1. Introduction
Mycobacterium tuberculosis, a highly pathogenic bacterium within the mycobacterium genus, is the causative agent of tuberculosis (TB), posing a serious threat to global health [
1]. The challenge posed by
M. tuberculosis remains formidable, with an estimated 10 million new TB cases and 1.5 million TB deaths annually [
2]. Misuse of antibiotics and poor patient compliance contribute significantly to the difficulty of completely eradicating this pathogenic bacterium [
3]. These challenges lead to the emergence of drug-resistant strains, further complicating this issue and rendering traditional therapeutic regimens ineffective [
4]. Consequently, the most crucial objective in the current treatment of drug-resistant TB is to search for more effective anti-
M. tuberculosis drugs and regimens.
In addition to
M. tuberculosis and
Mycobacterium leprae, there are approximately 150 species of opportunistic pathogenic non-tuberculous mycobacteria (NTM) that can cause lung diseases and skin infections in adults, as well as cervical lymphadenitis in children [
5]. NTM possess a distinctive cell wall structure characterized by a thin layer of peptidoglycan surrounded by a thick lipid-rich outer layer [
6]. This unique composition of the cell wall enables NTM to adhere to rough surfaces, resist antibiotics and disinfectants, and survive in low-oxygen and other stressful conditions [
5,
7]. Many NTM, such as
Mycobacterium abscessus, demonstrate intrinsic resistance to numerous clinical drugs, complicating and prolonging the treatment of NTM infections [
8]. Therefore, there is an urgent need for the discovery and development of effective drugs and regimens for clinical anti-NTM therapy. One practical approach is repurposing existing drugs that were originally designed for treating other diseases. These drugs have already been used clinically, which can significantly reduce the time required for them to be developed into the commercialized anti-mycobacterial drugs [
9]. Moreover, these drugs can also be further modified into new drugs against mycobacteria as lead compounds.
Allergic rhinitis is a commonly prevalent disease, affecting 10-25% of global population and significantly impacting patients’ daily lives [
10]. Histamine plays a crucial role in the development of allergic rhinitis primarily through its interaction with the histamine H1 receptor [
11]. Additionally, the platelet-activating factor is another significant inflammatory factor that promotes the release of histamine and vice versa in various tissues and cells [
11]. Rupatadine (RTD) is a second-generation H1-antihistamine agent with dual affinity for both histamine H1 and platelet-activating factor receptors, unlike most clinical drugs that only inhibit a single inflammatory factor [
12]. RTD is recommended for patients aged 12 years and older who suffer from seasonal allergic rhinitis, perennial allergic rhinitis, and chronic idiopathic urticaria due to its fast-acting and long-lasting effect [
13,
14]. Unlike first-generation H1-antihistamines, RTD does not cause side effects such as drowsiness, fatigue, headache, memory and learning difficulties, and visual disturbances [
15].
From the discovery of RTD to date, there have been no reports on the activity of RTD against mycobacteria. In this study, we discovered unexpected activity of RTD against mycobacteria, suggesting that RTD has great potential as an anti-mycobacterial drug candidate or as a lead compound for optimization.
2. Materials and Methods
2.1. Bacteria and Culture Conditions
The mycobacteria used in this study were preserved at -80℃ and cultured at 37℃ in 7H9 broth supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment medium, 0.2% glycerol, and 0.05% Tween 80. The autoluminescent mycobacterial strains, including autoluminescent
M. tuberculosis H37Ra (AlRa),
M. tuberculosis H37Rv (AlRv),
Mycobacterium marinum (AlMm),
M. abscessus (AlMab), and
Mycobacterium smegmatis (AlMs), were engineered and preserved by our laboratory [
16,
17,
18]. It has been verified that the insertion of the
lux gene cluster had no effect on drug susceptibility or growth rate of autoluminescent mycobacteria compared to their wild-type counterparts [
16,
17,
18]. Autoluminescent mycobacteria possess the distinctive ability to emit blue-green light without the supplement of additional substrate. This characteristic enables relative light units (RLUs) to serve as a surrogate for colony-forming units when evaluating drug activity against mycobacteria.
2.2 . Antimicrobials
RTD was bought from AiYan (Shanghai, China) with a specified purity of 98%. Amikacin (AMK), clofazimine (CLO), clarithromycin (CLR), isoniazid (INH), levofloxacin (LEV), linezolid (LZD), pretomanid (PTM), rifampicin (RIF), and streptomycin (STR) were bought from Meilun (Dalian, China). TB47 was synthesized by Boji (Guangzhou, China). INH, AMK, and STR were dissolved in sterile water, whereas the remaining drugs were dissolved in dimethyl sulfoxide (Xilong, Shantou, China). All drugs were diluted by their corresponding solvents and stored at -20℃ until use.
2.3. Evaluating Activity of RTD against Actively Growing Mycobacteria
The strains AlRa, AlRv, AlMm, AlMab, and AlMs were cultured until their optical density at 600 nm (OD
600) reached 0.8, with the luminescence peaking at approximately 5 × 10
6 RLUs/mL. The cultures were then diluted to 10
4 RLUs/mL using 7H9 without Tween 80. Following dilution, 196 µL of the bacterial culture and 4 µL of the drug solution were thoroughly mixed in the same sterile tube. The RLUs of the mixture were monitored at regular intervals over the following hours or days. Each experiment was conducted in triplicate. The minimal inhibitory concentration (MIC) based on the autoluminescence values is defined as the lowest drug concentration that reduces the RLUs to less than or equal to 10% of the RLUs detected in solvent-treated control group [
19].
To determine the MIC of RTD against non-luminescent
M. tuberculosis H37Rv, a microdilution method was employed in combination with a microplate Alamar Blue assay (MABA). Following a 7-day co-incubation, a mixture consisting of 20 µL of Alamar Blue and 12.5 µL of 20% Tween 80 was added into each well of 96-well plate. The plate was then incubated at 37℃ in a thermostat, and the color change was monitored after a 24- hour period. A transition from blue-purple to pink of Alamar Blue indicates the bacterial growth. Therefore, the MIC is identified as the lowest drug concentration corresponding to the blue-purple wells [
20].
2.4. Evaluating the Activity of RTD against Nonreplicating M. tuberculosis in Diverse Media
Following the protocols of the recently developed low-oxygen-recovery model, AlRa was cultured under aerobic conditions until the OD
600 reached 0.6-0.8, and RLUs increased to approximate 5 × 10
6/mL [
21]. The methylene blue was added into AlRa culture at a finial concentration of 6 μg/mL. The AlRa bacteria were then cultured in a low-oxygen incubator until the blue color of methylene blue indicator disappeared, indicating that the bacterial culture had achieved a nonreplicating state under anaerobic conditions. A mixture of 196 µL of 10-fold diluted nonreplicating AlRa (using 7H9 without Tween 80 and supplemented with glycerol or cholesterol) and 4 μL of RTD was added to the same sterile tube. After seven days of incubation, activated carbon (in a volume ratio of 1:5, 50 μL into 200 μL) was added to eliminate any potential carryover effects of residual drug. The tubes were transferred to aerobic conditions for recovery, and their RLUs were measured at 7-hour intervals over the subsequent 28 hours. The experiment was conducted in triplicate.
