Submitted:
07 June 2024
Posted:
10 June 2024
You are already at the latest version
Abstract
Keywords:
Introduction
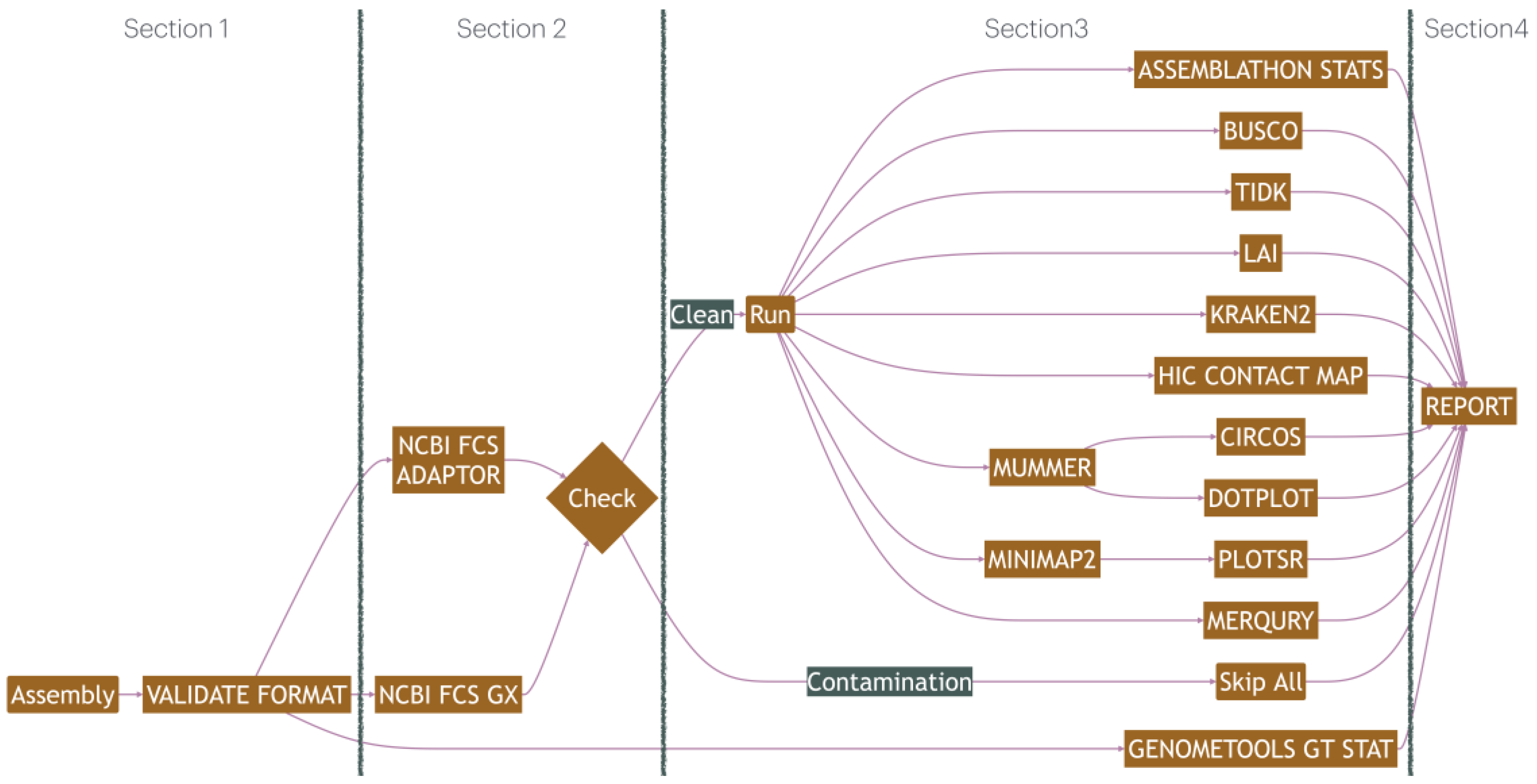
Materials and Methods
Design
Implementation Details
Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
References
- Agarwal, T., et al. Recent Advances in Gene and Genome Assembly: Challenges and Implications. In.; 2020.
- Andrews, S. FastQC: a quality control tool for high throughput sequence data. In.: Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom; 2010.
