Submitted:
24 June 2024
Posted:
25 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
- acquisition modality (CBCT, CT, MRI, PET/CT);
- number of patients or phantom;
- disease/s name (if appropriate);
- equipment vendor and model;
- presence of acquisition parameters;
- the total number of features;
- type of features subsampled in FO (first order), SM (shape metric), and TA textural features;
- type of software used in the radiomic feature extraction;
- image filtering used (Y/N, if Y the type was reported);
- voxel resampling;
- normalization process;
- discretization technique;
- retrospective study (Y/N or NA);
- statistical analysis: intraclass correlation coefficient (ICC), concordance correlation coefficient (CCC), area under the receiver operation curve (AUC), Mean, Average percentage difference, Relative difference, Spearman correlation, Kolmogorov Smirnov, double sample test, and two-way ANOVA;
- type of study (reproducibility/repeatability/both or best performance);
- main findings.
3. Results
3.1. Literature Search
3.2. Data Collection and Elaboration
3.2.1. Acquisition Parameters Presence and Voxel Resampling
3.2.2. Normalization Strategies
3.2.3. Discretization Strategies
3.2.4. Study Aims
3.2.5. Anatomic District
3.2.5. Comparison Metrics
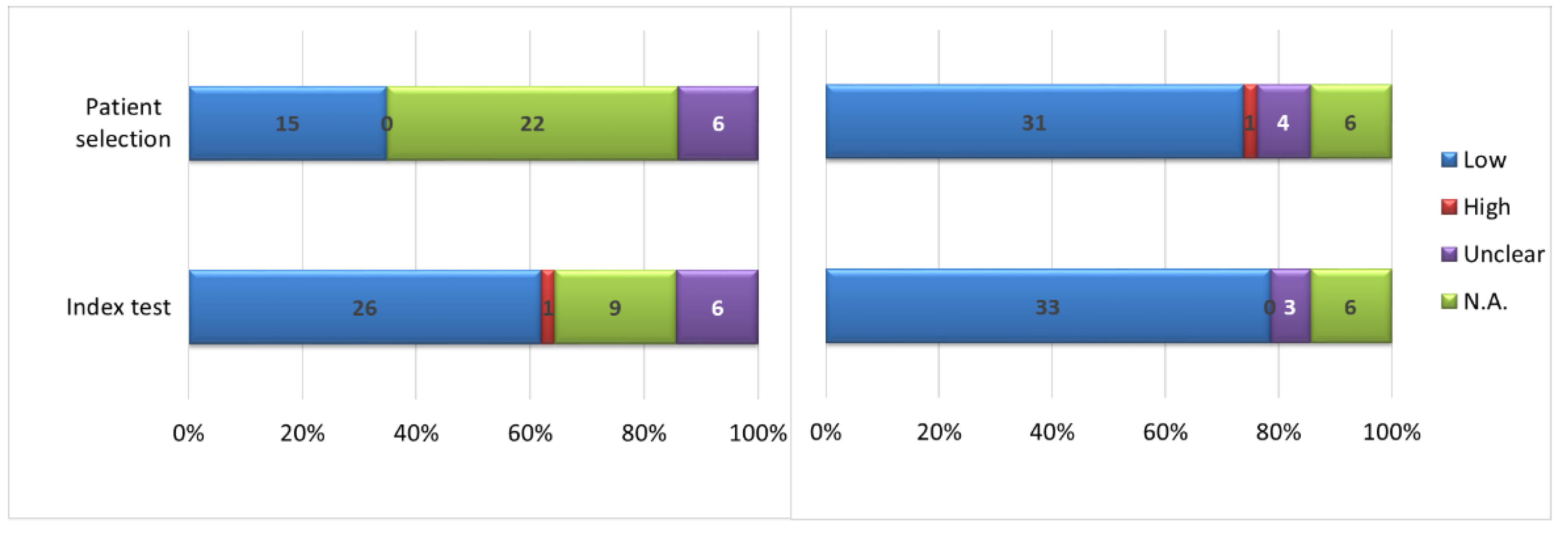
3.3. Risk of Bias Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; De Jong, E.E.C.; Van Timmeren, J.E.; Ibrahim, A.; Compter, I.; Peerlings, J.; Sanduleanu, S.; Refaee, T.; Keek, S.; Larue, R.T.H.M.; et al. Decision Support Systems in Oncology. JCO Clinical Cancer Informatics 2019, 1–9. [Google Scholar] [CrossRef]
- Shur, J.; Blackledge, M.; D’Arcy, J.; Collins, D.J.; Bali, M.; O’Leach, M.; Koh, D.-M. MRI Texture Feature Repeatability and Image Acquisition Factor Robustness, a Phantom Study and in Silico Study. Eur Radiol Exp 2021, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. European Journal of Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and Limitations of Radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Aerts, H.J.; Mak, R.H. Handcrafted versus Deep Learning Radiomics for Prediction of Cancer Therapy Response. The Lancet Digital Health 2019, 1, e106–e107. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat Commun 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology 2018, 289, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, A.; Khalvati, F.; Tyrrell, P.N.; Haider, M.A.; Tarique, U.; Jimenez-Juan, L.; Tjong, M.C.; Poon, I.; Eilaghi, A.; Ehrlich, L.; et al. Radiomics Analysis at PET/CT Contributes to Prognosis of Recurrence and Survival in Lung Cancer Treated with Stereotactic Body Radiotherapy. Sci Rep 2018, 8, 4003. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J.W.L. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front. Oncol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Blüthgen, C.; Patella, M.; Euler, A.; Baessler, B.; Martini, K.; Von Spiczak, J.; Schneiter, D.; Opitz, I.; Frauenfelder, T. Computed Tomography Radiomics for the Prediction of Thymic Epithelial Tumor Histology, TNM Stage and Myasthenia Gravis. PLoS ONE 2021, 16, e0261401. [Google Scholar] [CrossRef] [PubMed]
- E, L.; Lu, L.; Li, L.; Yang, H.; Schwartz, L.H.; Zhao, B. Radiomics for Classification of Lung Cancer Histological Subtypes Based on Nonenhanced Computed Tomography. Academic Radiology 2019, 26, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Stefan, P.-A.; Puscas, M.E.; Csuak, C.; Lebovici, A.; Petresc, B.; Lupean, R.; Mihu, C.M. The Utility of Texture-Based Classification of Different Types of Ascites on Magnetic Resonance. J BUON 2020, 25, 1237–1244. [Google Scholar] [PubMed]