2.5. Evaluating the Activities of RTD in Combination with Anti-TB Drugs
To assess the efficacy of the combination of RTD with other anti-TB drugs against
M. tuberculosis, a checkerboard assay was conducted. The drugs used in combination with RTD included LEV, AMK, INH, STR, PTM, RIF, CLO, LZD, and TB47. Each drug was evaluated at six different concentrations derived from their previously determined MICs. Specifically, 2 µL of RTD, 2 µL of the chosen drug solution, and 196 µL of a diluted bacterial culture at 1.5 × 10
5 RLUs/mL were added to individual wells of a 96-well plate. Subsequently, the plates were incubated at 37℃ for seven days. After incubation, the RLUs were measured using a luminometer. The fractional inhibitory concentration index (FICI) was determined by calculating the ratio of the MIC of RTD in combination to the MIC of RTD alone, plus the MIC of selected drugs in combination to the MIC of selected drug alone [
22]. The effects were classified into five groups based on the FICI: synergistic (FICI ≤ 0.5), partially synergistic (0.5 < FICI < 1), additive (FICI = 1), irrelevant (1 < FICI ≤ 4), and antagonistic (FICI > 4) [
22].
2.6. Evaluating the in vivo Anti-M. tuberculosis Activity of RTD
Female BALB/c mice, aged 6-8 weeks, underwent a five to seven-day acclimatization period prior to the initiation of experimental procedures. Subsequently, all mice were exposed to 10 mL of AlRv culture (106 RLUs / 200 μL) using an inhalation exposure system.
The in vivo activity of RTD against M. tuberculosis was initially assessed using oral administration. The dosages of RTD were set as 6.25 and 25 mg/kg. The combination of RTD and TB47 demonstrated a partial synergistic effect against M. tuberculosis in vitro, with the FICI being 0.75 assessed previously. Consequently, the in vivo efficacy of the combination of RTD with TB47 (25 mg/kg) was also evaluated. Mice were treated with drugs or solvent on the eighth day post-infection. The treatments were administered daily for eight days until the live mice RLUs of the solvent group exceeded 400. All mice were sacrificed for the measurement of lung suspension RLUs at the initiation and one day after completion of treatment.
Meanwhile, we also employed a recently developed non-invasive murine model of inhalable administration to assess the
in vivo anti-
M. tuberculosis activity of RTD [
23]. RTD is a less soluble compound that can reach a maximum concentration of 1.56 mg/mL. Briefly, the infected mice were administrated daily with either 4 mL of RTD at a concentration of 1.56 mg/mL or distilled water via inhalation [
23]. Meanwhile, the RIF was used as the positive control, with a concentration of 2 mg/mL. The duration of inhalable administration (4 mL) each time was approximately 25 minutes. The treatment duration lasted for 15 days. The
in vivo anti-
M. tuberculosis activity of RTD was determined by comparing the live mice RLUs and lung suspension RLUs of both RTD-treated and untreated groups.
2.7. The Transcriptomic Profiling of RTD-treated and Untreated M. tuberculosis
The AlRa was transferred into fresh 7H9 broth at a volume ratio of 1:10. Based on the MIC of RTD against M. tuberculosis, RTD was added to achieve a sub-inhibitory concentration concurrently. The bacteria were cultured with and without RTD and incubated in a shaker at 37℃ until the OD600 reached approximately 0.6-0.8. All samples were sent to Jingnuo (Shanghai, China) for transcriptome sequencing.
2.8. Statistical Analysis
Log-transformed data were analyzed for drug efficacy using GraphPad Prism version 8.3.0. A two-way analysis of variance was performed, with statistical significance set at P < 0.05.
3. Results
3.1. Anti-mycobacterial Activity of RTD
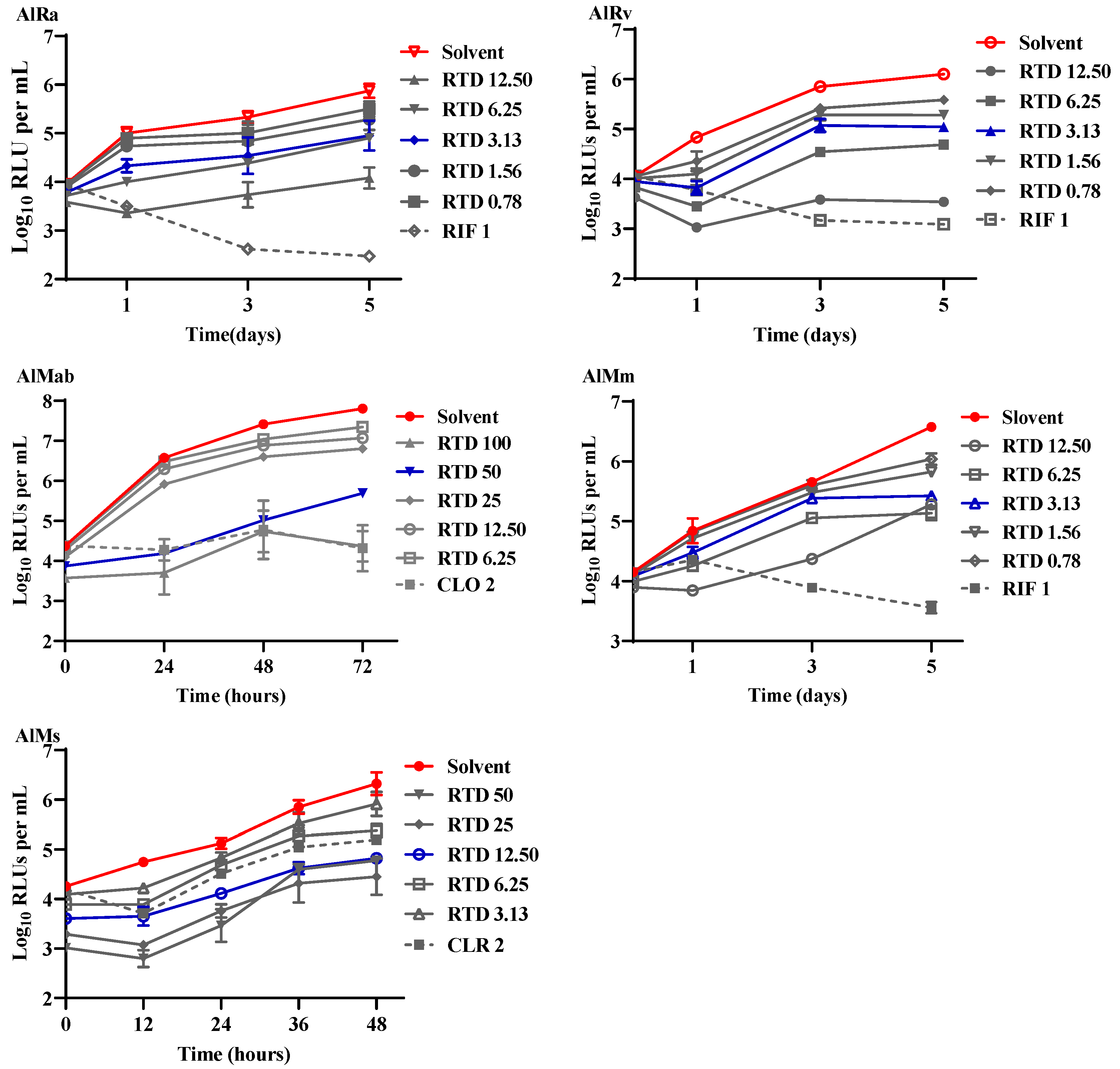
This research is the inaugural inquiry into the efficacy of RTD against mycobacteria, pioneering its exploration in this field. As depicted in
Figure 1, RTD exhibited substantial bacteriostatic activity against AlRa, AlMm, AlMab, and AlMs at concentrations of 3.13, 3.13, 50, and 12.5 µg/mL, respectively. The MICs of RTD against AlMab and AlMs were relatively higher compared to those against AlRa and AlMm. RTD potently inhibited the growth of AlRv, a virulent laboratory strain, at a remarkably low concentration of 3.13 μg/mL (
Figure 1). However, the MIC of RTD against non-luminescent
M. tuberculosis H37Rv was 25 μg/mL when assessed by MABA. Additionally, we also evaluated the activities of RTD against prevalent Gram-positive and Gram-negative bacteria, including
Klebsiella pneumoniae,
Pseudomonas aeruginosa,
Enterococcus faecalis, and
Staphylococcus aureus. Nevertheless, the MICs of RTD against all the mentioned Gram-positive and Gram-negative bacteria exceeded 100 μg/mL.
