Submitted:
05 June 2024
Posted:
07 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
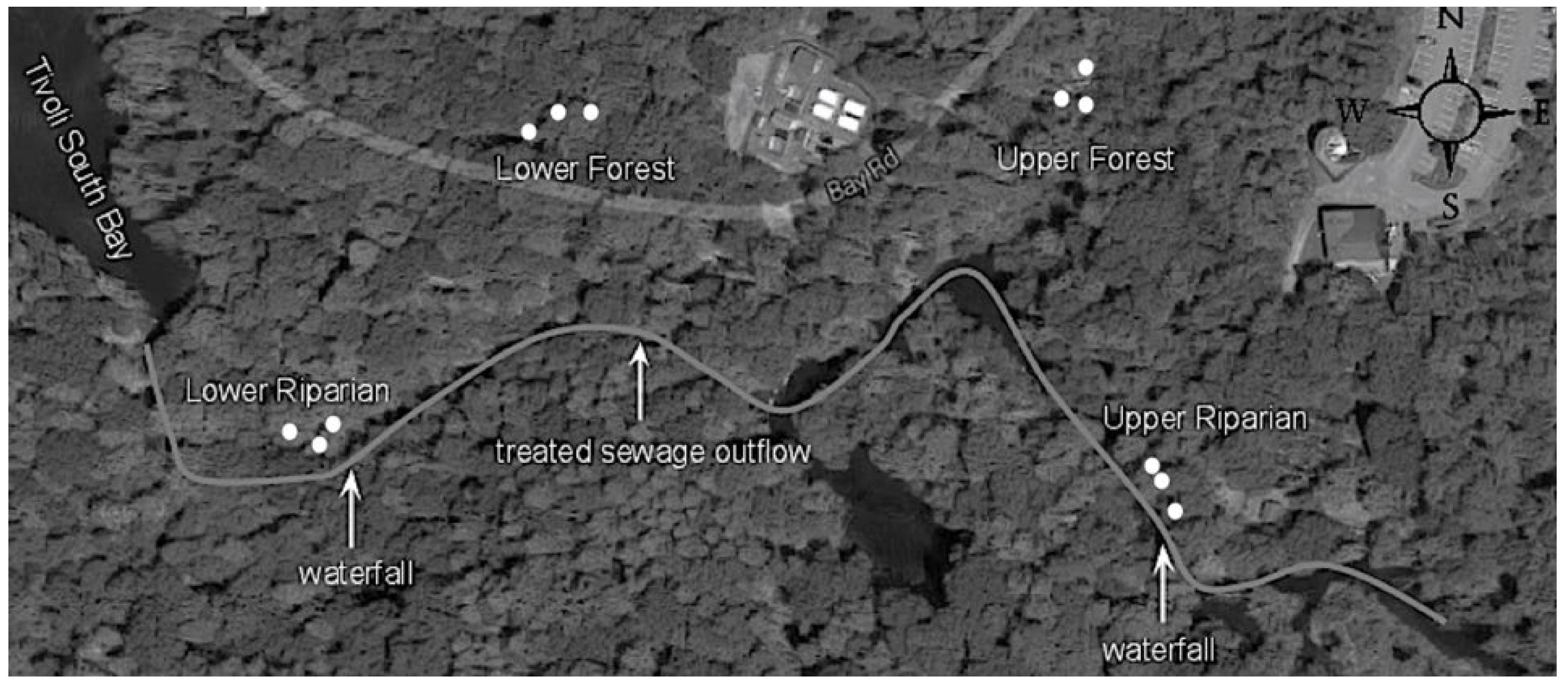
2.1. Study Sites
2.2. Surface water and phyllosphere sampling
2.3. Bacterial analyses
2.3.1. Culture-based methods
2.3.2. Culture-independent methods
2.4. Bioinformatic and Statistical analyses
3. Results
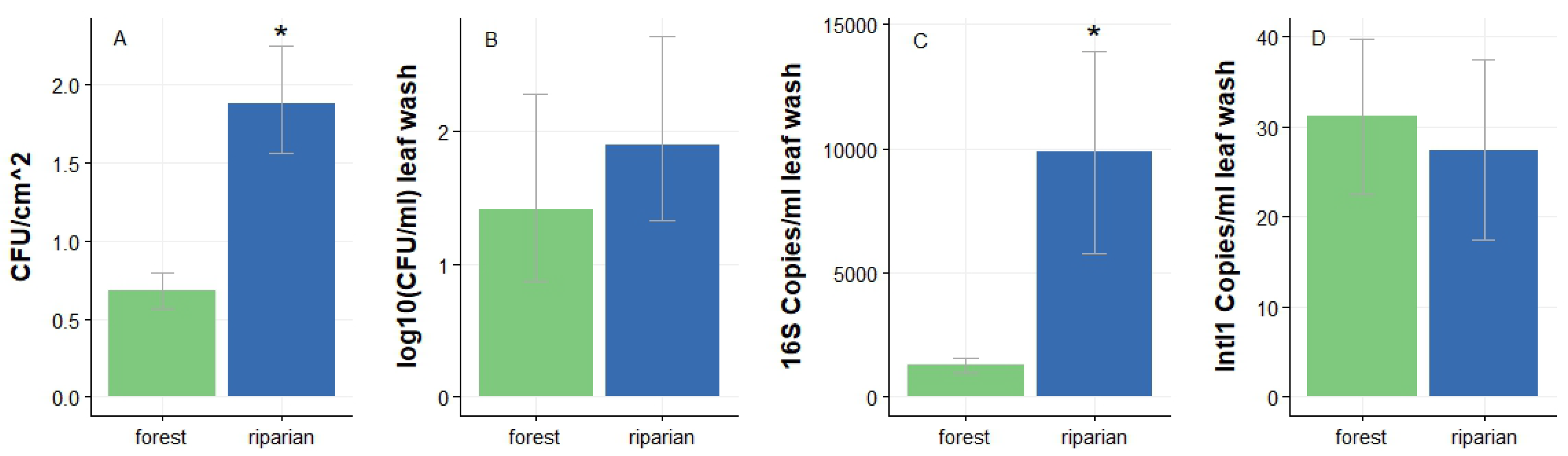