- Csutak, C.; Ștefan, P.-A.; Lupean, R.-A.; Lenghel, L.M.; Mihu, C.M.; Lebovici, A. Computed Tomography in the Diagnosis of Intraperitoneal Effusions: The Role of Texture Analysis. Bosn J Basic Med Sci 2021, 21, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Tharmalingam, H.; Tsang, Y.M.; Alonzi, R.; Beasley, W.; Taylor, N.J.; McWilliam, A.; Padhani, A.; Choudhury, A.; Hoskin, P.J. Changes in Magnetic Resonance Imaging Radiomic Features in Response to Androgen Deprivation Therapy in Patients with Intermediate- and High-Risk Prostate Cancer. Clin Oncol (R Coll Radiol) 2022, 34, e246–e253. [Google Scholar] [CrossRef] [PubMed]
- Ștefan, P.-A.; Lupean, R.-A.; Mihu, C.M.; Lebovici, A.; Oancea, M.D.; Hîțu, L.; Duma, D.; Csutak, C. Ultrasonography in the Diagnosis of Adnexal Lesions: The Role of Texture Analysis. Diagnostics (Basel) 2021, 11, 812. [Google Scholar] [CrossRef]
- Ștefan, R.-A.; Ștefan, P.-A.; Mihu, C.M.; Csutak, C.; Melincovici, C.S.; Crivii, C.B.; Maluțan, A.M.; Hîțu, L.; Lebovici, A. Ultrasonography in the Differentiation of Endometriomas from Hemorrhagic Ovarian Cysts: The Role of Texture Analysis. JPM 2021, 11, 611. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, P.; Bousabarah, K.; Hoevels, M.; Treuer, H. Radiomics in Radiation Oncology-Basics, Methods, and Limitations. Strahlenther Onkol 2020, 196, 848–855. [Google Scholar] [CrossRef]
- Ibrahim, A.; Primakov, S.; Beuque, M.; Woodruff, H.C.; Halilaj, I.; Wu, G.; Refaee, T.; Granzier, R.; Widaatalla, Y.; Hustinx, R.; et al. Radiomics for Precision Medicine: Current Challenges, Future Prospects, and the Proposal of a New Framework. Methods 2021, 188, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. JPM 2021, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Midya, A.; Chakraborty, J.; Gönen, M.; Do, R.K.G.; Simpson, A.L. Influence of CT Acquisition and Reconstruction Parameters on Radiomic Feature Reproducibility. J Med Imaging (Bellingham) 2018, 5, 011020. [Google Scholar] [CrossRef] [PubMed]
- Reiazi, R.; Abbas, E.; Famiyeh, P.; Rezaie, A.; Kwan, J.Y.Y.; Patel, T.; Bratman, S.V.; Tadic, T.; Liu, F.-F.; Haibe-Kains, B. The Impact of the Variation of Imaging Parameters on the Robustness of Computed Tomography Radiomic Features: A Review. Comput Biol Med 2021, 133, 104400. [Google Scholar] [CrossRef] [PubMed]
- Espinasse, M.; Pitre-Champagnat, S.; Charmettant, B.; Bidault, F.; Volk, A.; Balleyguier, C.; Lassau, N.; Caramella, C. CT Texture Analysis Challenges: Influence of Acquisition and Reconstruction Parameters: A Comprehensive Review. Diagnostics (Basel) 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B. Understanding Sources of Variation to Improve the Reproducibility of Radiomics. Front Oncol 2021, 11, 633176. [Google Scholar] [CrossRef] [PubMed]
- Shiri, I.; Abdollahi, H.; Shaysteh, S.; Rabi Mahdavi, S. Test-Retest Reproducibility and Robustness Analysis of Recurrent Glioblastoma MRI Radiomics Texture Features. Iran J Radiol 2017. Special iss. [Google Scholar] [CrossRef]
- Peerlings, J.; Woodruff, H.C.; Winfield, J.M.; Ibrahim, A.; Van Beers, B.E.; Heerschap, A.; Jackson, A.; Wildberger, J.E.; Mottaghy, F.M.; DeSouza, N.M.; et al. Stability of Radiomics Features in Apparent Diffusion Coefficient Maps from a Multi-Centre Test-Retest Trial. Sci Rep 2019, 9, 4800. [Google Scholar] [CrossRef] [PubMed]