3.2. Activity of RTD against Nonreplicating AlRa Under Diverse Conditions
Using the recently established low-oxygen-recovery model, we assessed the efficacy of RTD against AlRa under four distinct conditions: aerobic and anaerobic conditions using 7H9 medium supplemented with glycerol, or aerobic and anaerobic conditions using 7H9 medium supplemented with cholesterol [
21].
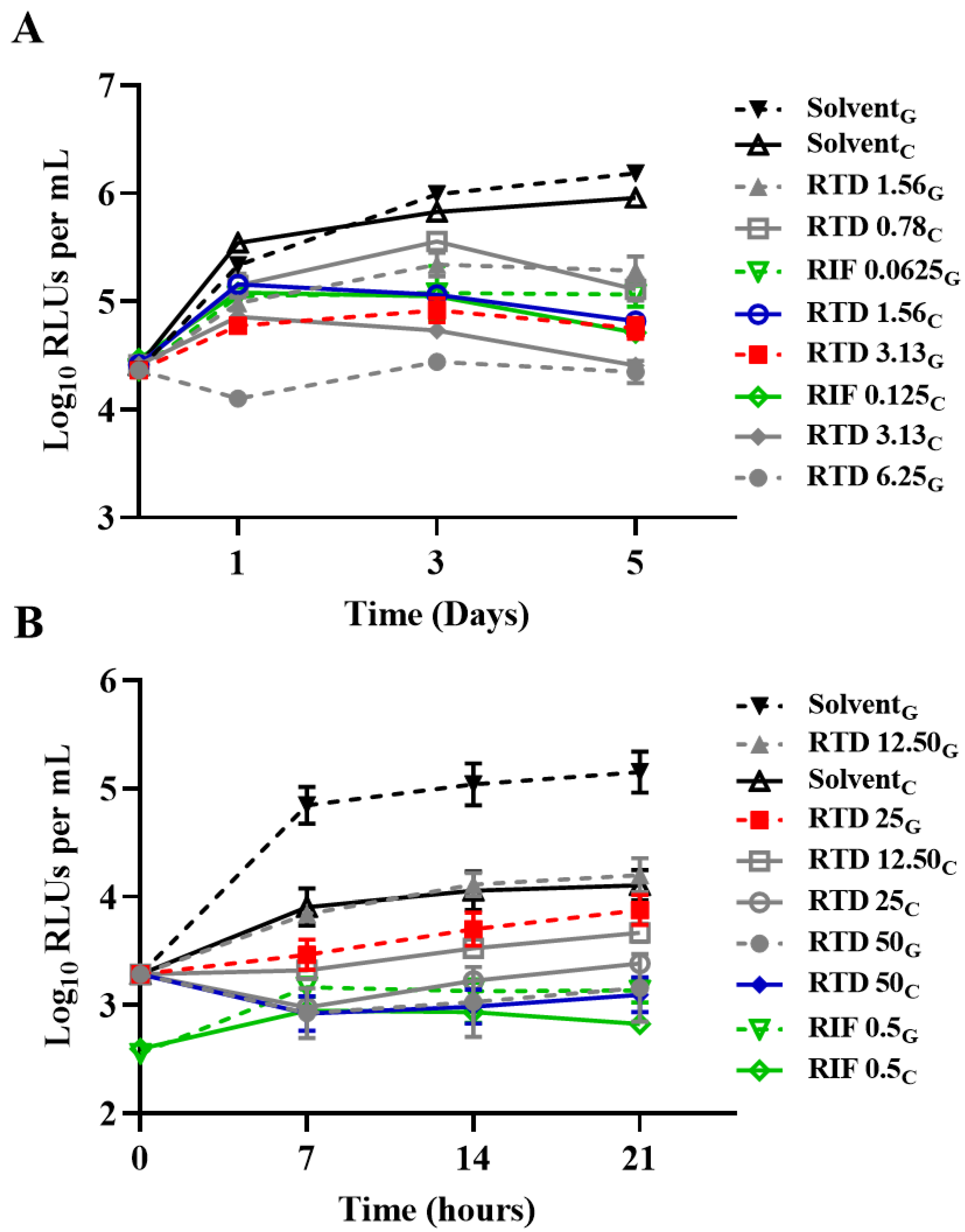
Figure 2 shows the activity of RTD against
M. tuberculosis under diverse conditions. The MICs of RTD were consistent under the aerobic conditions, whether the medium was enriched with glycerol or cholesterol, with respective MICs of 3.13 µg/mL and 1.56 µg/mL (
Figure 2A). However, under anaerobic conditions, the MICs were 25 µg/mL and 50 µg/mL when the liquid medium was supplemented with either glycerol or cholesterol, respectively (
Figure 2B). As expected, the MICs of RTD against nonreplicating AlRa were found to be higher than those against actively growing AlRa.
3.3. Partial Synergistic Effect of RTD in Combination with Several Anti-mycobacterial Drugs against M. tuberculosis
We investigated the potential synergistic activities of RTD in combination with various anti-mycobacterial drugs against
M. tuberculosis, including AMK, CLO, INH, LEV, LZD, PTM, RIF, STR, and TB47. As shown in
Table 1, RTD exhibited partial synergistic effects with PTM, CLO, and TB47, as evidenced by FICIs of 0.5625, 0.75, and 0.75, respectively (
Table 1). Notably, in the presence of sub-inhibitory concentrations of CLO, TB47, or PTM, the effective concentrations of RTD decreased to 1/4, 1/4, and 1/16 of its MIC, respectively (
Table 1). The combinations of RTD with the remaining six drugs exhibited either additive or indifferent effect against
M. tuberculosis, with all FICIs being equal to or greater than 1 (
Table 1). However, no antagonistic interactions were observed between RTD and the tested anti-mycobacterial drugs.