3.1. Culture-based and culture-independent quantification of leaf PMCs
3.2. Comparing bacterial communities in riparian and forest ecosystems
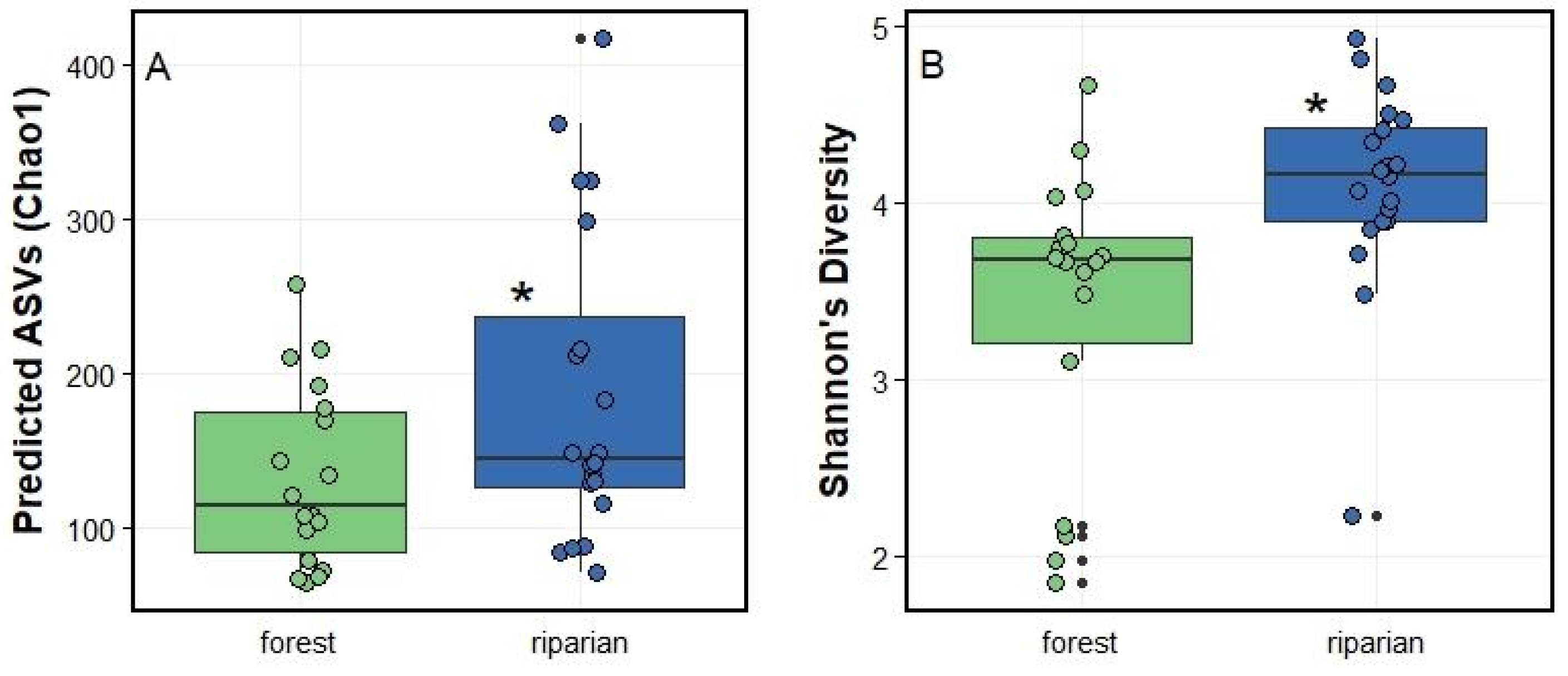
3.2.1. Riparian leaf PMCs show higher level of diversity
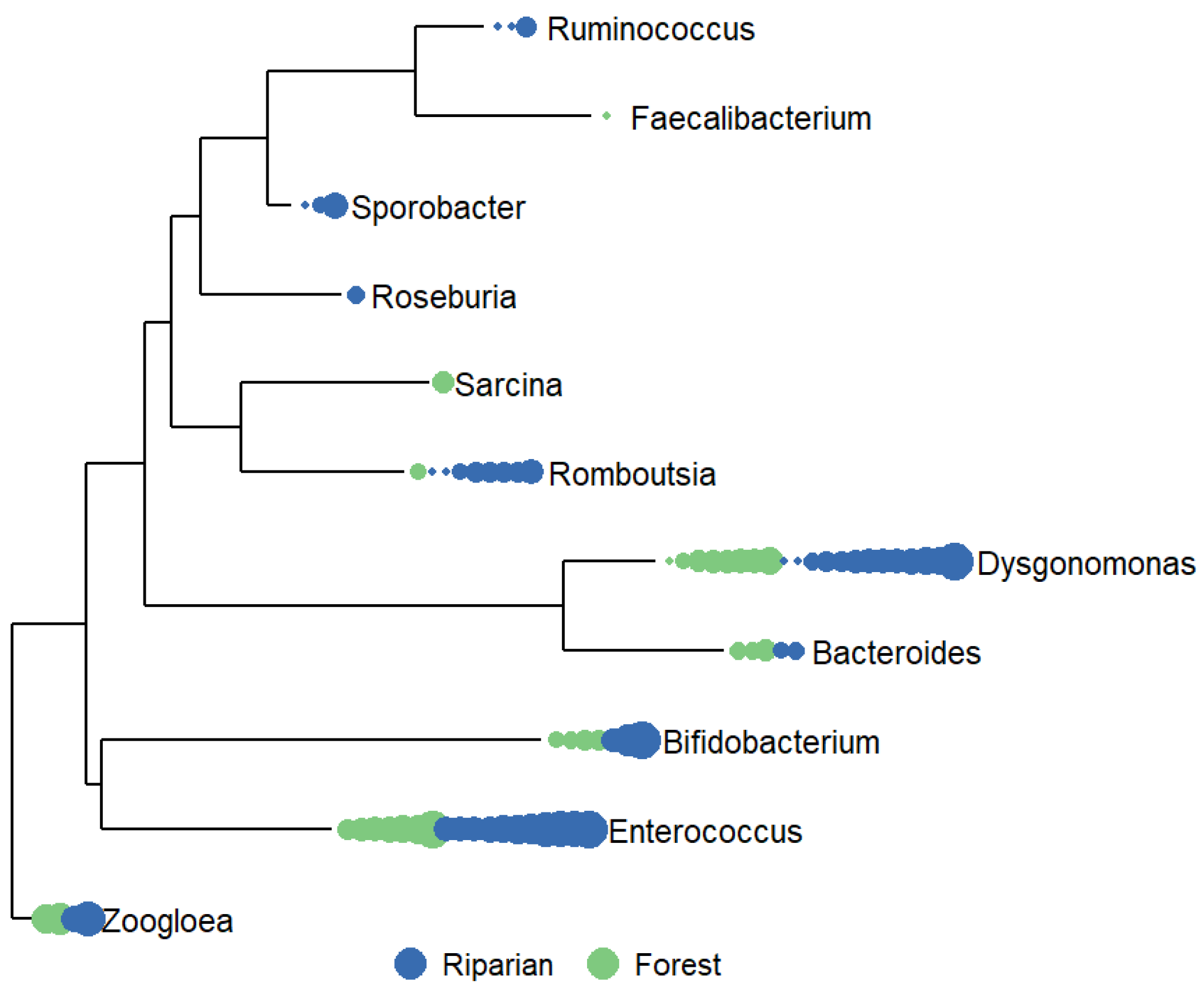
3.2.2. Bacterial communities in riparian ecosystems are distinct from forest ecosystems.
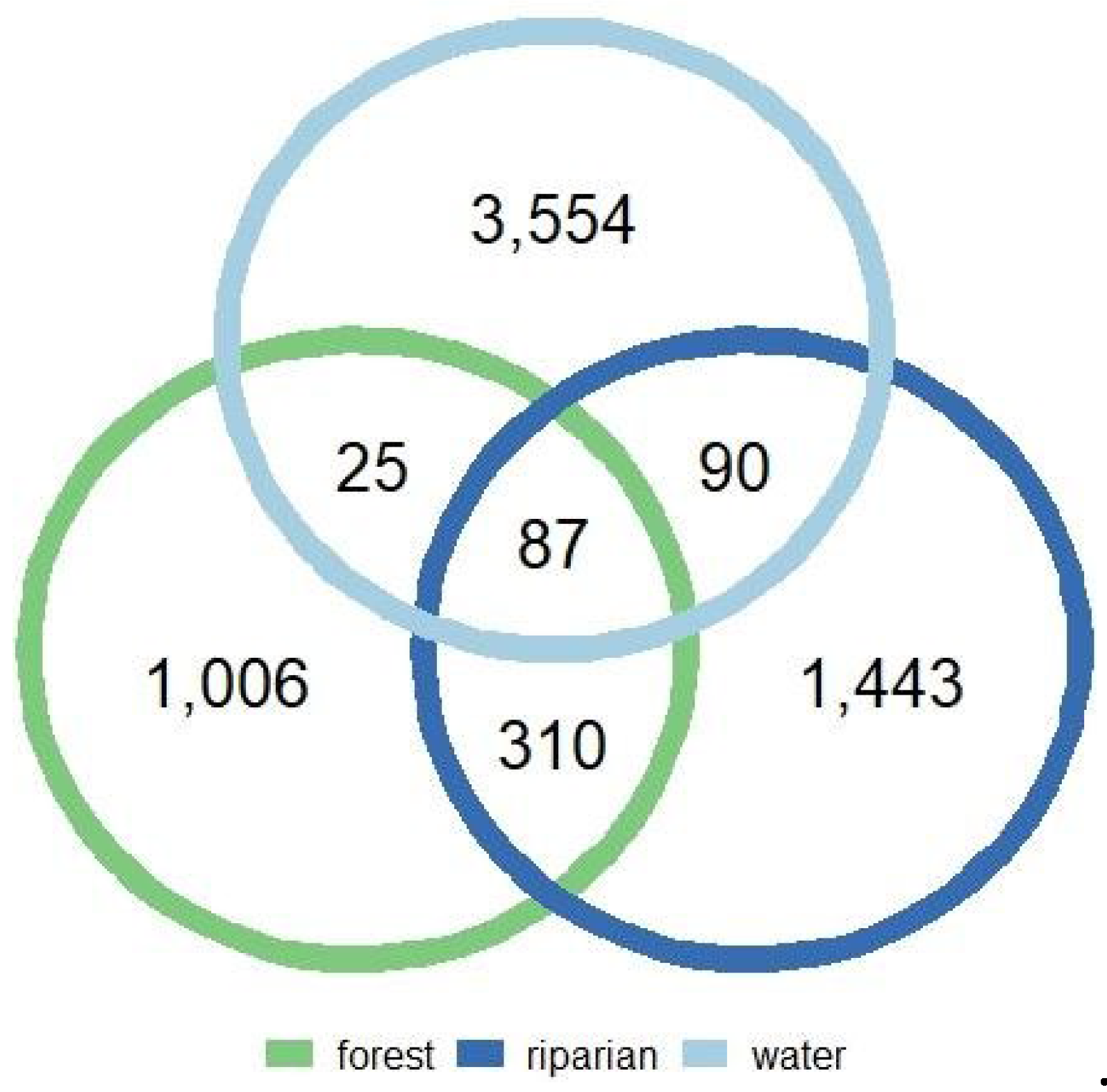
3.5. Water-phyllosphere connections