- Pfaehler, E.; Beukinga, R.J.; Jong, J.R.; Slart, R.H.J.A.; Slump, C.H.; Dierckx, R.A.J.O.; Boellaard, R. Repeatability of 18 F- FDG PET Radiomic Features: A Phantom Study to Explore Sensitivity to Image Reconstruction Settings, Noise, and Delineation Method. Med. Phys. 2019, 46, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Prayer, F.; Hofmanninger, J.; Weber, M.; Kifjak, D.; Willenpart, A.; Pan, J.; Röhrich, S.; Langs, G.; Prosch, H. Variability of Computed Tomography Radiomics Features of Fibrosing Interstitial Lung Disease: A Test-Retest Study. Methods 2021, 188, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tan, Y.; Tsai, W.-Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of Radiomics for Deciphering Tumor Phenotype with Imaging. Sci Rep 2016, 6, 23428. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Refaee, T.; Primakov, S.; Barufaldi, B.; Acciavatti, R.J.; Granzier, R.W.Y.; Hustinx, R.; Mottaghy, F.M.; Woodruff, H.C.; Wildberger, J.E.; et al. The Effects of In-Plane Spatial Resolution on CT-Based Radiomic Features’ Stability with and without ComBat Harmonization. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhovannik, I.; Bussink, J.; Traverso, A.; Shi, Z.; Kalendralis, P.; Wee, L.; Dekker, A.; Fijten, R.; Monshouwer, R. Learning from Scanners: Bias Reduction and Feature Correction in Radiomics. Clin Transl Radiat Oncol 2019, 19, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Refaee, T.; Primakov, S.; Barufaldi, B.; Acciavatti, R.J.; Granzier, R.W.Y.; Hustinx, R.; Mottaghy, F.M.; Woodruff, H.C.; Wildberger, J.E.; et al. Reply to Orlhac, F.; Buvat, I. Comment on “Ibrahim et al. The Effects of In-Plane Spatial Resolution on CT-Based Radiomic Features’ Stability with and without ComBat Harmonization. Cancers 2021, 13, 1848”. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Lu, L.; Yang, H.; Akin, O.; Schwartz, L.H.; Zhao, B. The Impact of Image Acquisition Parameters and ComBat Harmonization on the Predictive Performance of Radiomics: A Renal Cell Carcinoma Model. Appl Sci (Basel) 2022, 12, 9824. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J Radiol 2019, 20, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Ronald, J.; Vernuccio, F.; Nelson, R.C.; Ramirez-Giraldo, J.C.; Solomon, J.; Patel, B.N.; Samei, E.; Marin, D. Reproducibility of CT Radiomic Features within the Same Patient: Influence of Radiation Dose and CT Reconstruction Settings. Radiology 2019, 293, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Ehmke, R.C.; Schwartz, L.H.; Zhao, B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. PLoS One 2016, 11, e0166550. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Refaee, T.; Leijenaar, R.T.H.; Primakov, S.; Hustinx, R.; Mottaghy, F.M.; Woodruff, H.C.; Maidment, A.D.A.; Lambin, P. The Application of a Workflow Integrating the Variable Reproducibility and Harmonizability of Radiomic Features on a Phantom Dataset. PLoS One 2021, 16, e0251147. [Google Scholar] [CrossRef] [PubMed]
- Shafiq-ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic Dependencies of CT Radiomic Features on Voxel Size and Number of Gray Levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Wichtmann, B.D.; Harder, F.N.; Weiss, K.; Schönberg, S.O.; Attenberger, U.I.; Alkadhi, H.; Pinto Dos Santos, D.; Baeßler, B. Influence of Image Processing on Radiomic Features From Magnetic Resonance Imaging. Invest Radiol 2023, 58, 199–208. [Google Scholar] [CrossRef]