3.4. The in vivo Anti-M. tuberculosis Activity of RTD
In vivo evaluation of RTD was conducted using the oral administration method first as RTD is an oral drug, with the dosages of 6.25 and 25 mg/kg. However, the results indicated that RTD did not exhibit
in vivo activity against
M. tuberculosis even at the highest dosage of 25 mg/kg (
Figure 3A). The live mice RLUs of the combination of RTD with TB47 was slightly lower than those of the TB47 alone group (
Figure 3A,
P > 0.05). However, the RLUs of lung suspension showed no significant difference between the combination of RTD with TB47 and the TB47 alone groups (
Figure 3A,
P > 0.05).
To improve the lung local concentration of RTD, a non-invasive inhalation murine model was employed. The results demonstrated that the live mice RLUs and lung suspension RLUs of the RTD-treated group were not lower, but significantly higher than those of the solvent-treated group (
Figure 3B,
P < 0.05 and
P < 0.01, respectively). The RLUs of live mice and lung suspension of the RIF-treated group (2 mg/mL) were significantly lower than those of the solvent group (
Figure 3B, both
P < 0.0001). All results suggest that RTD at a concentration of 1.56 mg/mL did not exhibit significant
in vivo anti-
M. tuberculosis activity via inhalation delivery.
3.5. Transcriptome Profile of RTD-treated and Untreated M. tuberculosis
We conducted multiple trials (exceeding five biological replicates for each strain) to isolate spontaneous RTD-resistant mutants using both
M. tuberculosis and
M. marinum. Despite incorporating 5-Bromouracil into bacterial cultures to increase the spontaneous mutation rates and plating on agar containing RTD at concentrations equivalent to only four times of the MIC, our attempts were unsuccessful [
24]. Hence, we could not utilize whole genome sequencing of laboratory-generated drug-resistant mutants to determine the mechanism of action of RTD. We performed a transcriptome analysis of
M. tuberculosis in the presence and absence of RTD treatment to identify genes potentially associated with its mechanism of action. As shown in
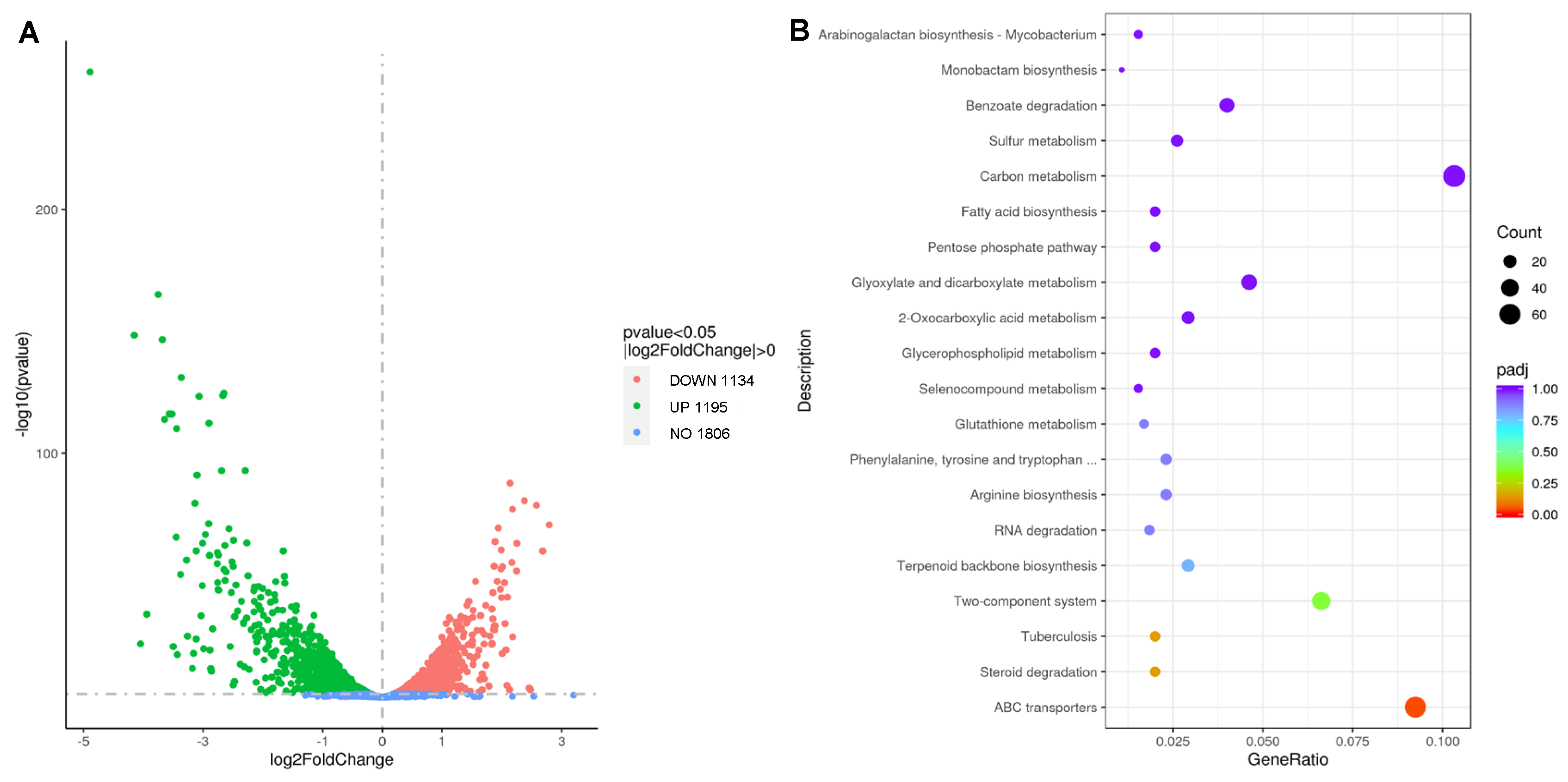
Figure 4A, out of the total genes analyzed, 1806 genes exhibited no significant changes, while the expressions of 1134 genes were upregulated and 1195 genes were downregulated.
Figure 4B highlights the top three categories with the highest number of genes, namely carbon metabolism pathways, ABC transporters, and the two-component systems.
All genes with a |log
2Foldchange| greater than 1 are listed in supplementary materials. Among these genes,
Rv0251c, a nonessential gene, exhibited the most significant change in expression. Rv0251 is believed to be involved in the initiation step of translation at high temperature [
25]. In the groups treated with RTD, the expression level of
Rv0251c was nearly 30 times higher compared to the groups without RTD treatment. Additionally, following RTD treatment, the expression of
Rv1405c and
Rv3161c was upregulated, with |log
2FoldChange| values of 4.15 and 4.05, respectively.