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morris CE, Kinkel LL, Lindow SE, Hecht-Poinar EI, Elliott VJ, editors. Fifty years of phyllosphere microbiology: significant contributions to research in related fields2002.
- Lighthart B, Shaffer BT. Increased airborne bacterial survival as a function of particle content and size. Aerosol Sci Technol. 1997;27(3):439-46. PubMed PMID: ISI:A1997XR30000011.
- Leveau JHJ, Beattie GA, Lindow SE, Mahaffee WF. Phyllosphere, Front and Center: Focus on a Formerly ‘Ecologically Neglected’ Microbial Milieu. Phytobiomes Journal. 2023;7(2):140-4. [CrossRef]
- Leveau J. A brief from the leaf: latest research to inform our understanding of the phyllosphere microbiome. Curr Opin Microbiol. 2019;49:41-9. [CrossRef]
- Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE. The Phyllosphere: Microbial Jungle at the Plant–Climate Interface. Annual Review of Ecology, Evolution and Systematics. 2016;47(Volume 47, 2016):1-24. [CrossRef]
- Bulgarelli D, Schlaeppi K Fau - Spaepen S, Spaepen S Fau - Ver Loren van Themaat E, Ver Loren van Themaat E Fau - Schulze-Lefert P, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. (1545-2123 (Electronic)).