- Altazi, B.A.; Zhang, G.G.; Fernandez, D.C.; Montejo, M.E.; Hunt, D.; Werner, J.; Biagioli, M.C.; Moros, E.G. Reproducibility of F18-FDG PET Radiomic Features for Different Cervical Tumor Segmentation Methods, Gray-level Discretization, and Reconstruction Algorithms. J Applied Clin Med Phys 2017, 18, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Bodet-Milin, C.; Couespel, S.; Necib, H.; Kraeber-Bodéré, F.; Ansquer, C.; Carlier, T. Revisiting the Robustness of PET-Based Textural Features in the Context of Multi-Centric Trials. PLoS ONE 2016, 11, e0159984. [Google Scholar] [CrossRef] [PubMed]
- Leijenaar, R.T.H.; Nalbantov, G.; Carvalho, S.; Van Elmpt, W.J.C.; Troost, E.G.C.; Boellaard, R.; Aerts, H.J.W.L.; Gillies, R.J.; Lambin, P. The Effect of SUV Discretization in Quantitative FDG-PET Radiomics: The Need for Standardized Methodology in Tumor Texture Analysis. Sci Rep 2015, 5, 11075. [Google Scholar] [CrossRef] [PubMed]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI Acquisition Protocols and Image Intensity Normalization Methods on Texture Classification. Magnetic Resonance Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities. Phys. Med. Biol. 2015, 60, 5471–5496. [Google Scholar] [CrossRef] [PubMed]
- Giannini, V.; Panic, J.; Regge, D.; Balestra, G.; Rosati, S. Could Normalization Improve Robustness of Abdominal MRI Radiomic Features? Biomed. Phys. Eng. Express 2023, 9, 055002. [Google Scholar] [CrossRef] [PubMed]
- Kremer, L.E.; Perri, N.; Sorber, E.; Chapman, A.; Armato, S.G. Normalization of MRI Signal Intensity in Polycystic Kidney Disease and the Effect on Radiomic Features. In Proceedings of the Medical Imaging 2022: Computer-Aided Diagnosis; Iftekharuddin, K.M., Drukker, K., Mazurowski, M.A., Lu, H., Muramatsu, C., Samala, R.K., Eds.; SPIE: San Diego, United States, April 4, 2022; p. 132. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, n71. [Google Scholar] [CrossRef]
- Whiting, P.F. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med 2011, 155, 529. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.T.H.M.; Van Timmeren, J.E.; De Jong, E.E.C.; Feliciani, G.; Leijenaar, R.T.H.; Schreurs, W.M.J.; Sosef, M.N.; Raat, F.H.P.J.; Van Der Zande, F.H.R.; Das, M.; et al. Influence of Gray Level Discretization on Radiomic Feature Stability for Different CT Scanners, Tube Currents and Slice Thicknesses: A Comprehensive Phantom Study. Acta Oncologica 2017, 56, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Mackin, D.; Fave, X.; Zhang, L.; Yang, J.; Jones, A.K.; Ng, C.S.; Court, L. Harmonizing the Pixel Size in Retrospective Computed Tomography Radiomics Studies. PLoS One 2017, 12, e0178524. [Google Scholar] [CrossRef] [PubMed]
- Stüber, A.T.; Coors, S.; Schachtner, B.; Weber, T.; Rügamer, D.; Bender, A.; Mittermeier, A.; Öcal, O.; Seidensticker, M.; Ricke, J.; et al. A Comprehensive Machine Learning Benchmark Study for Radiomics-Based Survival Analysis of CT Imaging Data in Patients With Hepatic Metastases of CRC. Invest Radiol 2023, 58, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Demircioğlu, A. The Effect of Preprocessing Filters on Predictive Performance in Radiomics. Eur Radiol Exp 2022, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Linsalata, S.; Borgheresi, R.; Marfisi, D.; Barca, P.; Sainato, A.; Paiar, F.; Neri, E.; Traino, A.C.; Giannelli, M. Radiomics of Patients with Locally Advanced Rectal Cancer: Effect of Preprocessing on Features Estimation from Computed Tomography Imaging. BioMed Research International 2022, 2022, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Au, R.C.; Tan, W.C.; Bourbeau, J.; Hogg, J.C.; Kirby, M. Impact of Image Pre-Processing Methods on Computed Tomography Radiomics Features in Chronic Obstructive Pulmonary Disease. Phys. Med. Biol. 2021, 66, 245015. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Shiri, I.; Zaidi, H. Radiomics-Based Machine Learning Model to Predict Risk of Death within 5-Years in Clear Cell Renal Cell Carcinoma Patients. Computers in Biology and Medicine 2021, 129, 104135. [Google Scholar] [CrossRef] [PubMed]