Rv1405c is known to participate in adaptive processes, while
Rv3161c primarily encodes a putative oxygenase [
26,
27]. Notably,
Rv0251c,
Rv1405c, and
Rv3161c are all nonessential genes. Given the low probability of these genes being the targets of RTD, we expanded our analysis to include additional genes by lowering the threshold of |log
2Foldchange| and emphasizing their essentiality as the criterion. Eight genes were filtered out based on the basic standards. They are
Rv0350,
Rv0351,
Rv0352,
Rv0384c,
Rv0440,
Rv2720,
Rv2827c, and
Rv3260c (
Table 2). Among them, only the expression of
Rv3260c gene was downregulated and the expression of remaining seven genes were upregulated (
Table 2). The gene cluster, encompassing
Rv0350, Rv0351 and
Rv0352, showed over 8-fold increase in expression level following treatment for
M. tuberculosis with RTD.
4. Discussion
Mycobacteria, recognized as some of the most virulent pathogens, are responsible for a wide range of human and animal diseases, including TB, leprosy, Buruli ulcer, and NTM infections [
28]. The emergence of drug-resistant mycobacteria urgently needs the development of novel and potent drugs or treatment regimens, which is imperative for both individuals and society [
29]. Repurposing existing drugs, originally intended for other diseases, presents a time-efficient, cost-effective, and labor-saving approach to overcome challenges associated with the screening and development of novel anti-mycobacterial agents.
RTD, a compound traditionally employed in the treatment of allergic rhinitis, has been identified in our study as a novel agent with potential for the development of anti-mycobacterial drugs. This is the first report to discover and document the efficacy of RTD against mycobacteria, including AlRa, AlRv, and AlMm, exhibiting MICs as low as 3.13 μg/mL (
Figure 1). However, the MIC of RTD against wild-type
M. tuberculosis H37Rv is 25 μg/mL determined by MABA. As described in the reference, the emission of blue-green light by AlRv is dependent on the presence of oxygen [
16]. Hence, the observed differences in the efficacy of RTD against luminescent and non-luminescent
M. tuberculosis H37Rv may be attributed to its impact on the respiration of bacteria or other substrates needed for the reaction catalyzed by LuxAB. These results suggest that the method based on RLU determination is more sensitive than MABA, thereby preventing the neglect of compounds with potential anti-
M. tuberculosis activity.
Notably, RTD displayed limited yet significant activity against nonreplicating
M. tuberculosis (
Figure 2). Additionally, partial synergistic effects were observed when RTD was combined with CLO, PTM, and TB47, resulting in enhanced inhibition of AlRa growth (
Table 1). These findings collectively suggest that RTD may be a promising candidate for anti-mycobacterial drug development. We employed gavage administration and observed that RTD (the highest dosage of 25 mg/kg) exhibited no activity against
M. tuberculosis and no obvious toxicity for mice (
Figure 3A). The combination of RTD and TB47 did not demonstrate an obvious synergistic effect against
M. tuberculosis in vivo via oral administration (
Figure 3A). Therefore, we aimed to evaluate the
in vivo efficacy of RTD using a recently developed murine model designed to enhance the pulmonary local concentration of drugs [
23]. Our findings revealed that RTD, at a concentration of 1.56 mg/mL, displayed no
in vivo anti-
M. tuberculosis activity when administered via inhalation for ~25 minutes (
Figure 3B). These results imply that RTD may serve as a lead compound for the development of derivatives with improved
in vivo anti-
M. tuberculosis activity.
Meanwhile, it proved challenging to obtain mycobacterial mutants resistant to RTD, highlighting its advantageous property in preventing the emergence of resistance (data not shown). To address the challenge of identifying mutants resistant to RTD, we altered our strategy and conducted transcriptome analysis of AlRa treated with RTD to identify potential targets of RTD. We found that a total of 2329 genes had significantly altered expression levels (
Figure 4A). Consequently, we established gene selection criteria based on a |log
2Foldchange| greater than 2 and essential genes, resulting in the identification of eight genes (
Table 2). Rv0351, along with Rv0352, act as cofactors that stimulate the ATPase activity of Rv0350 [
30,
31]. Rv0350 and Rv0440 play roles in maintaining
M. tuberculosis protein stability and sustaining long-term cell vitality under stressful conditions [
31]. Rv0384 is thought to be an ATPase subunit of an intracellular ATP-dependent protease crucial for
M. tuberculosis survival within macrophages and maintaining dormant states [
32]. Furthermore, Rv2720, involved in the SOS response, can bind to the operator region of multiple genes, inhibiting their expression during normal growth conditions [
33,
34]. However, Rv2720 may dissociate from these operators, triggering the activation of these genes to support DNA repair processes [
33]. The specific function of Rv2827, categorized as a conserved hypothetical protein, remains unknown. Some studies have revealed that Rv2827 may be a DNA-binding protein responsible for antitoxin expression required for growth [
35,
36]. A previous report has shown that Rv3260 functions as a transcriptional regulator associated with cell division [
37]. Notably, seven out of eight essential genes discussed herein exhibited upregulation following treatment of
M. tuberculosis with RTD, with the exception of
Rv3260c. Further comprehensive investigations are necessary to elucidate RTD's mechanism of action, primarily targeting the aforementioned eight genes.
5. Conclusions
For the first time, we have discovered that RTD exhibited activities against specific mycobacterial strains, particularly M. tuberculosis and M. marinum. RTD demonstrated a significant partial synergistic effect against M. tuberculosis in vitro when combined with CLO, PTM, and TB47, with FICIs ranging from 0.5 to 1. The screening for mycobacterial mutants resistant to RTD proved challenging during the experiment. This characteristic of RTD demonstrates favorable property as a clinical anti-mycobacterial agent that can prevent the emergence of drug-resistant strains. All results indicated that RTD provides fundamental information for drug repurposing or lead compound for optimization and offers alternative options for clinical treatment. Further experiments are necessary to elucidate the anti-mycobacterial activity of RTD-related derivatives in vivo using various mice model and uncover the corresponding mechanism.
6. Patents
Based on our experimental results, we have applied for a Chinese invention patent with patent number of 202310121229.X.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: The genes with a |log2Foldchange| greater than 1.
Author Contributions
Xirong Tian designed the experiment, performed the investigation, interpreted the results, and wrote and revised the manuscript. Wanli Ma evaluated the activity of RTD against nonreplicating M. tuberculosis. Buhari Yusuf and Biyi Su contributed to analysis of the data, reading MIC results and the revision of the manuscript. Jinxing Hu designed the experiment, analyzed the data, and modified the manuscript. Tianyu Zhang designed the experiment, interpreted the results, modified the manuscript, and conducted funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2021YFA1300900), and by the Guangzhou Science and Technology Planning Project (2023A03J0992).