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. Beyond biogeographic patterns: processes shaping the microbial landscape. Nature reviews Microbiology. 2012;10(7):497-506. Epub 05/15. [CrossRef]
- PubMed PMID: 22580365. [CrossRef]
- Finkel OM, Burch Ay Fau - Elad T, Elad T Fau - Huse SM, Huse Sm Fau - Lindow SE, Lindow Se Fau - Post AF, Post Af Fau - Belkin S, et al. Distance-decay relationships partially determine diversity patterns of phyllosphere bacteria on Tamarix trees across the Sonoran Desert [corrected]. (1098-5336 (Electronic)).
- Yan K, Han W, Zhu Q, Li C, Dong Z, Wang Y. Leaf surface microtopography shaping the bacterial community in the phyllosphere: evidence from 11 tree species. Microbiological Research. 2022;254:126897. [CrossRef]
- Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol. 2010;12(11):2885-93. PubMed PMID: WOS:000283737000002. [CrossRef]
- Lindow SE, Brandl MT. Microbiology of the phyllosphere. (0099-2240 (Print)).
- Lighthart B. The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol Ecol. 1997;23(4):263-74. PubMed PMID: ISI:A1997XU71100001.
- Warren SD. Microorganisms of the Phyllosphere: Origin, Transport, and Ecological Functions. Frontiers in Forests and Global Change. 2022;5. [CrossRef]
- Koskella B. The phyllosphere. (1879-0445 (Electronic)).
- Rastogi G, Coaker GL, Leveau JHJ. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett. 2013;348(1):1-10. [CrossRef]
- Schlechter RO, Miebach M, Remus-Emsermann MNP. Driving factors of epiphytic bacterial communities: A review. Journal of Advanced Research. 2019;19:57-65. [CrossRef]
- Burrows SM, Elbert W, Lawrence MG, Poschl U. Bacteria in the global atmosphere - Part 1: Review and synthesis of literature data for different ecosystems. Atmos Chem Phys. 2009;9(23):9263-80. PubMed PMID: ISI:000272689600013.
- Blanchard DC. The Ejection of Drops from the Sea and Their Enrichment with Bacteria and Other Materials - a Review. Estuaries. 1989;12(3):127-37. PubMed PMID: ISI:A1989AQ97400001.
- Blanchard DC. The production, distribution, and bacterial enrichment of the sea-salt aerosol. Air-sea exchange of gases and particles: Springer, Dordrecht; 1983. p. 407-54.
- Blanchard DC, Syzdek L. Bubbles and Water-to Air Transfer of Bacteria. Bull Amer Meteorol Soc. 1971;52(11):1136-41. PubMed PMID: ISI:A1971L248600015.
- Blanchard DC, Syzdek L. Mechanism for Water-to-Air Transfer and Concentration of Bacteria. Science. 1970;170(3958):626-&. PubMed PMID: ISI:A1970H672100024.
- Blanchard DC, Syzdek LD. Water-to-Air Transfer and Enrichment of Bacteria in Drops from Bursting Bubbles. Appl Environ Microbiol. 1982;43(5):1001-5. PubMed PMID: ISI:A1982NY69700005.
- Blanchard DC, Syzdek LD. Enrichment of Bacteria in Airborne Drops from Bubbles Bursting at the Surface of Bacterial-Laden Waters. Bull Amer Meteorol Soc. 1978;59(11):1516-. PubMed PMID: ISI:A1978GJ43200103.
- Blanchard DC, Syzdek LD. Concentration of Bacteria in Jet Drops from Bursting Bubbles. Journal of Geophysical Research. 1972;77(27):5087-&. PubMed PMID: ISI:A1972N516400004.
- Blanchard DC, Syzdek LD, Weber ME. Bubble Scavenging of Bacteria in Fresh-Water Quickly Produces Bacterial Enrichment in Airborne Jet Drops. Limnology and Oceanography. 1981;26(5):961-4. PubMed PMID: ISI:A1981MK65800016.
- Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J Aerosol Sci. 2005;36(5-6):801-12. PubMed PMID: ISI:000229348600018.
- Dueker ME, Weathers KC, O'Mullan GD, Juhl AR, Uriarte M. Environmental Controls on Coastal Coarse Aerosols: Implications for Microbial Content and Deposition in the Near-Shore Environment. Environ Sci Technol. 2011;45(8):3386-92. PubMed PMID: ISI:000289341300029.
- Dueker ME, O’Mullan GD, Martinez J, Juhl AR, Weathers KC. Onshore wind speed modulates microbial aerosols along an urban waterfront. Atmosphere. 2017;8(11):215. [CrossRef]
- Baylor ER, Baylor MB, Blanchard DC, Syzdek LD, Appel C. Virus transfer from surf to wind. Science. 1977;198(4317):575-80. PubMed PMID: ISI:A1977DZ50800006.
- Lavy A, McGrath DG, Matheus Carnevali PB, Wan J, Dong W, Tokunaga TK, et al. Microbial communities across a hillslope-riparian transect shaped by proximity to the stream, groundwater table, and weathered bedrock. Ecology and Evolution. 2019;9(12):6869-900. [CrossRef]
- Dodds WK, Zeglin LH, Ramos RJ, Platt TG, Pandey A, Michaels T, et al. Connections and Feedback: Aquatic, Plant, and Soil Microbiomes in Heterogeneous and Changing Environments. BioScience. 2020;70(7):548-62. [CrossRef]
- Marks R, Kruczalak K, Jankowska K, Michalska M. Bacteria and fungi in air over the Gulf of Gdansk and Baltic sea. J Aerosol Sci. 2001;32(2):237-50. PubMed PMID: ISI:000166719100005.
- Dueker ME, O'Mullan GD, Weathers KC, Juhl AR, Uriarte M. Coupling of fog and marine microbial content in the near-shore coastal environment. Biogeosciences. 2012;9(2):803-13. [CrossRef]
- Evans SE, Dueker ME, (co-lead authors), Logan JR, Weathers KC. The biology of fog: results from coastal Maine and Namib Desert reveal common drivers of fog microbial composition. Sci Total Environ. 2019;647:1547-56. [CrossRef]
- Dueker ME, French S, O’Mullan GD. Comparison of Bacterial Diversity in Air and Water of a Major Urban Center. Frontiers in Microbiology. 2018;9.
- Dueker ME, O'Mullan GD. Aeration remediation of a polluted waterway increases near-surface coarse and culturable microbial aerosols. The Science of the total environment. 2014;478:184-9. Epub 2014/02/18. PubMed PMID: 24531127. [CrossRef]
- Dueker ME, O'Mullan GD, Juhl AR, Weathers KC, Uriarte M. Local environmental pollution strongly influences culturable bacterial aerosols at an urban aquatic Superfund site. Environ Sci Technol. 2012;46(20):10926-32. [CrossRef]
- Adomako MO, Yu F-H. Potential effects of micro- and nanoplastics on phyllosphere microorganisms and their evolutionary and ecological responses. Sci Total Environ. 2023;884:163760. [CrossRef]
- Ivashchenko KV, Korneykova MV, Sazonova OI, Vetrova AA, Ermakova AO, Konstantinov PI, et al. Phylloplane Biodiversity and Activity in the City at Different Distances from the Traffic Pollution Source. Plants [Internet]. 2022; 11(3).