- Kolossváry, M.; Szilveszter, B.; Karády, J.; Drobni, Z.D.; Merkely, B.; Maurovich-Horvat, P. Effect of Image Reconstruction Algorithms on Volumetric and Radiomic Parameters of Coronary Plaques. Journal of Cardiovascular Computed Tomography 2019, 13, 325–330. [Google Scholar] [CrossRef]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Court, L.E. Impact of Image Preprocessing on the Volume Dependence and Prognostic Potential of Radiomics Features in Non-Small Cell Lung Cancer. Transl. Cancer Res. 2016, 5, 349–363. [Google Scholar] [CrossRef]
- Hunter, L.A.; Krafft, S.; Stingo, F.; Choi, H.; Martel, M.K.; Kry, S.F.; Court, L.E. High Quality Machine-Robust Image Features: Identification in Nonsmall Cell Lung Cancer Computed Tomography Images: Robust Quantitative Image Features. Med. Phys. 2013, 40, 121916. [Google Scholar] [CrossRef] [PubMed]
- Palani, D.; Ganesh, K.M.; Karunagaran, L.; Govindaraj, K.; Shanmugam, S. Statistical Analysis on Impact of Image Preprocessing of CT Texture Patterns and Its CT Radiomic Feature Stability: A Phantom Study. Asian Pac J Cancer Prev 2023, 24, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Delgadillo, R.; Spieler, B.O.; Ford, J.C.; Kwon, D.; Yang, F.; Studenski, M.; Padgett, K.R.; Abramowitz, M.C.; Dal Pra, A.; Stoyanova, R.; et al. Repeatability of CBCT Radiomic Features and Their Correlation with CT Radiomic Features for Prostate Cancer. Med. Phys. 2021, 48, 2386–2399. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ammari, S.; Balleyguier, C.; Lassau, N.; Chouzenoux, E. Impact of Preprocessing and Harmonization Methods on the Removal of Scanner Effects in Brain MRI Radiomic Features. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- McHugh, D.J.; Porta, N.; Little, R.A.; Cheung, S.; Watson, Y.; Parker, G.J.M.; Jayson, G.C.; O’Connor, J.P.B. Image Contrast, Image Pre-Processing, and T1 Mapping Affect MRI Radiomic Feature Repeatability in Patients with Colorectal Cancer Liver Metastases. Cancers 2021, 13, 240. [Google Scholar] [CrossRef] [PubMed]
- Carré, A.; Klausner, G.; Edjlali, M.; Lerousseau, M.; Briend-Diop, J.; Sun, R.; Ammari, S.; Reuzé, S.; Alvarez Andres, E.; Estienne, T.; et al. Standardization of Brain MR Images across Machines and Protocols: Bridging the Gap for MRI-Based Radiomics. Sci Rep 2020, 10, 12340. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Kazmierski, M.; Welch, M.L.; Weiss, J.; Fiset, S.; Foltz, W.D.; Gladwish, A.; Dekker, A.; Jaffray, D.; Wee, L.; et al. Sensitivity of Radiomic Features to Inter-Observer Variability and Image Pre-Processing in Apparent Diffusion Coefficient (ADC) Maps of Cervix Cancer Patients. Radiotherapy and Oncology 2020, 143, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Um, H.; Tixier, F.; Bermudez, D.; Deasy, J.O.; Young, R.J.; Veeraraghavan, H. Impact of Image Preprocessing on the Scanner Dependence of Multi-Parametric MRI Radiomic Features and Covariate Shift in Multi-Institutional Glioblastoma Datasets. Phys. Med. Biol. 2019, 64, 165011. [Google Scholar] [CrossRef] [PubMed]