Institutional Review Board Statement
The animal care and experimental protocols were approved by the Laboratory Animal Ethics Committee of the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors gratefully thank the thoughtful review and helpful comments of editors and reviewers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sia, J.K.; Rengarajan, J. Immunology of Mycobacterium tuberculosis infections. Microbiol Spectr. 2019, 7. [CrossRef]
- Urban, M.; Šlachtová, V.; Brulíková, L. Small organic molecules targeting the energy metabolism of Mycobacterium tuberculosis. Eur J Med Chem. 2021, 212, 113139. [CrossRef]
- Getahun, H.; Matteelli, A.; Abubakar, I.; Aziz, M.A.; Baddeley, A.; Barreira, D.; Den Boon, S.; Borroto Gutierrez, S.M.; Bruchfeld, J.; Burhan, E., et al. Management of latent Mycobacterium tuberculosis infection: WHO guidelines for low tuberculosis burden countries. Eur Respir J. 2015, 46, 1563-1576. [CrossRef]
- Mishra, A.; Surolia, A. Mycobacterium tuberculosis: Surviving and indulging in an unwelcoming host. IUBMB Life. 2018, 70, 917-925. [CrossRef]
- Sharma, S.K.; Upadhyay, V. Epidemiology, diagnosis & treatment of non-tuberculous mycobacterial diseases. Indian J Med Res. 2020, 152, 185-226. [CrossRef]
- Shaku, M.T.; Ocius, K.L.; Apostolos, A.J.; Pires, M.M.; VanNieuwenhze, M.S.; Dhar, N.; Kana, B.D. Amidation of glutamate residues in mycobacterial peptidoglycan is essential for cell wall cross-linking. Front Cell Infect Microbiol. 2023, 13, 1205829. [CrossRef]
- Falkinham, J.O. Challenges of NTM drug development. Front Microbiol. 2018, 9, 1613. [CrossRef]
- Story-Roller, E.; Maggioncalda, E.C.; Cohen, K.A.; Lamichhane, G. Mycobacterium abscessus and β-Lactams: Emerging insights and potential opportunities. Front Microbiol. 2018, 9, 2273. [CrossRef]
- Das, S.; Garg, T.; Chopra, S.; Dasgupta, A. Repurposing disulfiram to target infections caused by non-tuberculous mycobacteria. J Antimicrob Chemother. 2019, 74, 1317-1322. [CrossRef]
- Won, T.-B.; Kim, H.G.; Kim, J.-W.; Kim, J.K.; Kim, Y.H.; Kim, S.W.; Kim, H.Y.; Kim, D.W.; Kim, S.W.; Kim, C.-H., et al. Efficacy and safety of rupatadine fumarate in the treatment of perennial allergic rhinitis: A multicenter, double-blinded, randomized, placebo-controlled, bridging study in Koreans. Asian Pac J Allergy Immunol. 2021. [CrossRef]
- Bachert, C.; van Cauwenberge, P.; Khaltaev, N. Allergic rhinitis and its impact on asthma. In collaboration with the World Health Organization. Executive summary of the workshop report. 7-10 December 1999, Geneva, Switzerland. Allergy. 2002, 57, 841-855. [CrossRef]
- Molyva, D.; Kalokasidis, K.; Poulios, C.; Dedi, H.; Karkavelas, G.; Mirtsou, V.; Goulas, A. Rupatadine effectively prevents the histamine-induced up regulation of histamine H1R and bradykinin B2R receptor gene expression in the rat paw. Pharmacol Rep. 2014, 66, 952-955. [CrossRef]
- Mullol, J.; Bousquet, J.; Bachert, C.; Canonica, G.W.; Giménez-Arnau, A.; Kowalski, M.L.; Simons, F.E.R.; Maurer, M.; Ryan, D.; Scadding, G. Update on rupatadine in the management of allergic disorders. Allergy. 2015, 70 Suppl 100. [CrossRef]
- Keam, S.J.; Plosker, G.L. Rupatadine: a review of its use in the management of allergic disorders. Drugs. 2007, 67, 457-474. [CrossRef]
- Nettis, E.; Delle Donne, P.; Di Leo, E.; Calogiuri, G.F.; Ferrannini, A.; Vacca, A. Rupatadine for the treatment of urticaria. Expert Opin Pharmacother. 2013, 14, 1807-1813. [CrossRef]
- Zhang, T.; Li, S.-Y.; Nuermberger, E.L. Autoluminescent Mycobacterium tuberculosis for rapid, real-time, non-invasive assessment of drug and vaccine efficacy. PLoS One. 2012, 7, e29774. [CrossRef]
- Yang, F.; Njire, M.M.; Liu, J.; Wu, T.; Wang, B.; Liu, T.; Cao, Y.; Liu, Z.; Wan, J.; Tu, Z., et al. Engineering more stable, selectable marker-free autoluminescent mycobacteria by one step. PLoS One. 2015, 10, e0119341. [CrossRef]
- Liu, Y.; Tan, Y.; Islam, M.M.; Cao, Y.; Lu, X.; Zeng, S.; Hameed, H.M.A.; Zhou, P.; Cai, X.; Wang, S., et al. Assessment of clofazimine and TB47 combination activity against Mycobacterium abscessus using a bioluminescent approach. Antimicrob Agents Chemother. 2020, 64. [CrossRef]
- Liu, Y.; Gao, Y.; Liu, J.; Tan, Y.; Liu, Z.; Chhotaray, C.; Jiang, H.; Lu, Z.; Chiwala, G.; Wang, S., et al. The compound TB47 is highly bactericidal against Mycobacterium ulcerans in a Buruli ulcer mouse model. Nat Commun. 2019, 10, 524. [CrossRef]
- Cho, S.; Lee, H.S.; Franzblau, S. Microplate Alamar Blue assay (MABA) and low oxygen recovery assay (LORA) for Mycobacterium tuberculosis. Methods Mol Biol. 2015, 1285, 281-292. [CrossRef]
- Tian, X.; Ma, W.; Yusuf, B.; Li, C.; Hameed, H.M.A.; Wang, X.; Zhong, N.; Hu, J.; Zhang, T. High-throughput screening of compounds against autoluminescent nonreplicating Mycobacterium tuberculosis under diverse conditions. BioRxiv DOI: 10.1101/2024.03.10.584296.
- Liu, P.; Yang, Y.; Tang, Y.; Yang, T.; Sang, Z.; Liu, Z.; Zhang, T.; Luo, Y. Design and synthesis of novel pyrimidine derivatives as potent antitubercular agents. Eur J Med Chem. 2019, 163, 169-182. [CrossRef]
- Tian, X.; Gao, Y.; Ma, W.; Zhang, J.; Ju, Y.; Ding, J.; Zeng, S.; Hameed, H.M.A.; Zhong, N.; Cook, G.M., et al. Establishment of an inhalation administration non-invasive murine model for rapidly testing drug activity against Mycobacterium tuberculosis. BioRxiv DOI: 10.1101/2024.02.27.582260.