- Mandal M, Das S, Roy A, Rakwal R, Jones OAH, Popek R, et al. Interactive relations between plants, the phyllosphere microbial community, and particulate matter pollution. Sci Total Environ. 2023;890:164352. [CrossRef]
- Laforest-Lapointe I, Messier C, Kembel Steven W. Tree Leaf Bacterial Community Structure and Diversity Differ along a Gradient of Urban Intensity. mSystems. 2017;2(6):10.1128/msystems.00087-17. [CrossRef]
- Guo L, Maghirang RG. Numerical Simulation of Airflow and Particle Collection by Vegetative Barriers. Engineering Applications of Computational Fluid Mechanics. 2012;6(1):110-22. [CrossRef]
- Wu S, Bashir MA, Raza Q-U-A, Rehim A, Geng Y, Cao L. Application of riparian buffer zone in agricultural non-point source pollution control—A review. Frontiers in Sustainable Food Systems. 2023;7. [CrossRef]
- de Santana CO, Spealman P, Azulai D, Reid M, Dueker ME, Perron GG. Bacteria communities and water quality parameters in riverine water and sediments near wastewater discharges. Scientific Data. 2022;9(1). [CrossRef]
- Chen XY, Ran PX, Ho KF, Lu WJ, Li B, Gu ZP, et al. Concentrations and Size Distributions of Airborne Microorganisms in Guangzhou during Summer. Aerosol Air Qual Res. 2012;12(6):1336-44. PubMed PMID: WOS:000313100300030. [CrossRef]
- Tang G, Fan Y, Li X, Tian R, Tang R, Xu L, et al. Effects of leaf properties on the counts of microbes on the leaf surfaces of wheat, rye and triticale. FEMS Microbiol Ecol. 2023;99(4):fiad024. [CrossRef]
- Odds FC. Interactions among amphotericin B, 5-fluorocytosine, ketoconazole, and miconazole against pathogenic fungi in vitro. (0066-4804 (Print)).
- Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. Isme Journal. 2011;5(8):1253-61. PubMed PMID: WOS:000295782200003. [CrossRef]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6(8):1621-4. [CrossRef]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13(7):581-3. [CrossRef]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. Epub 05/01. [CrossRef]
- 10.1371/journal.pone.0061217. Print 2013. PubMed PMID: 23630581.
- McMurdie PJ, Holmes S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput Biol. 2014;10(4). [CrossRef]
- 10.1371/journal.pcbi.1003531. PubMed PMID: 24699258.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical.
- Computing, Vienna, Austria. 2018;URL https://www.R-project.org/.
- Laforest-Lapointe I, Messier C, Kembel SW. Tree phyllosphere bacterial communities: exploring the magnitude of intra- and inter-individual variation among host species. (2167-8359 (Print)).
- Laforest-Lapointe IA-O, Messier C, Kembel SW. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. (2049-2618 (Electronic)).
- Roldán DM, Kyrpides N, Woyke T, Shapiro N, Whitman WB, Králová S, et al. Hymenobacter artigasi sp. nov., isolated from air sampling in maritime Antarctica. Int J Syst Evol Microbiol. 2020;70(9):4935-41. [CrossRef]
- Kim Y, Subramanian P, Choi H, Weon HY, Kim S, Kwon SW, et al. Five novel Hymenobacter species isolated from air: Hymenobacter cellulosilyticus sp. nov., Hymenobacter cellulosivorans sp. nov., Hymenobacter aerilatus sp. nov., Hymenobacter sublimis sp. nov. and Hymenobacter volaticus sp. nov. LID. (1466-5034 (Electronic)). [CrossRef]
- Roldán DM, Kyrpides N, Woyke T, Shapiro N, Whitman WB, Králová S, et al. Hymenobacter caeli sp. nov., an airborne bacterium isolated from King George Island, Antarctica. LID (1466-5034 (Electronic)). [CrossRef]
- Erkorkmaz BA, Gat D, Rudich Y. Aerial transport of bacteria by dust plumes in the Eastern Mediterranean revealed by complementary rRNA/rRNA-gene sequencing. Communications Earth & Environment. 2023;4(1):24. [CrossRef]
- Xue H, Piao C-g, Wang X-z, Lin C-l, Guo M-w, Li Y. Sphingomonas aeria sp. nov., isolated from air. Int J Syst Evol Microbiol. 2018;68(9):2866-71. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