- Bleker, J.; Roest, C.; Yakar, D.; Huisman, H.; Kwee, T.C. The Effect of Image Resampling on the Performance of Radiomics-Based Artificial Intelligence in Multicenter Prostate MRI. Magnetic Resonance Imaging 2024, 59, 1800–1806. [Google Scholar] [CrossRef] [PubMed]
- Bologna, M.; Tenconi, C.; Corino, V.D.A.; Annunziata, G.; Orlandi, E.; Calareso, G.; Pignoli, E.; Valdagni, R.; Mainardi, L.T.; Rancati, T. Repeatability and Reproducibility of MRI-Radiomic Features: A Phantom Experiment on a 1.5 T Scanner. Med Phys 2023, 50, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Marzi, C.; Marfisi, D.; Barucci, A.; Del Meglio, J.; Lilli, A.; Vignali, C.; Mascalchi, M.; Casolo, G.; Diciotti, S.; Traino, A.C.; et al. Collinearity and Dimensionality Reduction in Radiomics: Effect of Preprocessing Parameters in Hypertrophic Cardiomyopathy Magnetic Resonance T1 and T2 Mapping. Bioengineering 2023, 10, 80. [Google Scholar] [CrossRef]
- Marfisi, D.; Tessa, C.; Marzi, C.; Del Meglio, J.; Linsalata, S.; Borgheresi, R.; Lilli, A.; Lazzarini, R.; Salvatori, L.; Vignali, C.; et al. Image Resampling and Discretization Effect on the Estimate of Myocardial Radiomic Features from T1 and T2 Mapping in Hypertrophic Cardiomyopathy. Sci Rep 2022, 12, 10186. [Google Scholar] [CrossRef] [PubMed]
- Koçak, B.; Yüzkan, S.; Mutlu, S.; Karagülle, M.; Kala, A.; Kadıoğlu, M.; Solak, S.; Sunman, Ş.; Temiz, Z.H.; Ganiyusufoğlu, A.K. Influence of Image Preprocessing on the Segmentation-Based Reproducibility of Radiomic Features: In Vivo Experiments on Discretization and Resampling Parameters. dir 2023, 0, 0–0. [Google Scholar] [CrossRef] [PubMed]
- Brynolfsson, P.; Nilsson, D.; Torheim, T.; Asklund, T.; Karlsson, C.T.; Trygg, J.; Nyholm, T.; Garpebring, A. Haralick Texture Features from Apparent Diffusion Coefficient (ADC) MRI Images Depend on Imaging and Pre-Processing Parameters. Sci Rep 2017, 7, 4041. [Google Scholar] [CrossRef] [PubMed]
- Abunahel, B.M.; Pontre, B.; Petrov, M.S. Effect of Gray Value Discretization and Image Filtration on Texture Features of the Pancreas Derived from Magnetic Resonance Imaging at 3T. J. Imaging 2022, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Toffoli, T.; Saut, O.; Etchegaray, C.; Jambon, E.; Le Bras, Y.; Grenier, N.; Marcelin, C. Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy. JPM 2023, 13, 1444. [Google Scholar] [CrossRef] [PubMed]
- Abunahel, B.M.; Pontre, B.; Ko, J.; Petrov, M.S. Towards Developing a Robust Radiomics Signature in Diffuse Diseases of the Pancreas: Accuracy and Stability of Features Derived from T1-Weighted Magnetic Resonance Imaging. Journal of Medical Imaging and Radiation Sciences 2022, 53, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Dewi, D.E.O.; Sunoqrot, M.R.S.; Nketiah, G.A.; Sandsmark, E.; Giskeødegård, G.F.; Langørgen, S.; Bertilsson, H.; Elschot, M.; Bathen, T.F. The Impact of Pre-Processing and Disease Characteristics on Reproducibility of T2-Weighted MRI Radiomics Features. Magn Reson Mater Phy 2023, 36, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Foltyn-Dumitru, M.; Schell, M.; Rastogi, A.; Sahm, F.; Kessler, T.; Wick, W.; Bendszus, M.; Brugnara, G.; Vollmuth, P. Impact of Signal Intensity Normalization of MRI on the Generalizability of Radiomic-Based Prediction of Molecular Glioma Subtypes. Eur Radiol 2023, 34, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Salome, P.; Sforazzini, F.; Grugnara, G.; Kudak, A.; Dostal, M.; Herold-Mende, C.; Heiland, S.; Debus, J.; Abdollahi, A.; Knoll, M. MR Intensity Normalization Methods Impact Sequence Specific Radiomics Prognostic Model Performance in Primary and Recurrent High-Grade Glioma. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Ubaldi, L.; Saponaro, S.; Giuliano, A.; Talamonti, C.; Retico, A. Deriving Quantitative Information from Multiparametric MRI via Radiomics: Evaluation of the Robustness and Predictive Value of Radiomic Features in the Discrimination of Low-Grade versus High-Grade Gliomas with Machine Learning. Physica Medica 2023, 107, 102538. [Google Scholar] [CrossRef] [PubMed]