- Astashyn, A., et al. Rapid and sensitive detection of genome contamination at scale with FCS-GX. bioRxiv 2023:2023.2006.2002.543519. [CrossRef]
- Brown, M., González De la Rosa, P. M. and Mark, B. A Telomere Identification Toolkit. In.; 2023.
- Cabanettes, F. and Klopp, C. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 2018;6:e4958. [CrossRef]
- Chen, S., et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34(17):i884-i890. [CrossRef]
- Cornet, L. and Baurain, D. Contamination detection in genomic data: more is not enough. Genome Biol 2022;23(1):60. [CrossRef]
- da Veiga Leprevost, F., et al. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 2017;33(16):2580-2582. [CrossRef]
- Danecek, P., et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10(2):giab008. [CrossRef]
- Di Tommaso, P., et al. Nextflow enables reproducible computational workflows. Nat Biotechnol 2017;35(4):316-319.
- Dida, F. and Yi, G. Empirical evaluation of methods for de novo genome assembly. PeerJ Comput Sci 2021;7:e636.
- Dudchenko, O., et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017;356(6333):92-95. [CrossRef]
- EBP. Report on Assembly Standards Version 5.0. In.: Earth Biogenome Project; 2023.
- Edwards, R. 2019. linsalrob/fasta_validator: Initial Release. Release v0.1. [CrossRef]
- Ellinghaus, D., Kurtz, S. and Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 2008;9(1):18. [CrossRef]
- Ewels, P.A., et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38(3):276-278. [CrossRef]
- Faust, G.G. and Hall, I.M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014;30(17):2503-2505. [CrossRef]
- Goel, M. and Schneeberger, K. plotsr: visualizing structural similarities and rearrangements between multiple genomes. Bioinformatics 2022;38(10):2922-2926. [CrossRef]
- Gremme, G., Steinbiss, S. and Kurtz, S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 2013;10(3):645-656. [CrossRef]
- Gruning, B., et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 2018;15(7):475-476. [CrossRef]
- Krzywinski, M., et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19(9):1639-1645. [CrossRef]
- Kurtzer, G.M., Sochat, V. and Bauer, M.W. Singularity: Scientific containers for mobility of compute. PLoS One 2017;12(5):e0177459. [CrossRef]
- Langer, B.E., et al. Empowering bioinformatics communities with Nextflow and nf-core. bioRxiv 2024:2024.2005.2010.592912.
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 2013.
- Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34(18):3094-3100. [CrossRef]
- Manchanda, N., et al. GenomeQC: a quality assessment tool for genome assemblies and gene structure annotations. BMC Genomics 2020;21(1):193. [CrossRef]
- Marcais, G., et al. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol 2018;14(1):e1005944. [CrossRef]
- Merkel, D. Docker: lightweight linux containers for consistent development and deployment. Linux j 2014;239(2):2.
- NCBI. NCBI Assembly Database. In.; 2024.
- NHGRI. DNA Sequencing Costs: Data. In.; 2023.
- Ondov, B.D., Bergman, N.H. and Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 2011;12(1):385. [CrossRef]
- Ou, S., Chen, J. and Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res 2018;46(21):e126. [CrossRef]
- Ou, S. and Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol 2018;176(2):1410-1422. [CrossRef]
- Ou, S. and Jiang, N. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob DNA 2019;10(1):48. [CrossRef]
- Rhie, A., et al. Towards complete and error-free genome assemblies of all vertebrate species. bioRxiv 2020:2020.2005.2022.110833. [CrossRef]
- Rhie, A., et al. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol 2020;21(1):245. [CrossRef]
- Robinson, J.T., et al. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst 2018;6(2):256-258 e251. [CrossRef]
- Seppey, M., Manni, M. and Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol Biol 2019;1962:227-245. [CrossRef]
- Shen, W., et al. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One 2016;11(10):e0163962. [CrossRef]
- Simao, F.A., et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31(19):3210-3212. [CrossRef]
- Sullivan, S. 2022. hic_qc. Release 6881c33. https://github.com/phasegenomics/hic_qc. (28 June 2023 date last accessed).
- Sun, Y., et al. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci 2022;27(4):391-401. [CrossRef]
- UCDAVIS-Bioinformatics. assemblathon2-analysis. In.; 2012.
- Wang, P. and Wang, F. A proposed metric set for evaluation of genome assembly quality. Trends Genet 2023;39(3):175-186. [CrossRef]
- Wood, D.E., Lu, J. and Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol 2019;20(1):257. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