- Makafe, G.G.; Hussain, M.; Surineni, G.; Tan, Y.; Wong, N.-K.; Julius, M.; Liu, L.; Gift, C.; Jiang, H.; Tang, Y., et al. Quinoline derivatives kill Mycobacterium tuberculosis by activating glutamate kinase. Cell Chem Biol. 2019, 26. [CrossRef]
- Wilkinson, K.A.; Stewart, G.R.; Newton, S.M.; Vordermeier, H.M.; Wain, J.R.; Murphy, H.N.; Horner, K.; Young, D.B.; Wilkinson, R.J. Infection biology of a novel alpha-crystallin of Mycobacterium tuberculosis: Acr2. J Immunol. 2005, 174, 4237-4243. [CrossRef]
- Healy, C.; Golby, P.; MacHugh, D.E.; Gordon, S.V. The MarR family transcription factor Rv1404 coordinates adaptation of Mycobacterium tuberculosis to acid stress via controlled expression of Rv1405c, a virulence-associated methyltransferase. Tuberculosis (Edinb). 2016, 97, 154-162. [CrossRef]
- Tükenmez, H.; Sarkar, S.; Anoosheh, S.; Kruchanova, A.; Edström, I.; Harrison, G.A.; Stallings, C.L.; Almqvist, F.; Larsson, C. Mycobacterium tuberculosis Rv3160c is a TetR-like transcriptional repressor that regulates expression of the putative oxygenase Rv3161c. Sci Rep. 2021, 11, 1523. [CrossRef]
- Larsen, M.H.; Lacourciere, K.; Parker, T.M.; Kraigsley, A.; Achkar, J.M.; Adams, L.B.; Dupnik, K.M.; Hall-Stoodley, L.; Hartman, T.; Kanipe, C., et al. The many hosts of mycobacteria 8 (MHM8): A conference report. Tuberculosis (Edinb). 2020, 121, 101914. [CrossRef]
- Togre, N.S.; Vargas, A.M.; Bhargavi, G.; Mallakuntla, M.K.; Tiwari, S. Fragment-based drug discovery against mycobacteria: The success and challenges. Int J Mol Sci. 2022, 23. [CrossRef]
- Kim, W.S.; Kim, J.-S.; Kim, H.M.; Kwon, K.W.; Eum, S.-Y.; Shin, S.J. Comparison of immunogenicity and vaccine efficacy between heat-shock proteins, HSP70 and GrpE, in the DnaK operon of Mycobacterium tuberculosis. Sci Rep. 2018, 8, 14411. [CrossRef]
- Trutneva, K.A.; Shleeva, M.O.; Demina, G.R.; Vostroknutova, G.N.; Kaprelyans, A.S. One-year old dormant, "Non-culturable" Mycobacterium tuberculosis preserves significantly diverse protein profile. Front Cell Infect Microbiol. 2020, 10, 26. [CrossRef]
- Tripathi, P.; Singh, L.K.; Kumari, S.; Hakiem, O.R.; Batra, J.K. ClpB is an essential stress regulator of Mycobacterium tuberculosis and endows survival advantage to dormant bacilli. Int J Med Microbiol. 2020, 310, 151402. [CrossRef]
- Chatterjee, C.; Majumdar, S.; Deshpande, S.; Pant, D.; Matheshwaran, S. Real-time kinetic studies of Mycobacterium tuberculosis LexA-DNA interaction. Biosci Rep. 2021, 41. [CrossRef]
- Chandran, A.V.; Srikalaivani, R.; Paul, A.; Vijayan, M. Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment. Acta Crystallogr D Struct Biol. 2019, 75, 41-55. [CrossRef]
- Beck, I.N.; Usher, B.; Hampton, H.G.; Fineran, P.C.; Blower, T.R. Antitoxin autoregulation of M. tuberculosis toxin-antitoxin expression through negative cooperativity arising from multiple inverted repeat sequences. Biochem J. 2020, 477, 2401-2419. [CrossRef]
- Janowski, R.; Panjikar, S.; Eddine, A.N.; Kaufmann, S.H.E.; Weiss, M.S. Structural analysis reveals DNA binding properties of Rv2827c, a hypothetical protein from Mycobacterium tuberculosis. J Struct Funct Genomics. 2009, 10, 137-150. [CrossRef]
- Konar, M.; Alam, M.S.; Arora, C.; Agrawal, P. WhiB2/Rv3260c, a cell division-associated protein of Mycobacterium tuberculosis H37Rv, has properties of a chaperone. FEBS J. 2012, 279, 2781-2792. [CrossRef]
Figure 1.
Time-killing curves of different mycobacteria treated with RTD. AlRa, autoluminescent M. tuberculosis H37Ra; AlRv, autoluminescent M. tuberculosis H37Rv; AlMm, autoluminescent M. marinum; AlMab, autoluminescent M. abscessus; AlMs, autoluminescent M. smegmatis; Solvent, dimethyl sulfoxide; RTD, rupatadine; RIF, rifampicin; CLO, clofazimine; CLR, clarithromycin. The numbers following compounds indicate the corresponding concentrations (μg/mL).
Figure 1.
Time-killing curves of different mycobacteria treated with RTD. AlRa, autoluminescent M. tuberculosis H37Ra; AlRv, autoluminescent M. tuberculosis H37Rv; AlMm, autoluminescent M. marinum; AlMab, autoluminescent M. abscessus; AlMs, autoluminescent M. smegmatis; Solvent, dimethyl sulfoxide; RTD, rupatadine; RIF, rifampicin; CLO, clofazimine; CLR, clarithromycin. The numbers following compounds indicate the corresponding concentrations (μg/mL).
Figure 2.
Time-killing curves of replicating AlRa or recovering curves of nonreplicating AlRa treated with RTD in diverse media. (A) aerobic conditions; (B) anaerobic conditions. Solvent, dimethyl sulfoxide; RTD, rupatadine; RIF, rifampicin; G, 7H9 enriched with glycerol; C, 7H9 enriched with cholesterol. The numbers following RTD or RIF indicate the corresponding concentrations (μg/mL).
Figure 2.
Time-killing curves of replicating AlRa or recovering curves of nonreplicating AlRa treated with RTD in diverse media. (A) aerobic conditions; (B) anaerobic conditions. Solvent, dimethyl sulfoxide; RTD, rupatadine; RIF, rifampicin; G, 7H9 enriched with glycerol; C, 7H9 enriched with cholesterol. The numbers following RTD or RIF indicate the corresponding concentrations (μg/mL).
Figure 3.
The in vivo activity of RTD against M. tuberculosis. A. The in vivo activity of RTD and the combination of RTD and TB47 using oral administration. Mice were infected with AlRv via aerosol. The treatments were administered daily from the eighth day post-infection and lasted for eight days. The RLUs of live mice and the lung suspension were detected at the initiation and one day after completion of treatment. The dosages of RTD were 6.25 and 25 mg/kg, and the dosage of TB47 was 25 mg/kg. B. The in vivo activity of RTD using inhalable administration. Mice were infected with AlRv via aerosol. The treatments were administered daily from the next day post-infection and lasted for 15 days. The RLUs of live mice and the lung suspension were detected at the initiation and one day after completion of treatment. The dosages of RTD and RIF were 1.56 and 2 mg/mL, respectively. The duration of inhalable administration (4 mL) each time was approximately 25 minutes. BT, before treatment; Solvent (A), sodium carboxymethyl cellulose; Solvent (B), distilled water; RTD, rupatadine; RIF, rifampicin; ns, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Figure 3.