- Granzier, R.W.Y.; Ibrahim, A.; Primakov, S.; Keek, S.A.; Halilaj, I.; Zwanenburg, A.; Engelen, S.M.E.; Lobbes, M.B.I.; Lambin, P.; Woodruff, H.C.; et al. Test–Retest Data for the Assessment of Breast MRI Radiomic Feature Repeatability. Magnetic Resonance Imaging 2022, 56, 592–604. [Google Scholar] [CrossRef]
- Hoebel, K.V.; Patel, J.B.; Beers, A.L.; Chang, K.; Singh, P.; Brown, J.M.; Pinho, M.C.; Batchelor, T.T.; Gerstner, E.R.; Rosen, B.R.; et al. Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma. Radiology: Artificial Intelligence 2021, 3, e190199. [Google Scholar] [CrossRef] [PubMed]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI Signal Normalization Affects Radiomics Features Reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Shiri, I.; Hajianfar, G.; Sohrabi, A.; Abdollahi, H.; Shayesteh, S.P.; Geramifar, P.; Zaidi, H.; Oveisi, M.; Rahmim, A. Repeatability of Radiomic Features in Magnetic Resonance Imaging of Glioblastoma: Test–Retest and Image Registration Analyses. Med. Phys. 2020, 47, 4265–4280. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.; Ford, J.C.; Llorente, R.; Portelance, L.; Yang, F.; Mellon, E.A.; Dogan, N. Impact of Quantization Algorithm and Number of Gray Level Intensities on Variability and Repeatability of Low Field Strength Magnetic Resonance Image-Based Radiomics Texture Features. Physica Medica 2020, 80, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Schwier, M.; Van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci Rep 2019, 9, 9441. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Kazmierski, M.; Shi, Z.; Kalendralis, P.; Welch, M.; Nissen, H.D.; Jaffray, D.; Dekker, A.; Wee, L. Stability of Radiomic Features of Apparent Diffusion Coefficient (ADC) Maps for Locally Advanced Rectal Cancer in Response to Image Pre-Processing. Physica Medica 2019, 61, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Lovinfosse, P.; Hermesse, J.; Decuypere, M.; Rousseau, C.; Lucia, F.; Schick, U.; Reinhold, C.; Robin, P.; Hatt, M.; et al. [18F]FDG PET Radiomics to Predict Disease-Free Survival in Cervical Cancer: A Multi-Scanner/Center Study with External Validation. Eur J Nucl Med Mol Imaging 2021, 48, 3432–3443. [Google Scholar] [CrossRef]
- Xu, H.; Lv, W.; Zhang, H.; Ma, J.; Zhao, P.; Lu, L. Evaluation and Optimization of Radiomics Features Stability to Respiratory Motion in 18 F-FDG 3D PET Imaging. Med. Phys. 2021, 48, 5165–5178. [Google Scholar] [CrossRef] [PubMed]
- Crandall, J.P.; Fraum, T.J.; Lee, M.; Jiang, L.; Grigsby, P.; Wahl, R.L. Repeatability of 18 F-FDG PET Radiomic Features in Cervical Cancer. J Nucl Med 2021, 62, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Ger, R.B.; Meier, J.G.; Pahlka, R.B.; Gay, S.; Mumme, R.; Fuller, C.D.; Li, H.; Howell, R.M.; Layman, R.R.; Stafford, R.J.; et al. Effects of Alterations in Positron Emission Tomography Imaging Parameters on Radiomics Features. PLoS ONE 2019, 14, e0221877. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ammari, S.; Balleyguier, C.; Lassau, N.; Chouzenoux, E. Impact of Preprocessing and Harmonization Methods on the Removal of Scanner Effects in Brain MRI Radiomic Features. Cancers 2021, 13, 3000. [Google Scholar] [CrossRef] [PubMed]
