of metastasis: a hallmark of cancer revisited. Sig Transduct Target Ther., 2020. 5: p. 28.
- Friedl, P. and S. Alexander, Cancer invasion and the microenvironment: plasticity and reciprocity. Cell, 2011. 147: p. 992-1009.
- Seyfried, T.N. and L.C. Huysentruyt, On the origin of cancer metastasis. Crit Rev Oncog., 2013. 18: p. 43-73.
- Guo, Z.-W., et al., Translated Long Non-Coding Ribonucleic Acid ZFAS1 Promotes Cancer Cell Migration by Elevating Reactive Oxygen Species Production in Hepatocellular Carcinoma. Front Genet., 2019. 10: p. 1111.
- Pauli, A., et al., Toddler: An Embryonic Signal That Promotes Cell Movement via Apelin Receptors. Science, 2014. 343: p. 1248636.
- Yi, Y., et al., APELA promotes tumor growth and cell migration in ovarian cancer in a p53-dependent manner. Gynecol Oncol., 2017. 147: p. 663-671.
- Meng, K., et al., LINC00493-encoded microprotein SMIM26 exerts anti-metastatic activity in renal cell carcinoma. EMBO Rep., 2023. 24: p. e56282.
- Geiger, T.R. and D.S. Peeper, Metastasis mechanisms. Biochim Biophys Acta., 2009. 1796: p. 293-308.
- Wu, S., et al., A micropeptide XBP1SBM encoded by lncRNA promotes angiogenesis and metastasis of TNBC via XBP1s pathway. Oncogene, 2022. 41: p. 2163-2172.
- Adam, J.K., B. Odhav, and K.D. Bhoola, Immune responses in cancer. Pharmacol Ther., 2003. 99: p. 113-132.
- Pei, X., et al., PC3-Secreted Microprotein Is a Novel Chemoattractant Protein and Functions as a High-Affinity Ligand for CC Chemokine Receptor 2. J Immunol., 2014. 192: p. 1878-1886.
- Jackson, R., et al., The translation of non-canonical open reading frames controls mucosal immunity. Nature 2018. 564: p. 434-438.
- Niu, L., et al., A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci Adv., 2020. 6: p. eaaz2059.
- Chong, C., et al., Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun, 2020. 11: p. 1293.
- Irajizad, E., et al., A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response. Int. J. Mol. Sci., 2022. 23: p. 8933.
- Slavoff, S., et al., A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J Biol Chem., 2014. 289: p. 10950-10957.
- Zhang, C., et al., Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol Cell., 2022. 82: p. 1297-1312.
- Deberardinis, R. and N. Chandel, Fundamentals of cancer metabolism. Sci Adv., 2016. 2: p. e1600200.
- Huang, N., et al., An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab., 2021. 33: p. 128-144.
- Zhu, K.-G., et al., The microprotein encoded by exosomal lncAKR1C2 promotes gastric cancer lymph node metastasis by regulating fatty acid metabolism. Cell Death Dis., 2023. 14: p. 708.
- Carracedo, A. L. Cantley, and P. Pandolfi, Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer., 2013. 13: p. 227-232.
- Posner, Z., I. Yannuzzi, and J.R. Prensner, Shining a light on the dark proteome: Non-canonical open reading frames and their encoded miniproteins as a new frontier in cancer biology. Protein Sci, 2023. 32(8): p. e4708.
- Hassel, K.R., O. Brito-Estrada, and C.A. Makarewich,, Microproteins: Overlooked regulators of physiology and disease. iScience, 2023. 26(6): p. 106781.
- Lee, Y.T., Y.J. Tan, and C.E. Oon, Molecular targeted therapy: treating cancer with specificity. Eur. J. Pharmacol., 2018. 834: p. 188-196.
- Wang, X., H. Zhang, and X. Chen, Drug resistance and combating drug resistance in cancer. Cancer Drug Resist., 2019. 2: p. 141-160.
- Huang, D., et al., Cisplatin resistance in gastric cancer cells is associated with HER2 upregulation-induced epithelial-mesenchymal transition. Sci. Rep., 2016. 6: p. 20502.
- Kurrey, N.K., et al., Snail and Slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells., 2009. 27: p. 2059-2068.
- Quintás-Cardama, A., H.M. Kantarjian, and J.E. Cortes, Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control., 2009. 16: p. 122-131.
- Zhai, S., et al., A microprotein N1DARP encoded by LINC00261 promotes Notch1 intracellular domain (N1ICD) degradation via disrupting USP10-N1ICD interaction to inhibit chemoresistance in Notch1-hyperactivated pancreatic cancer. Cell Discov., 2023. 9: p. 95.
| ncORFs | Cancer types | Biomarker | References |
|---|---|---|---|
| HOXB-AS3, SRSP, RGRP, ASAP | CRC | Prognosis | [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62] |
| ASRPS, CIP2A-BP | BC | Prognosis | [16,18] |
| pTINCR | BC, CSCC, LAC | Prognosis | |
| SHPRH-146aa, PINT87aa, FBXW7-185aa | GBM | Prognosis | [63,64,65] |
| SMIM30, MPM | HCC | Prognosis | [66,67] |
| DDUP | OC | Prognosis | [68,69] |
| SHLP2 | PC | Diagnosis | [70] |
| miPEP133 | NPC | Prognosis | [71] |
| APPLE/ASH1L-AS1 | AML | PrognosisDiagnosis | [17] |
| circMAPK1 | GC | Prognosis | [32] |
| MIAC | RCC | DiagnosisPrognosis | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