The in vivo activity of RTD against M. tuberculosis. A. The in vivo activity of RTD and the combination of RTD and TB47 using oral administration. Mice were infected with AlRv via aerosol. The treatments were administered daily from the eighth day post-infection and lasted for eight days. The RLUs of live mice and the lung suspension were detected at the initiation and one day after completion of treatment. The dosages of RTD were 6.25 and 25 mg/kg, and the dosage of TB47 was 25 mg/kg. B. The in vivo activity of RTD using inhalable administration. Mice were infected with AlRv via aerosol. The treatments were administered daily from the next day post-infection and lasted for 15 days. The RLUs of live mice and the lung suspension were detected at the initiation and one day after completion of treatment. The dosages of RTD and RIF were 1.56 and 2 mg/mL, respectively. The duration of inhalable administration (4 mL) each time was approximately 25 minutes. BT, before treatment; Solvent (A), sodium carboxymethyl cellulose; Solvent (B), distilled water; RTD, rupatadine; RIF, rifampicin; ns, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
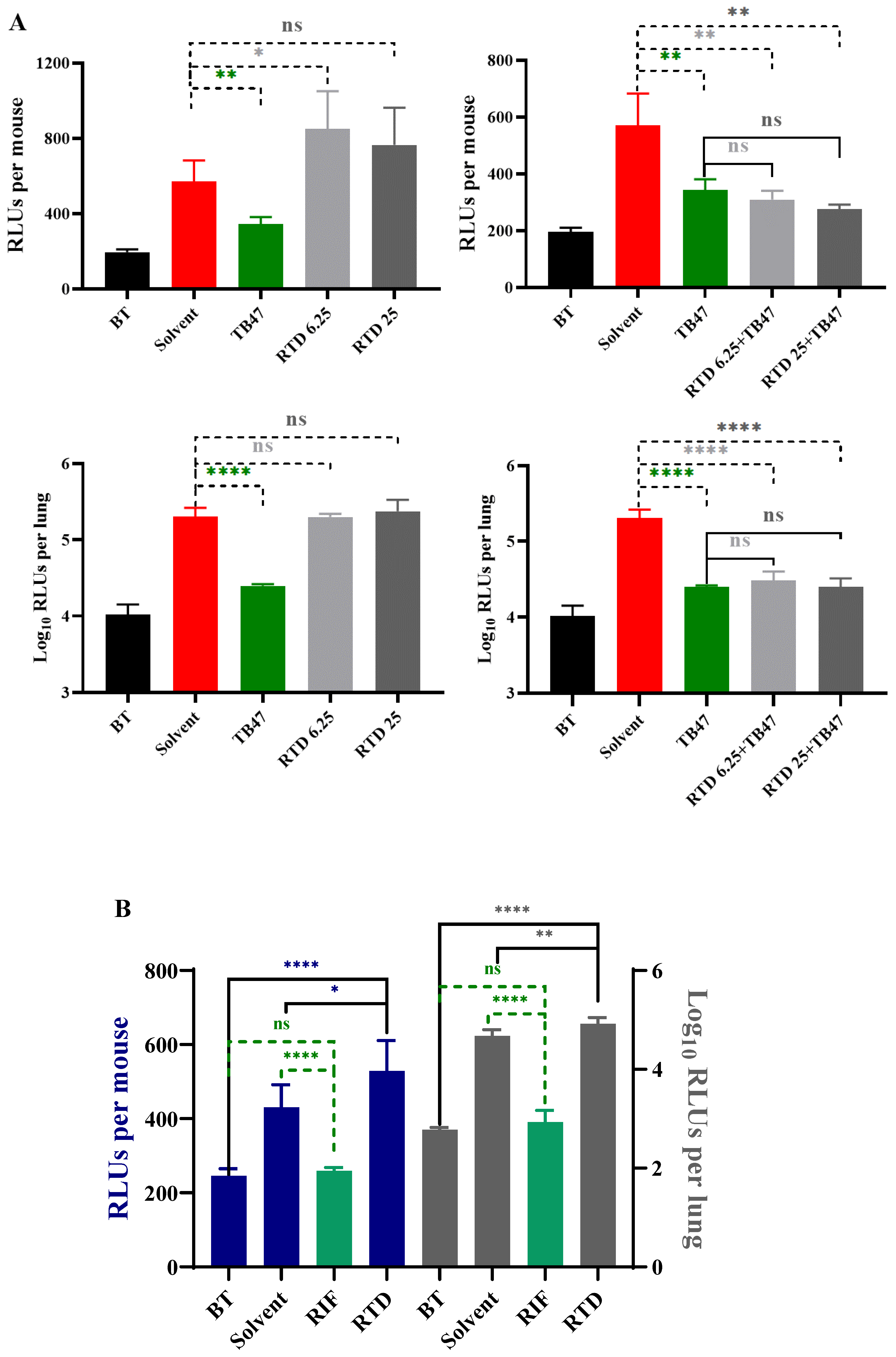
Figure 4.
The transcriptome profile of M. tuberculosis cocultured with or without RTD treatment. (A) The volcano plot of genes with |log2FoldChange| > 0 and P < 0.05; (B) The KEGG analysis of gene descriptions.
Figure 4.
The transcriptome profile of M. tuberculosis cocultured with or without RTD treatment. (A) The volcano plot of genes with |log2FoldChange| > 0 and P < 0.05; (B) The KEGG analysis of gene descriptions.
Table 1.
Activity of RTD in combination with anti-M. tuberculosis drugs against AlRa.
Table 1.
Activity of RTD in combination with anti-M. tuberculosis drugs against AlRa.
| Drug*
|
MIC# (µg/mL) |
FICI※
|
Effects |
| MICRTDC
|
MICRTDA
|
MICC
|
MICA
|
| AMK |
0.20 |
3.13 |
0.5 |
0.5 |
1.0625 |
Indifferent |
| CLO |
0.78 |
3.13 |
0.1 |
0.2 |
0.75 |
Partial synergistic |
| INH |
3.13 |
3.13 |
0.003125 |
0.05 |
1.0625 |
Indifferent |
| LEV |
0.20 |
3.13 |
0.125 |
0.125 |
1.0625 |
Indifferent |
| LZD |
3.13 |
3.13 |
0.5 |
1 |
1.5 |
Indifferent |
| PTM |
0.20 |
3.13 |
0.0625 |
0.125 |
0.5625 |
Partial synergistic |
| RIF |
3.13 |
3.13 |
0.0015625 |
0.0125 |
1.125 |
Indifferent |
| STR |
1.56 |
3.13 |
2 |
4 |
1 |
Additive |
| TB47 |
0.78 |
3.13 |
0.00075 |
0.0015 |
0.75 |
Partial synergistic |
Table 2.
The summary of essential genes with |log2FoldChange| exceeding 2.
Table 2.
The summary of essential genes with |log2FoldChange| exceeding 2.
| Gene_id |
log2FoldChange*
|
Gene_name |
Product/Function |
| Rv0350 |
3.68124885 |
dnaK |
Probable chaperone protein DnaK (heat shock protein 70); Acts as a chaperone. |
| Rv0351 |
3.750319008 |
grpE |
Probable GrpE protein (HSP-70 cofactor); Stimulates, jointly with Rv0352, the ATPase activity of Rv0350. |
| Rv0352 |
3.066383388 |
dnaJ1 |
Chaperone protein DnaJ (HSP-70 cofactor); Stimulates, jointly with Rv0351, the ATPase activity of Rv0350. |
| Rv0384c |
2.667760091 |
clpB |
Probable endopeptidase ATP binding protein ClpB (heat shock protein F84.1); Thought to be an ATPase subunit of an intracellular ATP-dependent protease. |
| Rv0440 |
2.448531361 |
groEL2 |
Molecular chaperone GroEL; Prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions. |
| Rv2720 |
2.27206078 |
lexA |
Repressor LexA; Involved in regulation of nucleotide excision repair and sos response. |
| Rv2827c |
2.142454829 |
Rv2827c |
Hypothetical protein; Unknown. |
| Rv3260c |
-2.134883679 |
whiB2 |
Probable transcriptional regulatory protein WhiB-like WhiB2; Involved in transcriptional mechanism. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).