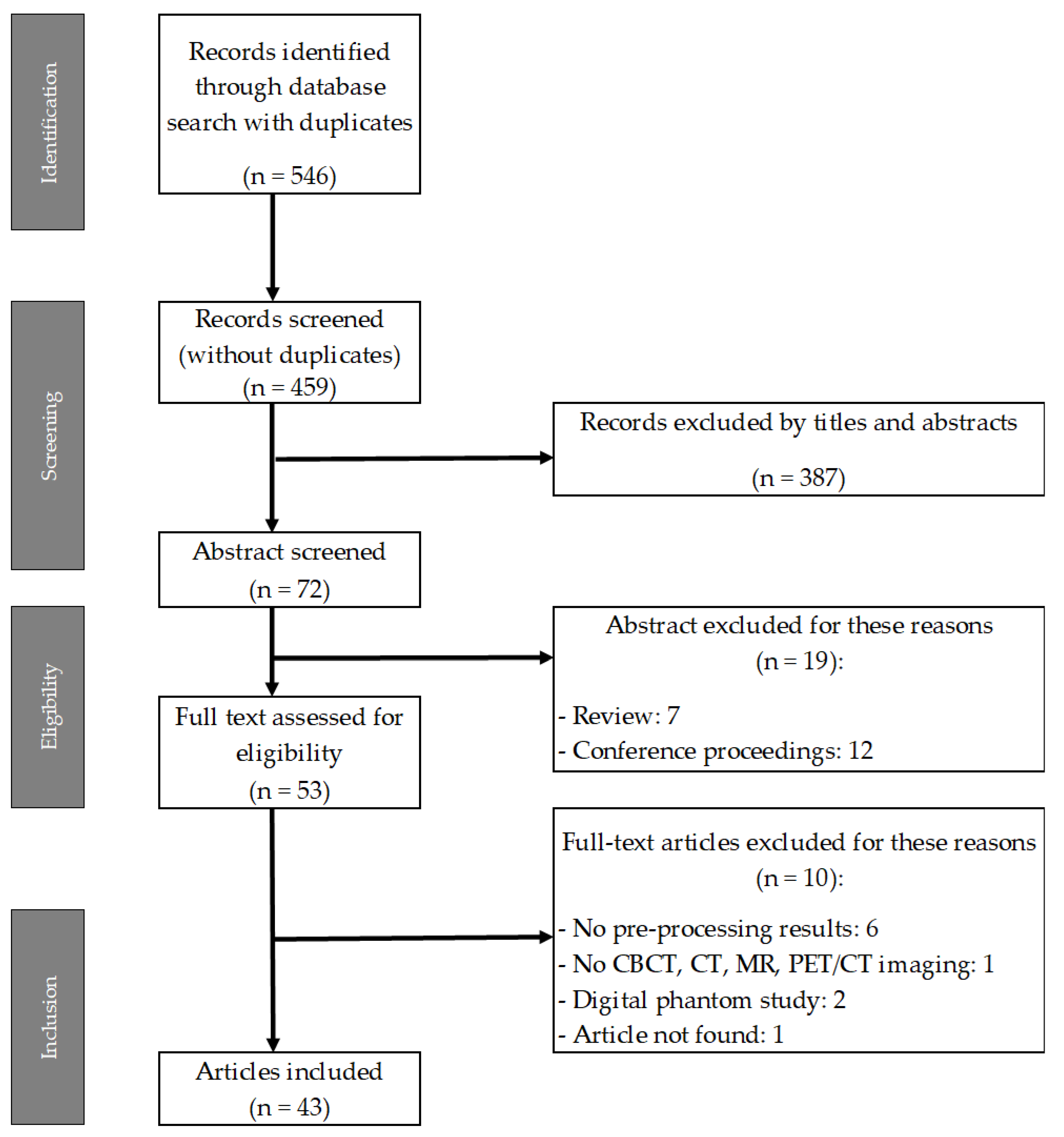
| Database (n° of record) | n° of total records (with duplicates) |
n° of total records (without duplicates) |
|||
|---|---|---|---|---|---|
| Medline | Embase | Cochrane | Scopus | ||
| 208 | 286 | 11 | 41 | 546 | 459 |
| Modality | Acquisition parameters reporting | Voxel resampling | ||||
|---|---|---|---|---|---|---|
| Isotropic | Multiple isotropic | Non-isotropic | N.A. | None | ||
| CT | 10 (83.3%) | 2 (16.7%) | 3 (25.0%) | 2 (16.7%) | 5 (41.7%) | 0 (0.0%) |
| Ref. | [54,55] | [53,56,57] | [39,52] | [58,59,60,61,62] | ||
| CBCT | 1 (100%) | 1 (100%) [63] | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Ref. | [63] | |||||
| MRI | 21 (80.8%) | 9 (34.6%) | 2 (7.7%) | 3 (11.5%) | 5 (19.2%) | 7 (26.9%) |
| Ref. | [55,64,65,66,67,68,69,70,71] | [72,73] | [74,75,76] | [77,78,79,80,81] | [82,83,84,85,86,87,88] | |
| PET/CT | 5 (100%) | 2 (40%) | 2 (40%) | 0 (0%) | 0 (0%) | 1 (20%) |
| Ref. | [89,90] | [28,91] | [92] | |||
| Modality | Absolute | Relative | Combination | None |
|---|---|---|---|---|
| CT | 0 (0%) | 1 (8.3%) | 1 (8.3%) | 10 (83.3%) |
| Ref. | [55] | [57] | [39,52,53,54,56,58,59,60,61,62] | |
| CBCT | 0 (0%) | 1 (100%) | 0 (0%) | 0 (0%) |
| Ref. | [63] | |||
| MRI | 0 (0%) | 14 (53.8%) | 4 (15.4%) | 8 (30.8%) |
| Ref. | [55,67,70,73,75,76,78,79,80,81,82,83,86,87] | [64,65,66,84] | [68,69,71,72,74,77,85,88] | |
| PET/CT | 0 (0%) | 1 (20%) | 1 (20%) | 3 (60%) |
| Ref. | [89] | [91] | [28,90,92] |
| BN | BW | BN + BW | None | |
|---|---|---|---|---|
| CT | 2 (16.7%) | 6 (50%) | 1 (8.3%) | 3 (25%) |
| Ref. | [39,58] | [52,54,55,56,57,60] | [59] | [53,61,62] |
| CBCT | 1 (100%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Ref. | [63] | |||
| MRI | 10 (38.5%) | 9 (36.6%) | 5 (19.2%) | 2 (7.7%) |
| Ref. | [64,68,70,74,80,81,83,84,85,86] | [55,65,71,72,75,76,82,87,88] | [66,67,73,77,78] | [69,79] |
| PET/CT | 1 (20%) | 0 (0%) | 4 (80%) | 0 (0%) |
| Ref. | [91] | [28,89,90,92] |
| Best performance | Repeatability | Reproducibility | Repeatability+reproducibility | |
|---|---|---|---|---|
| CT | 3 (25%) | 2 (16.7%) | 5 (41.6%) | 2 (16.7%) |
| Ref. | [54,55,58] | [53,61] | [39,56,57,59,60] | [52,62] |
| CBCT | 0.0% | 0.0% | 0.0% | 1 (100%) |
| Ref. | [63] | |||
| MRI | 8 (30.8%) | 7 (26.9%) | 6 (23.1%) | 5 (19.2%) |
| Ref. | [55,69,71,74,76,77,79,81] | [65,72,75,83,85,86,87] | [64,66,67,73,78,80] | [68,70,82,84,88] |
| PET/CT | 2 (40%) | 3 (60%) | 0 (0%) | 0 (0%) |
| Ref. | [89,90] | [28,91,92] |
| Abdomen | Brain | Thorax | Pelvis | N.A. | |
|---|---|---|---|---|---|
| CT | 1 (8.4%) | 0 | 3 (25%) | 4 (33.3%) | 4 (33.3%) |
| Ref. | [54] | [53,60,61] | [56,57,58,59] | [39,52,55,62] | |
| CBCT | 0 | 0 | 0.0% | 1 (100%) | |
| Ref. | [63] | ||||
| MRI | 2 (7.7%) | 10 (38.5%) | 2 (7.7%) | 9 (34.6%) | 4 (15.3%) |
| Ref. | [75,77] | [66,68,73,74,79,80,81,83,85,93] | [71,72] | [65,67,69,74,76,78,84,87,88] | [55,70,82,86] |
| PET/CT | 0 | 0 | 3 (60%) | 2 (40%) | 0 |
| Ref. | [28,90,92] | [89,91] |
| ICC | CCC | AUC | Other | |
|---|---|---|---|---|
| CT | 3 | 3 | 2 | 4 |
| Ref. | [56,57,59] | [52,53,61] | [55,58] | [39,54,60,62] |
| CBCT | 0 | 1 | 0 | 0 |
| Ref. | [63] | |||
| MRI | 15 | 7 | 5 | 9 |
| Ref. | [65,66,67,70,72,73,78,81,82,83,84,85,87,88,93] | [66,75,82,84,85,86,88] | [55,69,76,79,81] | [67,68,70,71,74,77,80,83,84] |
| PET/CT | 2 | 1 | 1 | 1 |
| Ref. | [28,92] | [91] | [89] | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





